Transcriptomic Analysis Reveals That Excessive Thyroid Hormone Signaling Impairs Phototransduction and Mitochondrial Bioenergetics and Induces Cellular Stress in Mouse Cone Photoreceptors
Abstract
:1. Introduction
2. Results
2.1. scRNAseq of Retinas Prepared from Control and T3-Treated Mice
2.2. Cone Transcriptomes Are at the Top among Retinal Cells after T3 Treatment
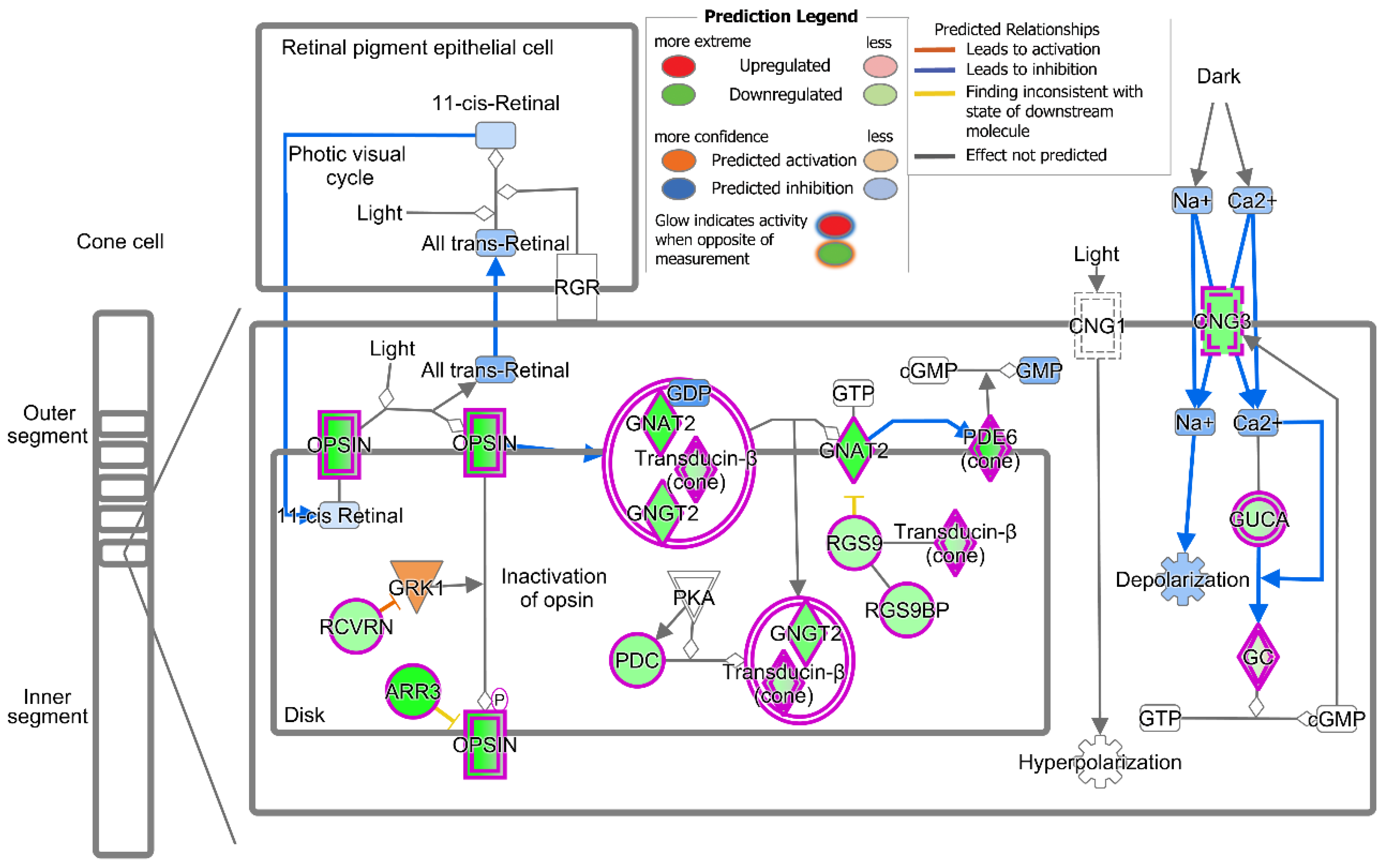
2.3. Treatment with T3 Suppresses the Cone Phototransduction Pathway
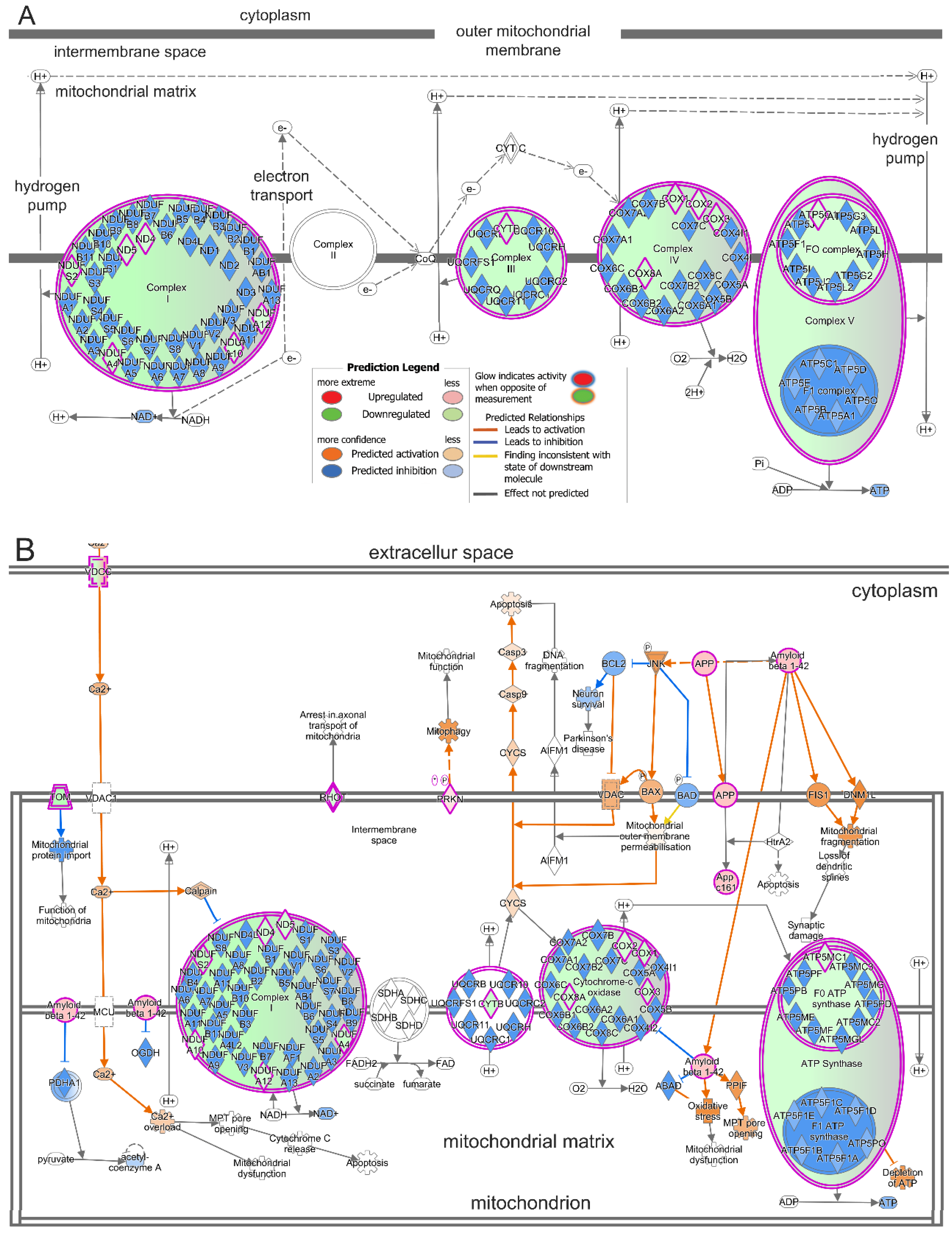
2.4. Treatment with T3 Suppresses Oxidative Phosphorylation in Cones and Induces Mitochondrial Dysfunction
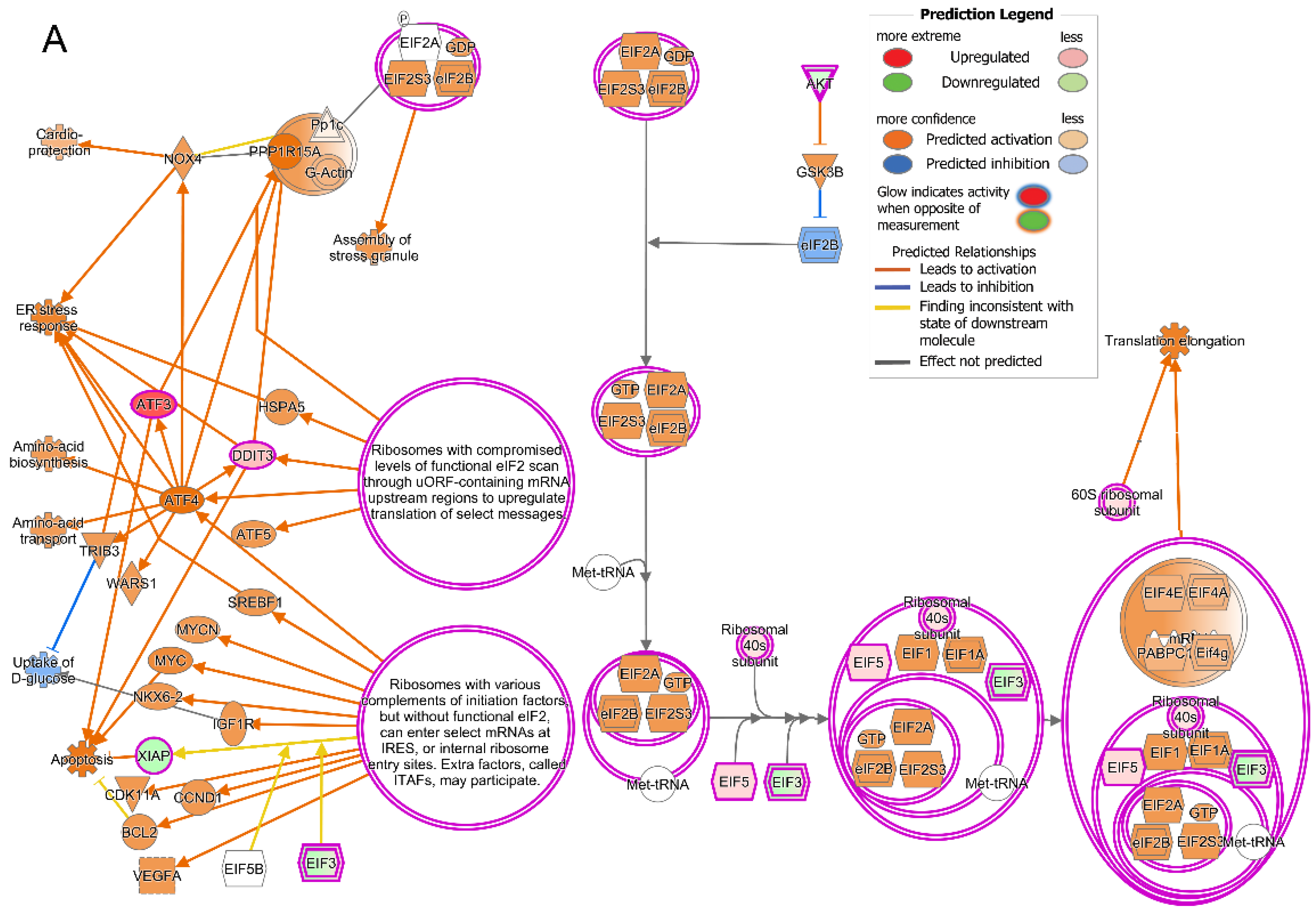
2.5. Treatment with T3 Induces Cellular Stress Pathways
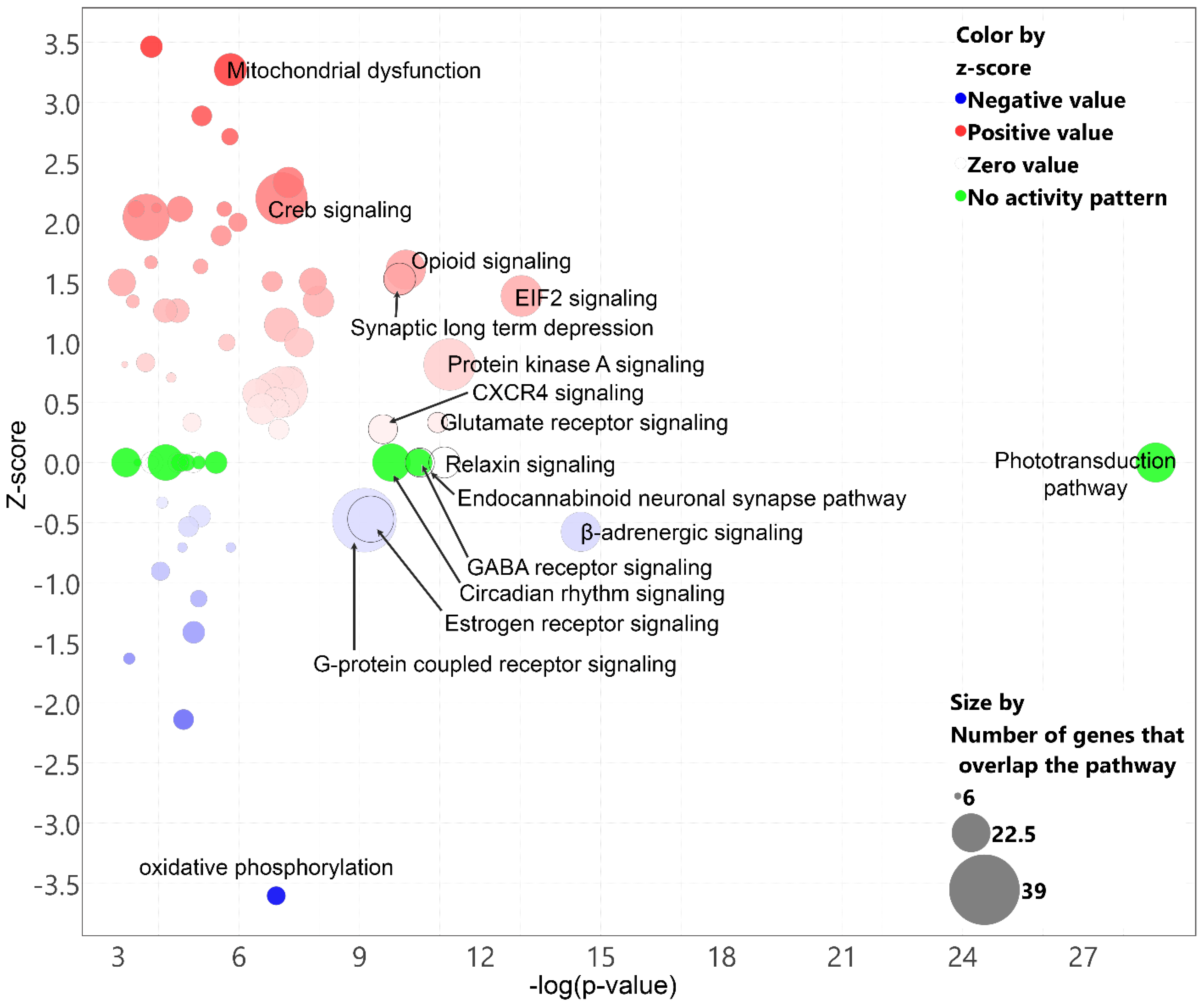
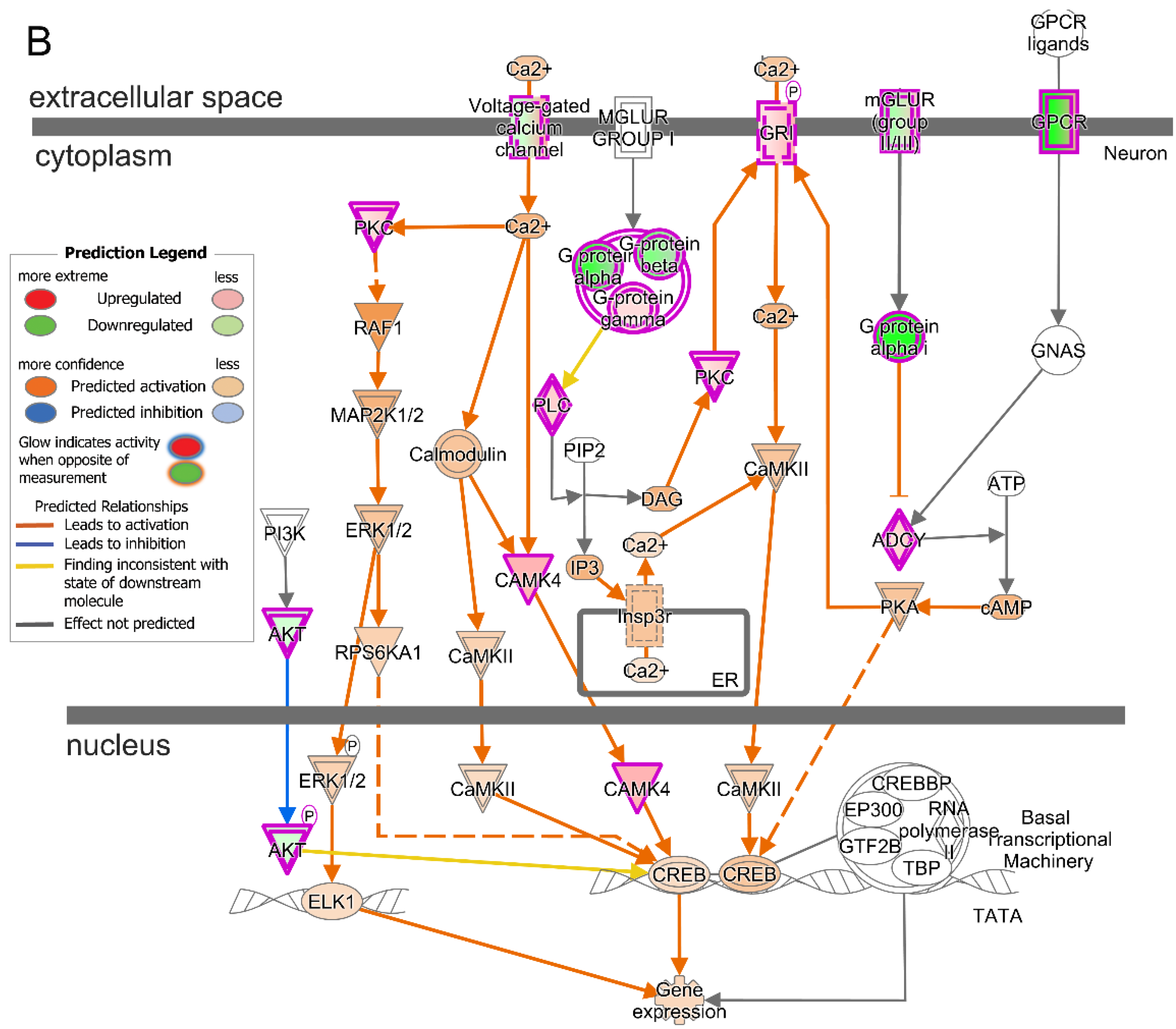
2.6. Prominent Pathway Networks and Upstream Regulators in Cones after T3 Treatment
2.7. Transcriptomic Alterations in Rods after T3 Treatment
3. Discussion
4. Materials and Methods
4.1. Thyroid Hormone Treatment
4.2. Retinal Dissection and Dissociation
4.3. Library Construction and scRNA-Seq
4.4. Data Preprocessing and Statistics
4.5. Cell Characterization and Identification of Differentially Expressed Genes
4.6. Ingenuity Pathway Analysis
4.7. RNA Isolation and Quantitative Real-Time PC
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brent, G.A. Mechanisms of thyroid hormone action. J. Clin. Investig. 2012, 122, 3035–3043. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.Y.; Leonard, J.L.; Davis, P.J. Molecular aspects of thyroid hormone actions. Endocr. Rev. 2010, 31, 139–170. [Google Scholar] [CrossRef] [PubMed]
- Forrest, D.; Reh, T.A.; Rusch, A. Neurodevelopmental control by thyroid hormone receptors. Curr. Opin. Neurobiol. 2002, 12, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.B.; Fu, L.; Hsia, S.C.; Tomita, A.; Buchholz, D. Thyroid hormone regulation of apoptotic tissue remodeling during anuran metamorphosis. Cell Res. 2001, 11, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Buchholz, D.R.; Tomita, A.; Fu, L.; Paul, B.D.; Shi, Y.B. Transgenic analysis reveals that thyroid hormone receptor is sufficient to mediate the thyroid hormone signal in frog metamorphosis. Mol. Cell Biol. 2004, 24, 9026–9037. [Google Scholar] [CrossRef] [PubMed]
- Mihara, S.; Suzuki, N.; Wakisaka, S.; Suzuki, S.; Sekita, N.; Yamamoto, S.; Saito, N.; Hoshino, T.; Sakane, T. Effects of thyroid hormones on apoptotic cell death of human lymphocytes. J. Clin. Endocrinol. Metab. 1999, 84, 1378–1385. [Google Scholar] [CrossRef] [PubMed]
- Sar, P.; Peter, R.; Rath, B.; Das Mohapatra, A.; Mishra, S.K. 3, 3’5 Triiodo L thyronine induces apoptosis in human breast cancer MCF-7 cells, repressing SMP30 expression through negative thyroid response elements. PLoS ONE 2011, 6, e20861. [Google Scholar] [CrossRef] [PubMed]
- Yamada-Okabe, T.; Satoh, Y.; Yamada-Okabe, H. Thyroid hormone induces the expression of 4-1BB and activation of caspases in a thyroid hormone receptor-dependent manner. Eur. J. Biochem. 2003, 270, 3064–3073. [Google Scholar] [CrossRef] [PubMed]
- Chiloeches, A.; Sanchez-Pacheco, A.; Gil-Araujo, B.; Aranda, A.; Lasa, M. Thyroid hormone-mediated activation of the ERK/dual specificity phosphatase 1 pathway augments the apoptosis of GH4C1 cells by down-regulating nuclear factor-kappaB activity. Mol. Endocrinol. 2008, 22, 2466–2480. [Google Scholar] [CrossRef] [PubMed]
- Ng, L.; Hernandez, A.; He, W.; Ren, T.; Srinivas, M.; Ma, M.; Galton, V.A.; St Germain, D.L.; Forrest, D. A protective role for type 3 deiodinase, a thyroid hormone-inactivating enzyme, in cochlear development and auditory function. Endocrinology 2009, 150, 1952–1960. [Google Scholar] [CrossRef] [PubMed]
- Peeters, R.P.; Hernandez, A.; Ng, L.; Ma, M.; Sharlin, D.S.; Pandey, M.; Simonds, W.F.; St Germain, D.L.; Forrest, D. Cerebellar abnormalities in mice lacking type 3 deiodinase and partial reversal of phenotype by deletion of thyroid hormone receptor alpha1. Endocrinology 2013, 154, 550–561. [Google Scholar] [CrossRef]
- Ceresini, G.; Lauretani, F.; Maggio, M.; Ceda, G.P.; Morganti, S.; Usberti, E.; Chezzi, C.; Valcavi, R.; Bandinelli, S.; Guralnik, J.M.; et al. Thyroid function abnormalities and cognitive impairment in elderly people: Results of the Invecchiare in Chianti study. J. Am. Geriatr. Soc. 2009, 57, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Kalmijn, S.; Mehta, K.M.; Pols, H.A.; Hofman, A.; Drexhage, H.A.; Breteler, M.M. Subclinical hyperthyroidism and the risk of dementia. The Rotterdam study. Clin. Endocrinol. 2000, 53, 733–737. [Google Scholar] [CrossRef] [PubMed]
- de Jong, F.J.; Masaki, K.; Chen, H.; Remaley, A.T.; Breteler, M.M.; Petrovitch, H.; White, L.R.; Launer, L.J. Thyroid function, the risk of dementia and neuropathologic changes: The Honolulu-Asia aging study. Neurobiol. Aging 2009, 30, 600–606. [Google Scholar] [CrossRef]
- Chaker, L.; Buitendijk, G.H.; Dehghan, A.; Medici, M.; Hofman, A.; Vingerling, J.R.; Franco, O.H.; Klaver, C.C.; Peeters, R.P. Thyroid function and age-related macular degeneration: A prospective population-based cohort study--the Rotterdam Study. BMC Med. 2015, 13, 94. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, B.; Liew, G.; Kifley, A.; Mitchell, P. Thyroid Dysfunction and Ten-Year Incidence of Age-Related Macular Degeneration. Investig. Ophthalmol. Vis. Sci. 2016, 57, 5273–5277. [Google Scholar] [CrossRef] [PubMed]
- Age-Related Eye Disease Study Research, G. Risk factors associated with age-related macular degeneration. A case-control study in the age-related eye disease study: Age-Related Eye Disease Study Report Number 3. Ophthalmology 2000, 107, 2224–2232. [Google Scholar]
- Lin, S.Y.; Hsu, W.H.; Lin, C.L.; Lin, C.C.; Lin, J.M.; Chang, Y.L.; Hsu, C.Y.; Kao, C.H. Evidence for an Association between Macular Degeneration and Thyroid Cancer in the Aged Population. Int. J. Environ. Res. Public Health 2018, 15, 902. [Google Scholar] [CrossRef] [PubMed]
- Chatziralli, I.; Mitropoulos, P.G.; Niakas, D.; Labiris, G. Thyroidopathy and Age-Related Macular Degeneration: Is There Any Correlation. Biomed. Hub. 2017, 2, 54706. [Google Scholar] [CrossRef] [PubMed]
- Hung, S.H.; Xirasagar, S.; Kuang, T.T.; Chang, W.W.; Cheng, Y.F.; Kuo, N.W.; Lin, H.C. Association of Age-Related Macular Degeneration with Prior Hyperthyroidism and Hypothyroidism: A Case-Control Study. J. Pers. Med. 2022, 12, 602. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Zhang, M.; Zhang, Q.; Xu, T.; Tao, L. Thyroid Disease Is Associated with Higher Age-Related Macular Degeneration Risk: Results from a Meta-Analysis of Epidemiologic Studies. Ophthalmic Res. 2021, 64, 696–703. [Google Scholar] [CrossRef] [PubMed]
- Farvardin, M.; Mousavi, S.E.; Zare, K.; Bazdar, S.; Farvardin, Z.; Johari, M. Thyroid Dysfunction as a Modifiable Risk Factor for Wet Type Age-Related Macular Degeneration: A Case-Control Study. J. Curr. Ophthalmol. 2021, 33, 449–452. [Google Scholar] [PubMed]
- Li, X.; Li, H.; Cheng, J.; Wang, M.; Zhong, Y.; Shi, G.; Yu, A.Y. Causal Associations of Thyroid Function and Age-Related Macular Degeneration: A Two-Sample Mendelian Randomization Study. Am. J. Ophthalmol. 2022, 239, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Abdelkader, M.; Abass, N. The Relation Between Age Related Macular Degeneration and Thyroid Disorders. Int. J. Ophthalmol. Vis. Sci. 2019, 4, 101–105. [Google Scholar] [CrossRef]
- Blum Meirovitch, S.; Leibovitch, I.; Kesler, A.; Varssano, D.; Rosenblatt, A.; Neudorfer, M. Retina and Nerve Fiber Layer Thickness in Eyes with Thyroid-Associated Ophthalmopathy. Isr. Med. Assoc. J. IMAJ 2017, 19, 277–281. [Google Scholar] [PubMed]
- Sayin, O.; Yeter, V.; Ariturk, N. Optic Disc, Macula, and Retinal Nerve Fiber Layer Measurements Obtained by OCT in Thyroid-Associated Ophthalmopathy. J. Ophthalmol. 2016, 2016, 9452687. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.R.; Srinivas, M.; Forrest, D.; Morreale de Escobar, G.; Reh, T.A. Making the gradient: Thyroid hormone regulates cone opsin expression in the developing mouse retina. Proc. Natl. Acad. Sci. USA 2006, 103, 6218–6223. [Google Scholar] [CrossRef] [PubMed]
- Glaschke, A.; Weiland, J.; Del Turco, D.; Steiner, M.; Peichl, L.; Glosmann, M. Thyroid hormone controls cone opsin expression in the retina of adult rodents. J. Neurosci. 2011, 31, 4844–4851. [Google Scholar] [CrossRef] [PubMed]
- Ng, L.; Hurley, J.B.; Dierks, B.; Srinivas, M.; Salto, C.; Vennstrom, B.; Reh, T.A.; Forrest, D. A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat. Genet. 2001, 27, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Thapa, A.; Morris, L.; Redmond, T.M.; Baehr, W.; Ding, X.Q. Suppressing thyroid hormone signaling preserves cone photoreceptors in mouse models of retinal degeneration. Proc. Natl. Acad. Sci. USA 2014, 111, 3602–3607. [Google Scholar] [CrossRef] [PubMed]
- Ng, L.; Liu, H.; St Germain, D.L.; Hernandez, A.; Forrest, D. Deletion of the Thyroid Hormone-Activating Type 2 Deiodinase Rescues Cone Photoreceptor Degeneration but Not Deafness in Mice Lacking Type 3 Deiodinase. Endocrinology 2017, 158, 1999–2010. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Ma, H.; Belcher, J.; Butler, M.R.; Redmond, T.M.; Boye, S.L.; Hauswirth, W.W.; Ding, X.Q. Targeting iodothyronine deiodinases locally in the retina is a therapeutic strategy for retinal degeneration. FASEB J. 2016, 30, 4313–4325. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Ma, H.; Boye, S.L.; Hauswirth, W.W.; Ding, X.Q. Overexpression of Type 3 Iodothyronine Deiodinase Reduces Cone Death in the Leber Congenital Amaurosis Model Mice. Adv. Exp. Med. Biol. 2018, 1074, 125–131. [Google Scholar] [PubMed]
- Ma, H.W.; Yang, F.; Butler, M.R.; Belcher, J.; Redmond, T.M.; Placzek, A.T.; Scanlan, T.S.; Ding, X.Q. Inhibition of thyroid hormone receptor locally in the retina is a therapeutic strategy for retinal degeneration. Faseb J. 2017, 31, 3425–3438. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Yang, F.; Ding, X.Q. Inhibition of thyroid hormone signaling protects retinal pigment epithelium and photoreceptors from cell death in a mouse model of age-related macular degeneration. Cell Death Dis. 2020, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Yang, F.; Ding, X.Q. Deficiency of thyroid hormone receptor protects retinal pigment epithelium and photoreceptors from cell death in a mouse model of age-related macular degeneration. Cell Death Dis. 2022, 13, 255. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Yang, F.; York, L.R.; Li, S.; Ding, X.Q. Excessive Thyroid Hormone Signaling Induces Photoreceptor Degeneration in Mice. eNeuro 2023, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Ng, L.; Lyubarsky, A.; Nikonov, S.S.; Ma, M.; Srinivas, M.; Kefas, B.; St Germain, D.L.; Hernandez, A.; Pugh, E.N., Jr.; Forrest, D. Type 3 deiodinase, a thyroid-hormone-inactivating enzyme, controls survival and maturation of cone photoreceptors. J. Neurosci. 2010, 30, 3347–3357. [Google Scholar] [CrossRef] [PubMed]
- Jeon, C.J.; Strettoi, E.; Masland, R.H. The major cell populations of the mouse retina. J. Neurosci. 1998, 18, 8936–8946. [Google Scholar] [CrossRef] [PubMed]
- Macosko, E.Z.; Basu, A.; Satija, R.; Nemesh, J.; Shekhar, K.; Goldman, M.; Tirosh, I.; Bialas, A.R.; Kamitaki, N.; Martersteck, E.M.; et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015, 161, 1202–1214. [Google Scholar] [CrossRef] [PubMed]
- Applebury, M.L.; Farhangfar, F.; Glosmann, M.; Hashimoto, K.; Kage, K.; Robbins, J.T.; Shibusawa, N.; Wondisford, F.E.; Zhang, H. Transient expression of thyroid hormone nuclear receptor TRbeta2 sets S opsin patterning during cone photoreceptor genesis. Dev. Dyn. 2007, 236, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Ma, H.; Yang, F.; Ding, X.-Q. Stimulation of Thyroid Hormone Signaling Induces Stress Responses in Mouse Retina. Adv. Exp. Med. Biol. 2024, in press. [Google Scholar]
- Wei, M.; Sun, Y.; Li, S.; Chen, Y.; Li, L.; Fang, M.; Shi, R.; Tong, D.; Chen, J.; Ma, Y.; et al. Single-cell profiling reveals Muller glia coordinate retinal intercellular communication during light/dark adaptation via thyroid hormone signaling. Protein Cell 2023, 14, 603–617. [Google Scholar] [CrossRef] [PubMed]
- Cioffi, F.; Senese, R.; Lanni, A.; Goglia, F. Thyroid hormones and mitochondria: With a brief look at derivatives and analogues. Mol. Cell Endocrinol. 2013, 379, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Psarra, A.M.; Sekeris, C.E. Steroid and thyroid hormone receptors in mitochondria. IUBMB Life 2008, 60, 210–223. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Chen, S.; Wu, H.; Song, H.; Wang, Y.; Shi, J.; Wu, Y. Congenital hypothyroidism impairs spine growth of dentate granule cells by downregulation of CaMKIV. Cell Death Discov. 2021, 7, 143. [Google Scholar] [CrossRef] [PubMed]
- Hamidi, S.; Aliesky, H.; Chen, C.R.; Rapoport, B.; McLachlan, S.M. Variable suppression of serum thyroxine in female mice of different inbred strains by triiodothyronine administered in drinking water. Thyroid 2010, 20, 1157–1162. [Google Scholar] [CrossRef]
- Isola, J.V.V.; Ocanas, S.R.; Hubbart, C.R.; Ko, S.; Mondal, S.A.; Hense, J.D.; Carter, H.N.C.; Schneider, A.; Kovats, S.; Alberola-Ila, J.; et al. A single-cell atlas of the aging murine ovary. bioRxiv 2023. [Google Scholar] [CrossRef]
- Menon, M.; Mohammadi, S.; Davila-Velderrain, J.; Goods, B.A.; Cadwell, T.D.; Xing, Y.; Stemmer-Rachamimov, A.; Shalek, A.K.; Love, J.C.; Kellis, M.; et al. Single-cell transcriptomic atlas of the human retina identifies cell types associated with age-related macular degeneration. Nat. Commun. 2019, 10, 4902. [Google Scholar] [CrossRef] [PubMed]
- Lukowski, S.W.; Lo, C.Y.; Sharov, A.A.; Nguyen, Q.; Fang, L.; Hung, S.S.; Zhu, L.; Zhang, T.; Grünert, U.; Nguyen, T.; et al. A single-cell transcriptome atlas of the adult human retina. Embo J. 2019, 38, e100811. [Google Scholar] [CrossRef] [PubMed]
- Rheaume, B.A.; Jereen, A.; Bolisetty, M.; Sajid, M.S.; Yang, Y.; Renna, K.; Sun, L.; Robson, P.; Trakhtenberg, E.F. Single cell transcriptome profiling of retinal ganglion cells identifies cellular subtypes. Nat. Commun. 2018, 9, 2759. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Thapa, A.; Morris, L.M.; Michalakis, S.; Biel, M.; Frank, M.B.; Bebak, M.; Ding, X.Q. Loss of cone cyclic nucleotide-gated channel leads to alterations in light response modulating system and cellular stress response pathways: A gene expression profiling study. Hum. Mol. Genet. 2013, 22, 3906–3919. [Google Scholar] [CrossRef] [PubMed]
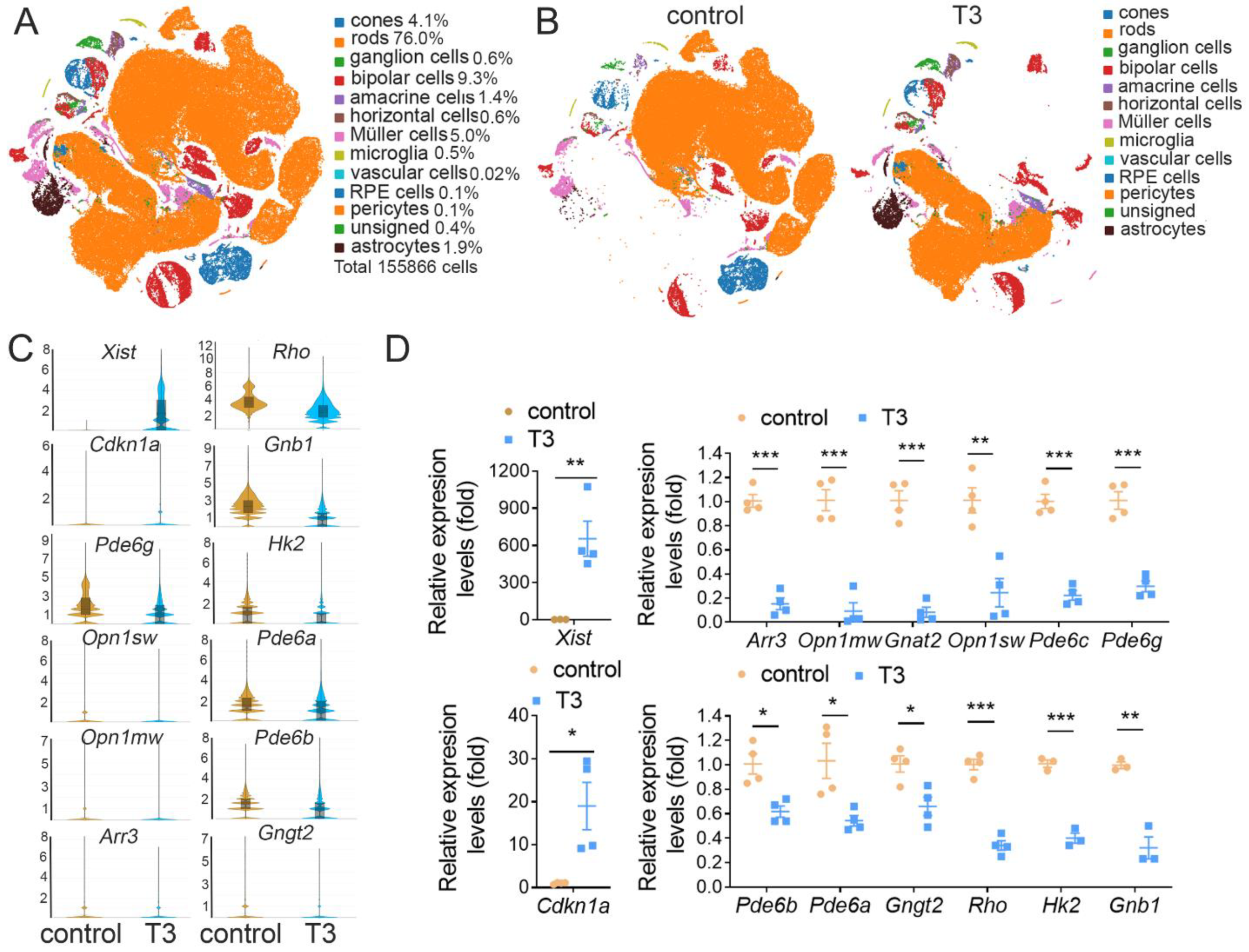


| Cell Type | Cell Number (%) | DEGs (%) | Marker Genes |
|---|---|---|---|
| Rods | 118381 (76.0) | 51 (4.4) | Rho, Cnga1, Pde6a, Ppef2, Nr2e3 |
| Cones | 6463 (4.1) | 450 (38.5) | Arr3, Opn1mw, Gnat2, Pde6h |
| Ganglion cells | 888 (0.6) | 101 (8.6) | Slc17a6, Nefm, Cncg, Nefl |
| Bipolar cells | 14531 (9.3) | 96 (8.2) | Camk2b, Trpm1, Tmem215, Grm6 |
| Amacrine cells | 2185 (1.4) | 10 (0.9) | Gad1, C1ql2, Calb1, Tmem215 |
| Müller cells | 7716 (5.0) | 179 (15.3) | Clue, Glul, Apoe, Rlbp1, Crabp1 |
| Horizontal cells | 979 (0.6) | 10 (0.9) | Lhx1, Onecut1, Onecut2, Calb1 |
| Microglia | 808 (0.5) | 100 (8.6) | C1qa, Tmem119, Aif1, Cd163, Apoe |
| Astrocytes | 2959 (1.9) | 163 (14.0) | Gfap, Apoe |
| RPE cells | 138 (0.1) | 5 (0.4) | Rlbp1, Apoe, Crabp1, Trpm1 |
| Pericytes | 184 (0.1) | 4 (0.3) | Acta2, Angpt2 |
| Vascular cells | 30 (0.02) | CD34, Adamts9, Rgs5, Cdh5 | |
| Unsigned | 604 (0.4) | ||
| Total cells | 155866 (100) |
| Ingenuity Canonical Pathways | −log(p-Value) | z-Score |
|---|---|---|
| Phototransduction pathway | 28.8 | |
| Oxidative phosphorylation | 6.93 | −3.6 |
| Mitochondrial dysfunction | 5.79 | 3.3 |
| Insulin secretion signaling pathway | 3.82 | 3.5 |
| Dopamine-DARPP32 feedback in cAMP signaling | 5.08 | 2.9 |
| CREB signaling in neurons | 7.06 | 2.2 |
| Sirtuin signaling pathway | 4.53 | 2.1 |
| Glutamate receptor signaling | 10.9 | 0.3 |
| Protein kinase A signaling | 11.2 | 0.8 |
| Signaling by Rho family GTPases | 4.47 | 1.3 |
| EIF2 Signaling | 13 | 1.4 |
| S100 family signaling pathway | 3.69 | 2.0 |
| Gustation pathway | 3.44 | 2.1 |
| Chemokine signaling | 3.95 | 2.1 |
| Renin-angiotensin signaling | 2.77 | 2.2 |
| Prolactin signaling | 2.11 | 2.2 |
| Granzyme A signaling | 2.61 | 2.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, H.; Stanford, D.; Freeman, W.M.; Ding, X.-Q. Transcriptomic Analysis Reveals That Excessive Thyroid Hormone Signaling Impairs Phototransduction and Mitochondrial Bioenergetics and Induces Cellular Stress in Mouse Cone Photoreceptors. Int. J. Mol. Sci. 2024, 25, 7435. https://doi.org/10.3390/ijms25137435
Ma H, Stanford D, Freeman WM, Ding X-Q. Transcriptomic Analysis Reveals That Excessive Thyroid Hormone Signaling Impairs Phototransduction and Mitochondrial Bioenergetics and Induces Cellular Stress in Mouse Cone Photoreceptors. International Journal of Molecular Sciences. 2024; 25(13):7435. https://doi.org/10.3390/ijms25137435
Chicago/Turabian StyleMa, Hongwei, David Stanford, Willard M. Freeman, and Xi-Qin Ding. 2024. "Transcriptomic Analysis Reveals That Excessive Thyroid Hormone Signaling Impairs Phototransduction and Mitochondrial Bioenergetics and Induces Cellular Stress in Mouse Cone Photoreceptors" International Journal of Molecular Sciences 25, no. 13: 7435. https://doi.org/10.3390/ijms25137435





