Proteomic Biomarkers Associated with Low Bone Mineral Density: A Systematic Review
Abstract
1. Introduction
2. Materials and Methods
2.1. Search Strategy, Eligibility Criteria, and Study Selection
2.2. Data Collection and Analysis
2.2.1. Data Extraction and Management
2.2.2. Risk of Bias
2.2.3. Data Synthesis
2.2.4. Network Analysis and Protein Enrichment
3. Results
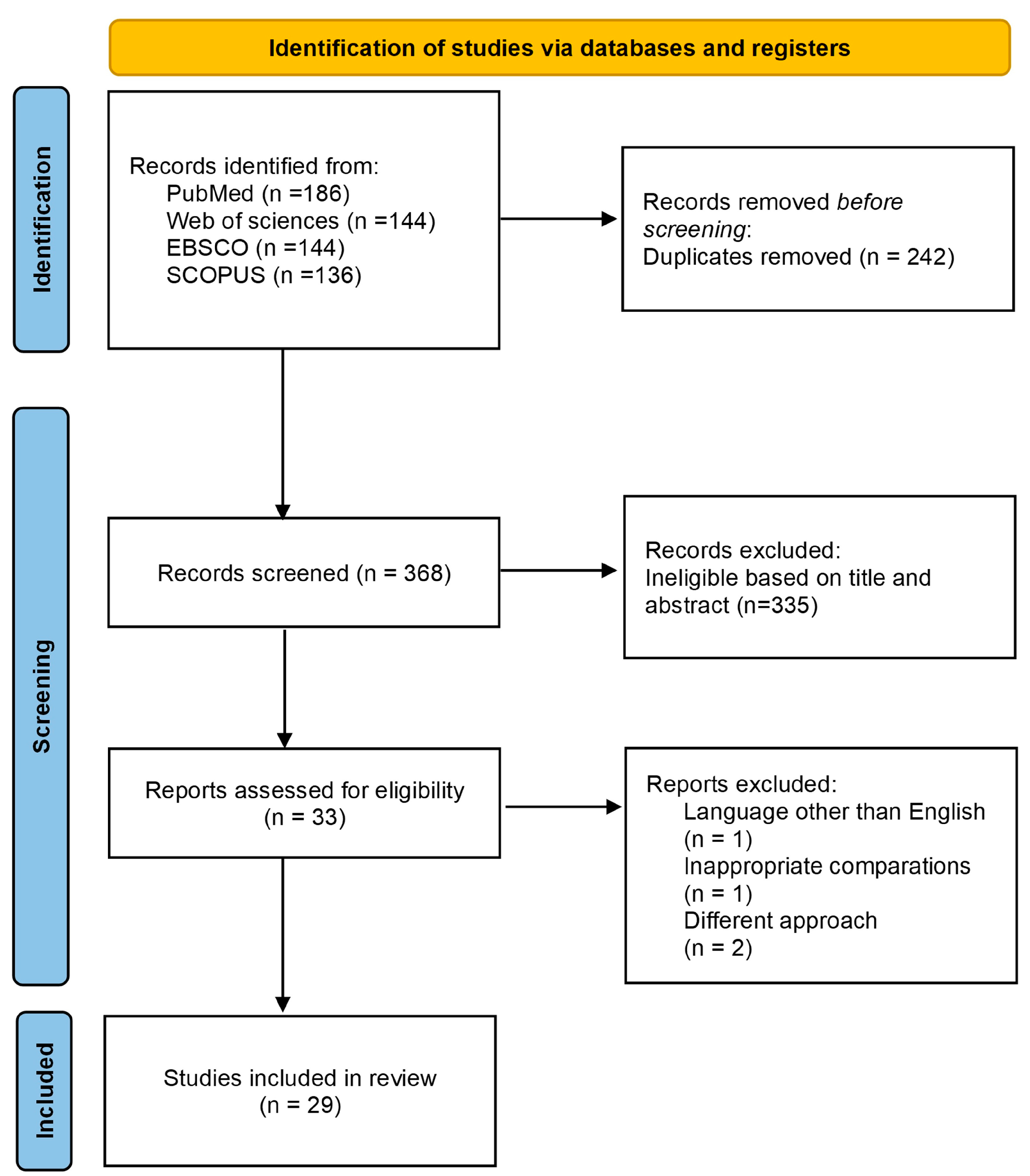
3.1. Systematic Research
3.2. Study Characteristics
3.3. Proteomic Techniques
3.4. Main Studies Performed
3.5. Risk of Bias
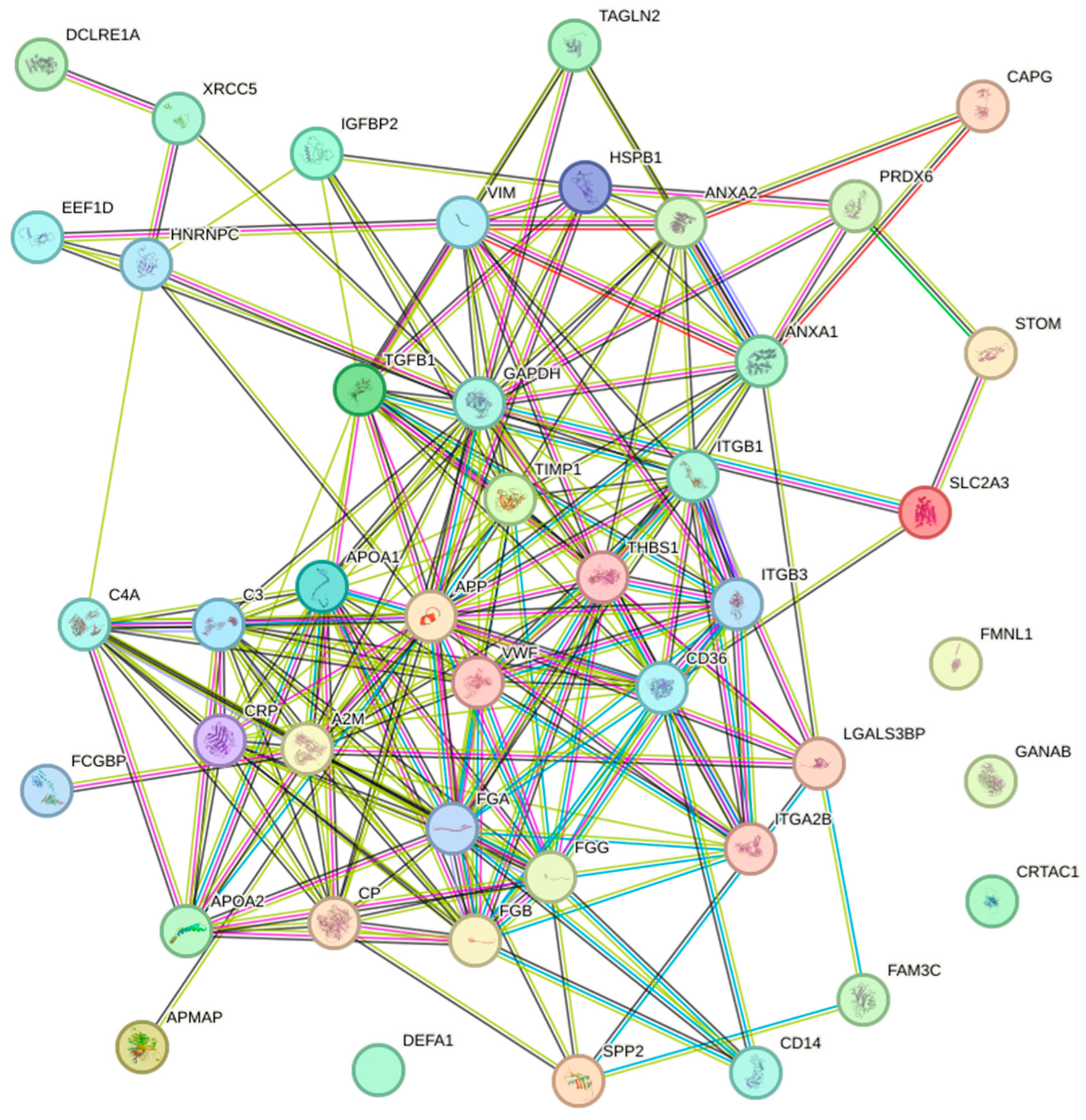
3.6. Potential Protein Biomarkers Found in Two or More Studies
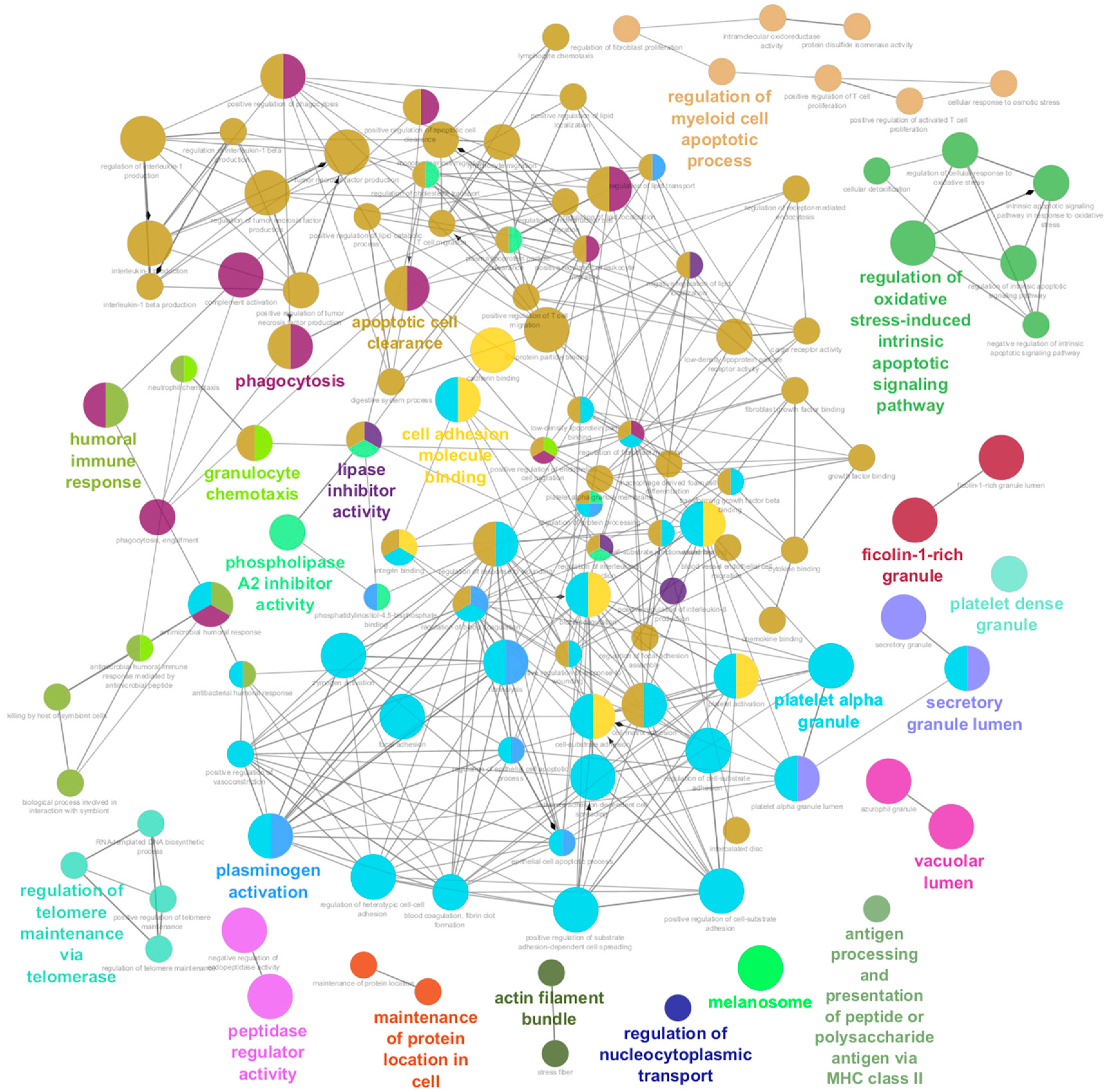
3.7. Pathways
4. Discussion
Challenges in Biomarker Research
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Salari, N.; Ghasemi, H.; Mohammadi, L.; Behzadi, M.H.; Rabieenia, E.; Shohaimi, S.; Mohammadi, M. The Global Prevalence of Osteoporosis in the World: A Comprehensive Systematic Review and Meta-Analysis. J. Orthop. Surg. Res. 2021, 16, 609. [Google Scholar] [CrossRef] [PubMed]
- Choksi, P.; Jepsen, K.J.; Clines, G.A. The Challenges of Diagnosing Osteoporosis and the Limitations of Currently Available Tools. Clin. Diabetes Endocrinol. 2018, 4, 12. [Google Scholar] [CrossRef] [PubMed]
- Sergio, R.O.; Nayelli, R.G.E. Evaluation of the Bone Mineral Density in the Mexican Female Population Using the Radiofrequency Echographic Multi Spectrometry (REMS) Technology. Arch. Osteoporos. 2022, 17, 43. [Google Scholar] [CrossRef] [PubMed]
- Patrick, G.; Sornay-Rendu, E.; Claustrat, B.; Delmas, P.D. Biochemical Markers of Bone Turnover, Endogenous Hormones and the Risk of Fractures in Postmenopausal Women: The OFELY Study. J. Bone Miner. Res. 2000, 15, 1526–1536. [Google Scholar] [CrossRef]
- Lee, J.; Vasikaran, S. Current Recommendations for Laboratory Testing and Use of Bone Turnover Markers in Management of Osteoporosis. Ann. Lab. Med. 2012, 32, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Lorentzon, M.; Branco, J.; Brandi, M.L.; Bruyère, O.; Chapurlat, R.; Cooper, C.; Cortet, B.; Diez-Perez, A.; Ferrari, S.; Gasparik, A.; et al. Algorithm for the Use of Biochemical Markers of Bone Turnover in the Diagnosis, Assessment and Follow-Up of Treatment for Osteoporosis. Adv. Ther. 2019, 36, 2811–2824. [Google Scholar] [CrossRef] [PubMed]
- Migliorini, F.; Maffulli, N.; Spiezia, F.; Tingart, M.; Maria, P.G.; Riccardo, G. Biomarkers as Therapy Monitoring for Postmenopausal Osteoporosis: A Systematic Review. J. Orthop. Surg. Res. 2021, 16, 318. [Google Scholar] [CrossRef] [PubMed]
- Bai, R.J.; Li, Y.S.; Zhang, F.J. Osteopontin, a Bridge Links Osteoarthritis and Osteoporosis. Front. Endocrinol. 2022, 13, 1012508. [Google Scholar] [CrossRef] [PubMed]
- Chiang, T.I.; Chang, I.C.; Lee, H.S.; Lee, H.; Huang, C.H.; Cheng, Y.W. Osteopontin Regulates Anabolic Effect in Human Menopausal Osteoporosis with Intermittent Parathyroid Hormone Treatment. Osteoporos. Int. 2011, 22, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.S.; Huang, L.; Tan, X.; Chen, Z.Q.; Chen, S.M.; Deng, W.M. Serum Osteopontin Levels in Relation to Bone Mineral Density and Bone Turnover Markers in Postmenopausal Women. Scand. J. Clin. Lab. Investig. 2016, 76, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Vancea, A.; Serban, O.; Fodor, D. Relationship between Osteopontin and Bone Mineral Density. Acta Endocrinol. 2021, 17, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Kumar, D.; Lal, A.K. Serum Osteocalcin as a Diagnostic Biomarker for Primary Osteoporosis in Women. J. Clin. Diagn. Res. 2015, 9, RC04–RC07. [Google Scholar] [CrossRef] [PubMed]
- Kalaiselvi, V.S.; Prabhu, K.; Ramesh, M.; Venkatesan, V. The Association of Serum Osteocalcin with the Bone Mineral Density in Post Menopausal Women. J. Clin. Diagn. Res. 2013, 7, 814–816. [Google Scholar] [CrossRef]
- Lee, N.K.; Sowa, H.; Hinoi, E.; Ferron, M.; Ahn, J.D.; Confavreux, C.; Dacquin, R.; Mee, P.J.; McKee, M.D.; Jung, D.Y.; et al. Endocrine Regulation of Energy Metabolism by the Skeleton. Cell 2007, 130, 456–469. [Google Scholar] [CrossRef] [PubMed]
- Kubota, K.; Wakabayashi, K.; Matsuoka, T. Proteome Analysis of Secreted Proteins during Osteoclast Differentiation Using Two Different Methods: Two-Dimensional Electrophoresis and Isotope-Coded Affinity Tags Analysis with Two-Dimensional Chromatography. Proteomics 2003, 3, 616–626. [Google Scholar] [CrossRef] [PubMed]
- Czupalla, C.; Mansukoski, H.; Pursche, T.; Krause, E.; Hoflack, B. Comparative Study of Protein and MRNA Expression during Osteoclastogenesis. Proteomics 2005, 5, 3868–3875. [Google Scholar] [CrossRef] [PubMed]
- Aasebø, E.; Brenner, A.K.; Hernandez-Valladares, M.; Birkeland, E.; Berven, F.S.; Selheim, F.; Bruserud, Ø. Proteomic Comparison of Bone Marrow Derived Osteoblasts and Mesenchymal Stem Cells. Int. J. Mol. Sci. 2021, 22, 5665. [Google Scholar] [CrossRef] [PubMed]
- Nakayasu, E.S.; Gritsenko, M.; Piehowski, P.D.; Gao, Y.; Orton, D.J.; Schepmoes, A.A.; Fillmore, T.L.; Frohnert, B.I.; Rewers, M.; Krischer, J.P.; et al. Tutorial: Best Practices and Considerations for Mass-Spectrometry-Based Protein Biomarker Discovery and Validation. Nat. Protoc. 2021, 16, 3737–3760. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Hwang, S. Identification of Osteoporosis-Associated Protein Biomarkers from Ovariectomized Rat Urine. Curr. Proteom. 2017, 14, 130–137. [Google Scholar] [CrossRef]
- Martínez-Aguilar, M.M.; Aparicio-Bautista, D.I.; Ramírez-Salazar, E.G.; Reyes-Grajeda, J.P.; De la Cruz-Montoya, A.H.; Antuna-Puente, B.; Hidalgo-Bravo, A.; Rivera-Paredez, B.; Ramírez-Palacios, P.; Quiterio, M.; et al. Serum Proteomic Analysis Reveals Vitamin D-Binding Protein (VDBP) as a Potential Biomarker for Low Bone Mineral Density in Mexican Postmenopausal Women. Nutrients 2019, 11, 2853. [Google Scholar] [CrossRef] [PubMed]
- Porcelli, T.; Pezzaioli, L.; Delbarba, A.; Maffezzoni, F.; Cappelli, C.; Ferlin, A. Protein Markers in Osteoporosis. Protein Pept. Lett. 2020, 27, 1253–1259. [Google Scholar] [CrossRef] [PubMed]
- Daswani, B.; Gupta, M.K.; Gavali, S.; Desai, M.; Sathe, G.J.; Patil, A.; Parte, P.; Sirdeshmukh, R.; Khatkhatay, M.I. Monocyte Proteomics Reveals Involvement of Phosphorylated HSP27 in the Pathogenesis of Osteoporosis. Dis. Markers 2015, 2015, 196589. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, Y.Z.; Zeng, Y.; Zhu, W.; Zhao, Y.C.; Zhang, J.G.; Zhu, J.Q.; He, H.; Shen, H.; Tian, Q.; et al. Network-Based Proteomic Analysis for Postmenopausal Osteoporosis in Caucasian Females. Proteomics 2016, 16, 12–28. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Zhang, L.; Zhu, W.; Xu, C.; He, H.; Zhou, Y.; Liu, Y.Z.; Tian, Q.; Zhang, J.G.; Deng, F.Y.; et al. Quantitative Proteomics and Integrative Network Analysis Identified Novel Genes and Pathways Related to Osteoporosis. J. Proteom. 2016, 142, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Shen, H.; Zhang, J.G.; Zhang, L.; Zeng, Y.; Huang, H.L.; Zhao, Y.C.; He, H.; Zhou, Y.; Wu, K.H.; et al. Cytosolic Proteome Profiling of Monocytes for Male Osteoporosis. Osteoporos. Int. 2017, 28, 1035–1046. [Google Scholar] [CrossRef] [PubMed]
- Huo, C.; Li, Y.; Qiao, Z.; Shang, Z.; Cao, C.; Hong, Y.; Xiao, H. Comparative Proteomics Analysis of Microvesicles in Human Serum for the Evaluation of Osteoporosis. Electrophoresis 2019, 40, 1839–1847. [Google Scholar] [CrossRef] [PubMed]
- Nielson, C.M.; Wiedrick, J.; Shen, J.; Jacobs, J.; Baker, E.S.; Baraff, A.; Piehowski, P.; Lee, C.G.; Baratt, A.; Petyuk, V.; et al. Identification of Hip BMD Loss and Fracture Risk Markers Through Population-Based Serum Proteomics. J. Bone Miner. Res. 2017, 32, 1559–1567. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.H.; Sun, H.; Yan, G.L.; Han, Y.; Wang, X.J. Serum Proteomics in Biomedical Research: A Systematic Review. Appl. Biochem. Biotechnol. 2013, 170, 774–786. [Google Scholar] [CrossRef] [PubMed]
- Al-Ansari, M.M.; Aleidi, S.M.; Masood, A.; Alnehmi, E.A.; Abdel Jabar, M.; Almogren, M.; Alshaker, M.; Benabdelkamel, H.; Abdel Rahman, A.M. Proteomics Profiling of Osteoporosis and Osteopenia Patients and Associated Network Analysis. Int. J. Mol. Sci. 2022, 23, 10200. [Google Scholar] [CrossRef]
- Xu, J.; Cai, X.; Miao, Z.; Yan, Y.; Chen, D.; Yang, Z.; Yue, L.; Hu, W.; Zhuo, L.; Wang, J.; et al. Proteome-Wide Profiling Reveals Dysregulated Molecular Features and Accelerated Aging in Osteoporosis: A 9.8-Year Prospective Study. Aging Cell 2023, 23, e14035. [Google Scholar] [CrossRef] [PubMed]
- Shamseer, L.; Moher, D.; Clarke, M.; Ghersi, D.; Liberati, A.; Petticrew, M.; Shekelle, P.; Stewart, L.A.; Altman, D.G.; Booth, A.; et al. Preferred Reporting Items for Systematic Review and Meta-Analysis Protocols (PRISMA-P) 2015: Elaboration and Explanation. BMJ 2015, 350, g7647. [Google Scholar] [CrossRef] [PubMed]
- Moola, S.; Munn, Z.; Sears, K.; Sfetcu, R.; Currie, M.; Lisy, K.; Tufanaru, C.; Qureshi, R.; Mattis, P.; Mu, P. Conducting Systematic Reviews of Association (Etiology): The Joanna Briggs Institute’s Approach. Int. J. Evid. Based Healthc. 2015, 13, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape Plug-in to Decipher Functionally Grouped Gene Ontology and Pathway Annotation Networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Li, Y.; Lv, H.; Yin, P.; Zhang, L.; Tang, P. Quantitative Proteomics and Reverse Engineer Analysis Identified Plasma Exosome Derived Protein Markers Related to Osteoporosis. J. Proteom. 2020, 228, 103940. [Google Scholar] [CrossRef] [PubMed]
- Deng, F.Y.; Liu, Y.Z.; Li, L.M.; Jiang, C.; Wu, S.; Chen, Y.; Jiang, H.; Yang, F.; Xiong, J.X.; Xiao, P.; et al. Proteomic Analysis of Circulating Monocytes in Chinese Premenopausal Females with Extremely Discordant Bone Mineral Density. Proteomics 2008, 8, 4259–4272. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Wang, Y.; Lv, J.; Yan, Y.; Hu, Y.; Liu, C.; Zhang, F.; Wang, J.; Hao, D. Proteomic Profiling Analysis of Postmenopausal Osteoporosis and Osteopenia Identifies Potential Proteins Associated with Low Bone Mineral Density. PeerJ 2020, 8, e9009. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Pan, H.; Liu, W.; Jin, G.; Liu, W.; Liang, C.; Jiang, X. Discovery of Novel Serum Biomarkers for Diagnosing and Predicting Postmenopausal Osteoporosis Patients by 4D-Label Free Protein Omics. J. Orthop. Res. 2023, 41, 2713–2720. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.L.; Li, C.W.; Liang, B.C.; He, K.H.; Li, X.Y. Weak Cation Magnetic Separation Technology and MALDI-TOF-MS in Screening Serum Protein Markers in Primary Type I Osteoporosis. Genet. Mol. Res. 2015, 14, 15285–15294. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Liang, B.; Shi, Z.; Wang, B.; Wu, P.; Kong, L.; Yao, J.; Li, C. Discovery and Identification of Serum Biomarkers for Postmenopausal Osteoporosis Based on TMT Labeling and HPLC-MS/MS Technology. Int. J. Clin. Exp. Med. 2017, 10, 334–346. [Google Scholar]
- Xie, Y.; Gao, Y.; Zhang, L.; Chen, Y.; Ge, W.; Tang, P. Involvement of Serum-Derived Exosomes of Elderly Patients with Bone Loss in Failure of Bone Remodeling via Alteration of Exosomal Bone-Related Proteins. Aging Cell 2018, 17, e12758. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Xu, L.; He, P.; Sun, Y.-H.; Lu, X.; Lei, S.-F.; Deng, F.-Y. Quantitative Proteomic Study of Peripheral Blood Monocytes Identified Novel Genes Involved in Osteoporosis. Curr. Proteom. 2020, 18, 467–479. [Google Scholar] [CrossRef]
- Zhang, L.L.; Li, C.W.; Liu, K.; Liu, Z.; Liang, B.C.; Yang, Y.R.; Shi, X.L. Discovery and Identification of Serum Succinyl-Proteome for Postmenopausal Women with Osteoporosis and Osteopenia. Orthop. Surg. 2019, 11, 784–793. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, C.H.; He, P.; Wu, L.F.; Lu, X.; Lei, S.F.; Deng, F.Y. Abl Interactor 1: A Novel Biomarker for Osteoporosis in Chinese Elderly Men. J. Proteom. 2019, 207, 103440. [Google Scholar] [CrossRef]
- He, W.T.; Liang, B.C.; Shi, Z.Y.; Li, X.Y.; Li, C.W.; Shi, X.L. Weak Cation Exchange Magnetic Beads Coupled with Matrix-Assisted Laser Desorption Ionization-Time of Flight-Mass Spectrometry in Screening Serum Protein Markers in Osteopenia. Springerplus 2016, 5, 679. [Google Scholar] [CrossRef] [PubMed]
- He, W.-T.; Wang, B.; Li, M.; Liang, B.-C.; Shi, Y.-Z.; Li, X.-Y.; Li, C.-W.; Shi, X.-L. Weak Cation Exchange Magnetic Beads Coupled with MALDI-TOF-MS in Screening Serum Markers in Perimenopausal Women with Osteopenia. Int. J. Clin. Exp. Med. 2016, 9, 8136. [Google Scholar]
- Zhou, Q.; Xie, F.; Zhou, B.; Wang, J.; Wu, B.; Li, L.; Kang, Y.; Dai, R.; Jiang, Y. Differentially Expressed Proteins Identified by TMT Proteomics Analysis in Bone Marrow Microenvironment of Osteoporotic Patients. Osteoporos. Int. 2019, 30, 1089–1098. [Google Scholar] [CrossRef] [PubMed]
- Deng, F.Y.; Lei, S.F.; Zhang, Y.; Zhang, Y.L.; Zheng, Y.P.; Zhang, L.S.; Pan, R.; Wang, L.; Tian, Q.; Shen, H.; et al. Peripheral Blood Monocyte-Expressed ANXA2 Gene Is Involved in Pathogenesis of Osteoporosis in Humans. Mol. Cell. Proteom. 2011, 10, M111.011700. [Google Scholar] [CrossRef] [PubMed]
- Deng, F.Y.; Zhu, W.; Zeng, Y.; Zhang, J.G.; Yu, N.; Liu, Y.Z.; Liu, Y.J.; Tian, Q.; Deng, H.W. Is GSN Significant for Hip BMD in Female Caucasians? Bone 2014, 63, 69–75. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zeng, Y.; Zhang, L.; Zhu, W.; He, H.; Sheng, H.; Tian, Q.; Deng, F.Y.; Zhang, L.S.; Hu, H.G.; Deng, H.W. Network Based Subcellular Proteomics in Monocyte Membrane Revealed Novel Candidate Genes Involved in Osteoporosis. Osteoporos. Int. 2017, 28, 3033–3042. [Google Scholar] [CrossRef] [PubMed]
- Pepe, J.; Rossi, M.; Battafarano, G.; Vernocchi, P.; Conte, F.; Marzano, V.; Mariani, E.; Mortera, S.L.; Cipriani, C.; Rana, I.; et al. Characterization of Extracellular Vesicles in Osteoporotic Patients Compared to Osteopenic and Healthy Controls. J. Bone Miner. Res. 2022, 37, 2186–2200. [Google Scholar] [CrossRef] [PubMed]
- Terracciano, R.; Migliaccio, V.; Savino, R.; Pujia, A.; Montalcini, T. Association between Low Bone Mineral Density and Increased α-Defensin in Salivary Fluid among Postmenopausal Women. Menopause 2013, 20, 1275–1279. [Google Scholar] [CrossRef] [PubMed]
- Grgurevic, L.; Macek, B.; Durdevic, D.; Vukicevic, S. Detection of Bone and Cartilage-Related Proteins in Plasma of Patients with a Bone Fracture Using Liquid Chromatography-Mass Spectrometry. Int. Orthop. 2007, 31, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Qundos, U.; Drobin, K.; Mattsson, C.; Hong, M.G.; Sjöberg, R.; Forsström, B.; Solomon, D.; Uhlén, M.; Nilsson, P.; Michaëlsson, K.; et al. Affinity Proteomics Discovers Decreased Levels of AMFR in Plasma from Osteoporosis Patients. Proteom. Clin. Appl. 2016, 10, 681–690. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, S.; Siegel, E.R.; Achenbach, S.J.; Khosla, S.; Suva, L.J. Serum Biomarker Profile Associated with High Bone Turnover and BMD in Postmenopausal Women. J. Bone Miner. Res. 2008, 23, 1106–1117. [Google Scholar] [CrossRef]
- Blaine, J.; Dylewski, J. Regulation of the Actin Cytoskeleton in Podocytes. Cells 2020, 9, 1700. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Luo, X.; Lin, J.; Fu, S.; Feng, P.; Su, H.; He, X.; Liang, X.; Liu, K.; Deng, W. Gelsolin Promotes Cancer Progression by Regulating Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma and Correlates with a Poor Prognosis. J. Oncol. 2020, 2020, 1980368. [Google Scholar] [CrossRef]
- Chellaiah, M.; Kizer, N.; Silva, M.; Alvarez, U.; Kwiatkowski, D.; Hruska, K.A. Gelsolin Deficiency Blocks Podosome Assembly and Produces Increased Bone Mass and Strength. J. Cell Biol. 2000, 148, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, A.; Bydoun, M.; Holloway, R.; Waisman, D. Annexin A2 Heterotetramer: Structure and Function. Int. J. Mol. Sci. 2013, 14, 6259–6305. [Google Scholar] [CrossRef] [PubMed]
- Kirsch, T.; Harrison, G.; Golub, E.E.; Nah, H.D. The Roles of Annexins and Types II and X Collagen in Matrix Vesicle-Mediated Mineralization of Growth Plate Cartilage. J. Biol. Chem. 2000, 275, 35577–35583. [Google Scholar] [CrossRef] [PubMed]
- Denis, C.V. Molecular and Cellular Biology of von Willebrand Factor. Int. J. Hematol. 2002, 75, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Gebetsberger, J.; Schirmer, M.; Wurzer, W.J.; Streif, W. Low Bone Mineral Density in Hemophiliacs. Front. Med. 2022, 9, 794456. [Google Scholar] [CrossRef] [PubMed]
- Simonet, W.S.; Lacey, D.L.; Dunstan, C.R.; Kelley, M.; Chang, M.S.; Lüthy, R.; Nguyen, H.Q.; Wooden, S.; Bennett, L.; Boone, T.; et al. Osteoprotegerin: A Novel Secreted Protein Involved in the Regulation of Bone Density. Cell 1997, 89, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Shahbazi, S.; Lenting, P.J.; Fribourg, C.; Terraube, V.; Denis, C.V.; Christophe, O.D. Characterization of the Interaction between von Willebrand Factor and Osteoprotegerin. J. Thromb. Haemost. 2007, 5, 1956–1962. [Google Scholar] [CrossRef] [PubMed]
- Benham, A.M. The Protein Disulfide Isomerase Family: Key Players in Health and Disease. Antioxid. Redox Signal. 2012, 16, 781–789. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhao, D.; Zheng, W.; Wang, O.; Jiang, Y.; Xia, W.; Xing, X.; Li, M. A Novel Missense Mutation in P4HB Causes Mild Osteogenesis Imperfecta. Biosci. Rep. 2019, 39, BSR20182118. [Google Scholar] [CrossRef] [PubMed]
- Rauch, F.; Fahiminiya, S.; Majewski, J.; Carrot-Zhang, J.; Boudko, S.; Glorieux, F.; Mort, J.S.; Bächinger, H.P.; Moffatt, P. Cole-Carpenter Syndrome Is Caused by a Heterozygous Missense Mutation in P4HB. Am. J. Hum. Genet. 2015, 96, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.; Li, L.; Li, D.; Wu, R.; Zhu, W.; Wang, J.; Ye, L.; Han, P. Prolyl 4-Hydroxylase Subunit Beta (P4HB) Could Serve as a Prognostic and Radiosensitivity Biomarker for Prostate Cancer Patients. Eur. J. Med. Res. 2023, 28, 245. [Google Scholar] [CrossRef] [PubMed]
- Hughes, D.E.; Salter, D.M.; Dedhar, S.; Simpson, R. Integrin Expression in Human Bone. J. Bone Miner. Res. 1993, 8, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Tasca, A.; Astleford, K.; Lederman, A.; Jensen, E.D.; Lee, B.S.; Gopalakrishnan, R.; Mansky, K.C. Regulation of Osteoclast Differentiation by Myosin X. Sci. Rep. 2017, 7, 7603. [Google Scholar] [CrossRef] [PubMed]
- Blair, H.C.; Kalyvioti, E.; Papachristou, N.I.; Tourkova, I.L.; Syggelos, S.A.; Deligianni, D.; Orkoula, M.G.; Kontoyannis, C.G.; Karavia, E.A.; Kypreos, K.E.; et al. Apolipoprotein A-1 Regulates Osteoblast and Lipoblast Precursor Cells in Mice. Lab. Investig. 2016, 96, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; James, A.W.; Kwak, J.H.; Shen, J.; Yokoyama, K.K.; Ting, K.; Soo, C.B.; Chiu, R.H. Cyclophilin A (CypA) Plays Dual Roles in Regulation of Bone Anabolism and Resorption. Sci. Rep. 2016, 6, 22378. [Google Scholar] [CrossRef][Green Version]
- Dar, H.Y.; Singh, A.; Shukla, P.; Anupam, R.; Mondal, R.K.; Mishra, P.K.; Srivastava, R.K. High Dietary Salt Intake Correlates with Modulated Th17-Treg Cell Balance Resulting in Enhanced Bone Loss and Impaired Bone-Microarchitecture in Male Mice. Sci. Rep. 2018, 8, 2503. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.C.; Schindler, S.E.; McCue, L.M.; Moulder, K.L.; Benzinger, T.L.S.; Cruchaga, C.; Fagan, A.M.; Grant, E.; Gordon, B.A.; Holtzman, D.M.; et al. Assessment of Racial Disparities in Biomarkers for Alzheimer Disease. JAMA Neurol. 2019, 76, 264–273. [Google Scholar] [CrossRef] [PubMed]
- Gijsberts, C.M.; den Ruijter, H.M.; Asselbergs, F.W.; Chan, M.Y.; de Kleijn, D.P.V.; Hoefer, I.E. Biomarkers of Coronary Artery Disease Differ between Asians and Caucasians in the General Population. Glob. Heart 2015, 10, 301–311.e11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Gao, R.; Rong, X.; Zhu, S.; Cui, Y.; Liu, H.; Li, M. Immunoporosis: Role of Immune System in the Pathophysiology of Different Types of Osteoporosis. Front. Endocrinol. 2022, 13, 965258. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.K.; Dar, H.Y.; Mishra, P.K. Immunoporosis: Immunology of Osteoporosis-Role of T Cells. Front. Immunol. 2018, 9, 657. [Google Scholar] [CrossRef] [PubMed]
- Dar, H.Y.; Shukla, P.; Mishra, P.K.; Anupam, R.; Mondal, R.K.; Tomar, G.B.; Sharma, V.; Srivastava, R.K. Lactobacillus Acidophilus Inhibits Bone Loss and Increases Bone Heterogeneity in Osteoporotic Mice via Modulating Treg-Th17 Cell Balance. Bone Rep. 2018, 8, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Adamopoulos, I.E.; Chao, C.C.; Geissler, R.; Laface, D.; Blumenschein, W.; Iwakura, Y.; McClanahan, T.; Bowman, E.P. Interleukin-17A Upregulates Receptor Activator of NF-KappaB on Osteoclast Precursors. Arthritis Res. Ther. 2010, 12, R29. [Google Scholar] [CrossRef]
- Okamoto, K.; Takayanagi, H. Effect of T Cells on Bone. Bone 2023, 168, 116675. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Liu, G.; Hu, S. Osteoporosis: Interferon-Gamma-Mediated Bone Remodeling in Osteoimmunology. Front. Immunol. 2024, 15, 1396122. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Li, L.; Sun, Y.; Wang, W.; Wang, X.; Ye, Y.; Chen, X.; Xu, Y. IL-17 Regulates the Expressions of RANKL and OPG in Human Periodontal Ligament Cells via TRAF6/TBK1-JNK/NF-ΚB Pathways. Immunology 2014, 144, 472–485. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.S.; Goyal, N. Estrogens as Regulator of Hematopoietic Stem Cell, Immune Cells and Bone Biology. Life Sci. 2021, 269, 119091. [Google Scholar] [CrossRef] [PubMed]
- Barczyk, M.; Carracedo, S.; Gullberg, D. Integrins. Cell Tissue Res. 2010, 339, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.H.; Wu, J.J.; Guo, D.Y.; Li, Y.Y.; Chen, M.N.; Zhang, Z.Y.; Yuan, Z.D.; Zhang, K.W.; Chen, W.W.; Tian, F.; et al. Physiological Functions of Podosomes: From Structure and Function to Therapy Implications in Osteoclast Biology of Bone Resorption. Ageing Res. Rev. 2023, 85, 101842. [Google Scholar] [CrossRef] [PubMed]
- Brommage, R.; Liu, J.; Vogel, P.; Mseeh, F.; Thompson, A.Y.; Potter, D.G.; Shadoan, M.K.; Hansen, G.M.; Jeter-Jones, S.; Cui, J.; et al. NOTUM Inhibition Increases Endocortical Bone Formation and Bone Strength. Bone Res. 2019, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Menale, C.; Tabacco, G.; Naciu, A.M.; Schiavone, M.L.; Cannata, F.; Morenghi, E.; Sobacchi, C.; Palermo, A. Dipeptidyl Peptidase 3 Activity as a Promising Biomarker of Bone Fragility in Postmenopausal Women. Molecules 2022, 27, 3929. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xiao, Z.; Quarles, L.D.; Li, W. Osteoporosis: Mechanism, Molecular Target and Current Status on Drug Development. Curr. Med. Chem. 2021, 28, 1489–1507. [Google Scholar] [CrossRef] [PubMed]
- Kavanagh, K.L.; Guo, K.; Dunford, J.E.; Wu, X.; Knapp, S.; Ebetino, F.H.; Rogers, M.J.; Russell, R.G.G.; Oppermann, U. The Molecular Mechanism of Nitrogen-Containing Bisphosphonates as Antiosteoporosis Drugs. Proc. Natl. Acad. Sci. USA 2006, 103, 7829–7834. [Google Scholar] [CrossRef] [PubMed]
- Altindag, O.; Erel, O.; Soran, N.; Celik, H.; Selek, S. Total Oxidative/Anti-Oxidative Status and Relation to Bone Mineral Density in Osteoporosis. Rheumatol. Int. 2008, 28, 317–321. [Google Scholar] [CrossRef] [PubMed]
- Manolagas, S.C. From Estrogen-Centric to Aging and Oxidative Stress: A Revised Perspective of the Pathogenesis of Osteoporosis. Endocr. Rev. 2010, 31, 266–300. [Google Scholar] [CrossRef] [PubMed]
- Malekian, S.; Mirghafourvand, M.; Najafipour, F.; Ostadrahimi, A.; Ghassab-Abdollahi, N.; Farshbaf-Khalili, A. The Associations between Bone Mineral Density and Oxidative Stress Biomarkers in Postmenopausal Women. Korean J. Fam. Med. 2023, 44, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Rodríguez, M.A.; Ruiz-Ramos, M.; Correa-Muñoz, E.; Mendoza-Núñez, V.M. Oxidative Stress as a Risk Factor for Osteoporosis in Elderly Mexicans as Characterized by Antioxidant Enzymes. BMC Musculoskelet. Disord. 2007, 8, 124. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Yang, J.; Zhuge, A.; Li, L.; Ni, S. Gut Microbiota Modulates Osteoclast Glutathione Synthesis and Mitochondrial Biogenesis in Mice Subjected to Ovariectomy. Cell Prolif. 2022, 55, e13194. [Google Scholar] [CrossRef]
- León-Reyes, G.; Argoty-Pantoja, A.D.; Becerra-Cervera, A.; López-Montoya, P.; Rivera-Paredez, B.; Velázquez-Cruz, R. Oxidative-Stress-Related Genes in Osteoporosis: A Systematic Review. Antioxidants 2023, 12, 915. [Google Scholar] [CrossRef] [PubMed]
- Le, L.; Sirés-Campos, J.; Raposo, G.; Delevoye, C.; Marks, M.S. Melanosome Biogenesis in the Pigmentation of Mammalian Skin. Integr. Comp. Biol. 2021, 61, 1517–1545. [Google Scholar] [CrossRef] [PubMed]
- Wiriyasermkul, P.; Moriyama, S.; Nagamori, S. Membrane Transport Proteins in Melanosomes: Regulation of Ions for Pigmentation. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183318. [Google Scholar] [CrossRef]
- Hanel, A.; Carlberg, C. Skin Colour and Vitamin D: An Update. Exp. Dermatol. 2020, 29, 864–875. [Google Scholar] [CrossRef] [PubMed]
- Batai, K.; Cui, Z.; Arora, A.; Shah-Williams, E.; Hernandez, W.; Ruden, M.; Hollowell, C.M.P.; Hooker, S.E.; Bathina, M.; Murphy, A.B.; et al. Genetic Loci Associated with Skin Pigmentation in African Americans and Their Effects on Vitamin D Deficiency. PLoS Genet. 2021, 17, e1009319. [Google Scholar] [CrossRef]
- Adhikari, K.; Mendoza-Revilla, J.; Sohail, A.; Fuentes-Guajardo, M.; Lampert, J.; Chacón-Duque, J.C.; Hurtado, M.; Villegas, V.; Granja, V.; Acuña-Alonzo, V.; et al. A GWAS in Latin Americans Highlights the Convergent Evolution of Lighter Skin Pigmentation in Eurasia. Nat. Commun. 2019, 10, 358. [Google Scholar] [CrossRef] [PubMed]
- Webb, A.R.; Kazantzidis, A.; Kift, R.C.; Farrar, M.D.; Wilkinson, J.; Rhodes, L.E. Colour Counts: Sunlight and Skin Type as Drivers of Vitamin D Deficiency at UK Latitudes. Nutrients 2018, 10, 457. [Google Scholar] [CrossRef] [PubMed]
- Sobsey, C.A.; Ibrahim, S.; Richard, V.R.; Gaspar, V.; Mitsa, G.; Lacasse, V.; Zahedi, R.P.; Batist, G.; Borchers, C.H. Targeted and Untargeted Proteomics Approaches in Biomarker Development. Proteomics 2020, 20, e1900029. [Google Scholar] [CrossRef] [PubMed]
- Deracinois, B.; Flahaut, C.; Duban-Deweer, S.; Karamanos, Y. Comparative and Quantitative Global Proteomics Approaches: An Overview. Proteomes 2013, 1, 180–218. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Cheng, C.; Zhang, L. High-Throughput Proteomics: A Methodological Mini-Review. Lab. Investig. 2022, 102, 1170–1181. [Google Scholar] [CrossRef] [PubMed]
- Anwardeen, N.R.; Diboun, I.; Mokrab, Y.; Althani, A.A.; Elrayess, M.A. Statistical Methods and Resources for Biomarker Discovery Using Metabolomics. BMC Bioinform. 2023, 24, 250. [Google Scholar] [CrossRef] [PubMed]
- Frantzi, M.; Bhat, A.; Latosinska, A. Clinical Proteomic Biomarkers: Relevant Issues on Study Design & Technical Considerations in Biomarker Development. Clin. Transl. Med. 2014, 3, e7. [Google Scholar] [CrossRef]



| Author, Year | Country | Ethnicity of Analyzed Population | Study Design | Sample Size (W/M) | Number of Cases | Number of Controls | Mean Age (Years) | Measurement Site (BMD) | Outcome Definition | Confounders |
|---|---|---|---|---|---|---|---|---|---|---|
| Al-Ansari et al., 2022 [29] | Saudi Arabia | Saudi Arabian | Case-control study | 69 (52 W/ 17 M) | 47 (OP: 25, OS: 22) 39 W/8 M | 22 (13 W/9 M) | Case: (OP: 66.16 ± 1.78; OS: 64.64 ± 1.72) Control: 54.82± 1.03 | LS, FN | OS a, OP a | T2DM, thyroid disease, gender, and medication |
| Chen et al., 2020 [34] | China | Chinese | Case-control study | 30 (26 W/4 M) | 20 (OP: 10 W/0 M, OS: 9 W/1 M) | 10 (7 W/3 M) | Case: (OP: 81 ± 9; OS: 73 ± 11) Control: 76 ± 14 | LS, TH | OS a, OP a | Age, BMI, and gender |
| Daswani et al., 2015 [22] | India | Indian | Case-control study | 40 W | 10 PEW LBMD (OS: 10) 10 POW LBMD (OS: 10, OP: 9) | 20 | PEW LBMD: 36.1 ± 1.2 PEW HBMD: 36 ± 1.1, POW LBMD: 55.7 ± 1.1 POW LBMD: 53.8 ± 0. | TH, FN, LS | OS a, OP a | Age, BMI |
| Deng et al., 2008 [35] | China | Chinese | Case-control study | 30 W | 15 | 15 | 27.3 ± 5.0 | TH, FN, (combined value of TR, IR) | BMD b | NR |
| Deng et al., 2011 [47] | USA | Caucasian | Case-control study | 28 W | 14 | 14 | LBMD: 67.7 ± 1.7 HBMD: 68.7 ± 1.1 | TH | BMD c | Age, gender, height, and weight |
| Deng et al., 2014 [48] | USA/China | Caucasian | Case-control study | 34 W | 17 | 17 | LBMD: 50.2 ±1.9 HBMD: 51.8 ± 2.2 | TH, FN, (combined value of TR, IR) | BMD c | NR |
| He et al., 2016 [44] | China | Chinese | Case-control study | 20 W | 10 | 10 | Case: 56.3 ± 3.61 Normal: 55.0 ± 3.48 | LS | OS a | Age, height, and weight |
| He et al., 2016 [45] | China | Chinese | Case-control study | 20 W | 10 | 10 | Case: 53.32 ± 2.61 Normal 52.35 ± 1.94 | LS | OS a | Age, height, and weight |
| Huang et al., 2020 [36] | China | Chinese | Case-control study | 54 W | OP: 18 OS: 18 | 18 | Case: (OP: 58.33 ± 5.40; OS: 56.72 ± 4.92) Control: 55.22 ± 5.31 | LS, TH | OS a, OP a | Age, BMI |
| Huo et al., 2019 [26] | China | Chinese | Case-control study | 84 (61 W/23 M) | OP: 28 (26 W/2 M), OS: 28 (20 W/8 M) | 28 (15 W/13 M) | Case: (OP: 73.29 ± 5.25; OS: 67.96 ± 6.28) Control: 68.11 ± 7.56, | NR | OS a, OP a | NR |
| Li et al., 2023 [37] | China | Chinese | Case-control study | 16 W | 10 | 6 | Case: 71 ± 1 Control: 65 ± 12 | NR | NR | Age, BMI |
| Martínez-Aguilar et al.,2019 [20] | Mexico | Mexican-Mestizo | Case-control study | 30 W | OP: 10, OS: 10, | 10 | Case: (OP: 75 ± 4; OS: 74 ± 3) Control: 73 ± 2 | LS, TH | OS a, OP a | Age, height, weight and BMI |
| Pepe et al., 2022 [50] | Italy | Italian | Case-control study | 24 W | OP: 9, OS: 9 | 9 | Case: (OP: 64.5 ± 9.8; OS: 62.2 ± 7.9) Control: 61.9 ± 6.8 | LS, FN | OS a, OP a | Age, BMI |
| Qundos et al., 2016 [53] | Sweden | Swedish | Case-control study | 25 W | 16 | 6 | 59 to 70 | LS, TH | OP a. | NR |
| Shi et al., 2015 [38] | China | Chinese | Case-control study | 25 W | 16 | 9 | Case: 61.32 Control: 58 | LS | OP a | Age, height, and weight |
| Shi et al., 2017 [39] | China | Chinese | Case-control study | 20 W | 10 | 10 | Case: 55.2 ± 2.35 Control: 54.4 ± 2.07 | LS | OP a | Age |
| Xie et al., 2018 [40] | China | Chinese | Case-control study | 139 (68 W/71 M) | OP: 31 (23 w/8 m), OS: 46 (21 w/25 m) | 26 YN (9 W/17 M) 36 AN (15 W/21 M) | Control: (YN: 34.6 ± 7.4, AN: 64 ± 3.8) Case: (OS: 63 ± 5.3, OP: 63.8 ± 4) | One-third radius site | OS a, OP a | NR |
| Xu et al., 2020 [41] | China | Chinese | Case-control study | 42 (24 W/18 M) | 12 W S1/ 9 M S2 | 12 W S1/ 9 M S2 | NR | TH, FN, (combined value of TR, IR) | BMD d | NR |
| Zeng et al., 2016 [24] | USA | Caucasian | Case-control study | 33 W | 17 | 16 | LBMD: 50.3 ± 1.86 HBDM: 51.8 ± 2.27 | LS, TH (combined value of FN, TR, IR) | BMD e | Age, height, and weight |
| Zeng et al., 2017 [49] | USA | Caucasian | Case-control study | 59 M | 29 M | 30 M | LBMD: 40.3 ±7.6 HBDM: 41.1 ±7.5 | TH, FN, (combined value TR, IR) | BMD f | Age, height, and weight |
| Zhang et al., 2019 [42] | China | Chinese | Case-control study | 30 W | OP: 10, OS: 10 | 10 | 63.28 ± 5.78 | LS | OS a, OP a | Age |
| Zhang et al., 2016 [23] | China | Caucasian | Case-control study | 42 W | 21 | 21 | LBMD: 62.43 ± 9.3 HBDM: 63.95 ± 8.39 | LS, TH (combined value of FN, TR, IR) | BMD g | Age, height, and weight |
| Zhou et al., 2019 [46] | China | Chinese | Case-control study | 16 (12 W/4 M) | 4 (3 W/1 M) | 4 (3 M/1 W) | Case: 56.3 ± 2.3 Control: 54.0 ± 1.1 | LS, TH, FN | OP a, non-OP h | Age, BMI |
| Zhou et al., 2019 [43] | China | Chinese | Case-control study | 36 M | LBMD: 9 M OF: 18 M | 9 M | OF: 77.3 ± 12.0 LBMD: 70.0 ± 5.4 HBMD: 75.3 ± 7.1 | TH (combined value of FN, TR, IR) | OF, BMD i | Age, height, and weight |
| Zhu et al., 2017 [25] | USA | Caucasian | Case-control study | 59 M | 29 M | 30 M | LBMD: 40.3 ± 7.6 HBDM: 41.1 ± 7.5 | TH (combined value of FN, TR, IR) | BMD k | Age, weight, and, height |
| Nielson et al., 2017 [27] | USA | non-Hispanic white | Cohort | 2473 M | accelerated loss n = 237 M | BMD maintenance n = 453 M | 73.6 ± 5.8 | TH | BMD m | Age, BMI |
| Bhattacharyya et al., 2008 [54] | USA | NR | Cross-sectional study | 58 W | 49 (OP: 28, OS: 21) | 8 | High turnover group: 80.5 low/normal turnover group: 70.8 | LS, TH, mid-distal radius and ulna | Bone turnover | Age, NTX |
| Grgurevic et al., 2007 [52] | Croatia | Croatian | Cross-sectional study | 25 W * | 25 | - | 21 to 60 | NR | Acute bone fracture | Age, BMI |
| Terracciano et al., 2013 [51] | Italy | Italian | Cross-sectional study | 61 W | 43 | 18 | 61.6 ± 9 | FN | BMD a | Age, height |
| Author, Year | Specimen Type | Proteomic Approach | Statistical Analysis/Fold Change Cut-Off | Number of DEPs | Main Findings |
|---|---|---|---|---|---|
| Al-Ansari et al., 2022 [29] | Serum | Nano-LC-ESI-MS/MS | ANOVA using post-hoc Tukey’s analysis method, FC >1.5 and <0.67, FDR p < 0.05 | 219 | DEPs were associated with humoral immune response, inflammatory response, LXR/RXR activation, FXR/RXR activation, and hematopoiesis. Dysregulation of inflammatory signaling pathways in the LBMD patients. |
| Chen et al., 2020 [34] | Serum-exosomes | Nano-LC-MS/MS | Mann–Whitney U test p < 0.05, FC > 1.2 A | 45 LH | Pathways involved with degenerative diseases (Parkinson’s disease and Alzheimer’s disease), and the neuromuscular process of controlling balance. |
| Daswani et al., 2015 [22] | Peripheral blood monocyte | 4—plex iTRAQ LC-MS/MS | Student’s t-test p < 0.05, FC ≥ 1.5 | 45 LH | Effect of pHSP27 in monocyte migration towards bone milieu can result in increased osteoclast formation and, thus, contribute to pathogenesis osteoporosis. |
| Deng et al., 2008 [35] | Peripheral blood monocyte | 2DE-MALDI-TOF/TOF | Student’s t-test or Kruskal–Wallis test p < 0.05, FC ≥ 0.52 | 38 LH | DEPs might affect CMCs’ trans-endothelium, differentiation, and/or downstream osteoclast functions, thus contribute to differential osteoclastogenesis. |
| Deng et al., 2011 [47] | Peripheral blood monocyte | LC–nano-ESI-MSE | Kruskal-Wallis Test p < 0.05 | 6 LH | ANXA2 protein significantly promoted monocyte migration across an endothelial barrier in vitro. |
| Deng et al., 2014 [48] | Peripheral blood monocyte | LC–nano-ESI-MSE | Student’s t-test p < 0.05 | 57 LH | Using a proteomics-based multi-disciplinary and integrative study strategy, GSN was significantly down-regulated in premenopausal Caucasians with low vs. high hip BMD. |
| He et al., 2016 [44] | Serum | WCX-MALDI-TOF-MS | Youden Index, p < 0.05 | 10 OSN | A strategy for screening serum proteins <20 kDa to analyze serum profiles and find potential biomarkers for osteopenia. |
| He et al., 2016 [45] | Serum | WCX-MALDI-TOF-MS | Youden Index, p < 0.05 | 2 OSN | New serological method for discovering serum protein markers to screen and diagnose osteopenia. |
| Huang et al., 2020 [36] | Plasma | TMT-LC-MS/MS | Student’s t-test, FC > 1.2 and <0.83, p < 0.05 | 208 | The differentially abundant proteins exhibited binding, molecular function regulator, transporter and molecular transducer activity, and were involved in metabolic and cellular processes, stimulus response, biological regulation, and immune system processes. |
| Huo et al., 2019 [26] | Serum-microvesicles | Nano-LC MS/MS | ANOVA using post-hoc Tukey’s analysis method, FC > 2 and <0.5, p < 0.05 | 24 LH | Bone homeostasis-related novel MVs proteins and signaling pathways demonstrated that “integrin signaling pathway” were enriched for osteoporosis. Profilin 1 is verified as a valuable diagnostic indicator for the evaluation of osteoporosis disease. |
| Li et al., 2023 [37] | Serum | 4D-LC-MS/MS | Student’s t-test or Mann–Whitney U tests, FC ≥ 2 and ≤0.5, p < 0.05 | 293 OPN | The most significantly enriched GO terms and pathway that the DEPs involved in includes the PI3K–Akt signaling pathway, ECM-receptor interaction, platelet activation, neutrophil extracellular trap formation, as well as complement and coagulation cascades. |
| Martínez-Aguilar et al., 2019 [20] | Serum | 2D DIGE -MALDI TOF/TOF | Student’s t-test, FC ≥ 1.5 and ≤1.5, FDR p < 0.05 | 39 | VDBP could be considered as a novel biomarker for the early detection of osteoporosis. |
| Pepe et al., 2022 [50] | Extracellular vesicles blood | Nano-LC-ESI-MS/MS | Unpaired t-test or Mann–Whitney U test, p ≤ 0.05 | 140 | Bioinformatic analysis revealed the four most represented biological processes, including blood coagulation, gonadropin-releasing hormone receptor, inflammation mediated by chemokine and cytokine signaling, and plasminogen activate cascade pathways. |
| Qundos et al., 2016 [53] | Plasma | Antibody arrays | Linear model and Wilcoxon rank sum test, p < 0.001 | 7 OPN | AMFR is a potential marker in plasma to differentiate women diagnosed with osteoporosis compared to controls. A decreased gene and protein expression of AMFR may further reflect a lower level of physical activity in osteoporotic patients, when considering that transcripts were abundant in skeletal muscle and mirroring a reduced turnaround in muscle proteins. |
| Shi et al., 2015 [38] | Serum | MALDI-TOF-MS | Wilcoxon tests using the Youden index, p ≤ 0.05 | 16 OPN | New serological method for the screening and diagnosis of primary type I osteoporosis using serum protein markers. |
| Shi et al., 2017 [39] | Serum | TMT-LC-ESI-MS/MS | Student’s t-test p < 0.01, FC ≥ 1.5 and ≤0.67 | 87 OPN | According to the molecular functions, most of the differentially expressed proteins were involved in binding, catalytic activity and enzyme regulator activity. Candidate biomarkers of postmenopausal osteoporosis were associated with the bone remodeling. |
| Xie et al., 2018 [40] | Serum exosomes | TMT-LC-MS/MS | One-way ANOVA with a post hoc test p < 0.05, FC > 20 and <5 | 401 | Serum-derived exosomes (SDEs) from aged normal volunteers might play a protective role in bone health through facilitating adhesion of bone cells and suppressing aging-associated oxidative stress. |
| Xu et al., 2020 [41] | Peripheral blood monocyte | LC-MS/MS | Student’s t-test p < 0.05 | 331 LH | WNK1, SHTN1, and DPM1 were found differentially expressed between low BMD and high BMD subjects in both genders. |
| Zeng et al., 2016 [24] | Peripheral blood monocyte | LC-nano-ESI-MS | Student’s t-test p < 0.05, FC > 1 and <1 | 30 LH | The contribution of the genes ITGA2B, GSN, and RHOA and the pathways regulation of actin cytoskeleton and leukocyte transendothelial migration to osteoporosis risk. |
| Zeng et al., 2017 [49] | Peripheral blood monocyte | 2D-nano-LC-ESI-MS/MS | Student’s t-test p < 0.05 | 35 LH | Numerous pathways/modules including response to elevated platelet cytosolic Ca2+, the adherens junction pathway and the leukocyte transendothelial migration pathway, which are thought to be related to osteogenesis, bone formation, and resorption. |
| Zhang et al., 2019 [42] | Serum | LC-MS/MS | FC > 1.2 and <1/1.2 | 77 OPN 77 OSN 68 OPOS | ApoA-I, Apo A-II, and haptoglobin were mediated with receptors, factors, mechanisms, that related to bone metabolism, while HBD was valuable for diagnosis of osteopenia. |
| Zhang et al., 2016 [23] | Peripheral blood monocyte | LC-nano-ESI-MSE | Mann–Whitney U test, FC > 1.5 and <1.5 | 7 LH | Network analysis showed that the module including the annexin gene family was significantly correlated with low BMD, and the lipid-binding and regulating pro-inflammatory cytokines activities were enriched. |
| Zhou et al., 2019 [46] | Vertebral body-derived bone marrow supernatant fluid | TMT-LC-MS/MS | Wilcoxon-test p < 0.05/FC > 1.3 | 219 OPN | Upregulated proteins were mainly associated with the regulation of transcription and protein metabolism, and downregulated proteins were involved in immune response and movements of the cell and cellular components. |
| Zhou et al., 2019 [43] | Peripheral blood monocyte | LC-MS/MS | Student’s t-test or Unpaired t-test with Welch’s correction p < 0.05 | 253 OFN 13 LH 8 OLH | ABI1 protein, via promoting osteoblast growth, differentiation and activity, and attenuating monocyte trans-endothelial migration and osteoclast differentiation, influences BMD variation and fracture risk in humans. |
| Zhu et al., 2017 [25] | Peripheral blood monocyte | LC-nano-ESI-MSE | Student’s t-test p <0.05 | 16 LH | ALDOA, MYH14, and Rap1B were identified based on multiple omics evidence, and they may influence the pathogenic mechanisms of osteoporosis by regulating the proliferation, differentiation, and migration of monocytes. |
| Nielson et al., 2017 [27] | Serum | LC-MS-MS | Markov Chain Monte Carlo meta-fold > 1.1, and meta p < 0.1 | 237 had accelerated hip BMD loss, and 453 maintained hip BMD | CD14 and SHBG were associated with fracture risk; B2MG and TIMP1 have biological role in cellular senescence and aging, and CO7, CO9, CFAD has documented in complement activation and innate immunity functions. |
| Bhattacharyya et al., 2008 [54] | Serum | LC-MS | Wilcoxon rank-sum test/Student’s t-test, FC > 1.5 | 11 | ITIH4 is stored within the bone matrix and is a substrate for enzymatic degradation by osteoclast. |
| Grgurevic et al., 2007 [52] | Plasma | LC-MS/MS | No comparisons | 12 | A significant proportion of proteins were of extracellular origin and was involved in the cell growth and proliferation, transport, and coagulation. Several proteins have not been previously identified in the plasma, including: TGF-β-induced protein IG-H3, cartilage acidic protein 1, procollagen C proteinase enhancer protein, and TGF-β receptor III. |
| Terracciano et al., 2013 [51] | Salivary fluid | MALDI TOF/TOF | NR | α-defensin HNP-1 could be a novel biomarker for osteoporosis. |
| Protein | Sample Type | Direction of Differential Expression in OP/OS/LBMD | Reference |
|---|---|---|---|
| GSN | PBM | ↑LBMD | Deng et al., 2008 [35] |
| PBM | ↓LBMD | Deng et al., 2014 [48] | |
| PBM | ↓LBMD | Zeng et al., 2016 [24] | |
| Serum | ↓OP/OS | Martínez-Aguilar et al., 2019 [20] | |
| ANXA2 | PBM | ↑LBMD | Deng et al., 2011 [47] |
| PBM | ↓LBMD | Daswani et al., 2015 [22] | |
| PBM | ↑LBMD | Zhang et al., 2016 [23] | |
| APOA1 | EVB | ↑OS/OP | Pepe et al., 2022 [50] |
| Serum | ↓OS—↑OP | Zhang et al., 2016 [23] | |
| PPIA | PBM | ↓LBMD | Deng et al., 2011 [47] |
| PBM | ↓LBMD | Zhang et al., 2016 [23] | |
| P4HB | PBM | ↓LBMD | Deng et al., 2008 [35] |
| PBM M | ↓LBMD | Zeng et al., 2017 [49] | |
| ITGB1 | Serum exosomes WM | ↓OP | Zeng et al., 2017 [49] |
| PBM M | ↑LBMD | Xie et al., 2018 [40] | |
| ITGA2B | PBM | ↑LBMD | Deng et al., 2014 [48] |
| PBM | ↓LBMD | Zeng et al., 2016 [24] | |
| MYH14 | PBM M | ↑LBMD | Zhu et al., 2017 [25] |
| Serum WM | ↓OP | Al-Ansari et al., 2022 [29] | |
| VWF | EVB | ↓OS/OP | Pepe et al., 2022 [50] |
| Serum | ↑OP | Li et al., 2023 [37] | |
| LOC654188 | PBM | ↓LBMD | Deng et al., 2011 [47] |
| PBM | ↓LBMD | Zhang et al., 2016 [23] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Becerra-Cervera, A.; Argoty-Pantoja, A.D.; Aparicio-Bautista, D.I.; López-Montoya, P.; Rivera-Paredez, B.; Hidalgo-Bravo, A.; Velázquez-Cruz, R. Proteomic Biomarkers Associated with Low Bone Mineral Density: A Systematic Review. Int. J. Mol. Sci. 2024, 25, 7526. https://doi.org/10.3390/ijms25147526
Becerra-Cervera A, Argoty-Pantoja AD, Aparicio-Bautista DI, López-Montoya P, Rivera-Paredez B, Hidalgo-Bravo A, Velázquez-Cruz R. Proteomic Biomarkers Associated with Low Bone Mineral Density: A Systematic Review. International Journal of Molecular Sciences. 2024; 25(14):7526. https://doi.org/10.3390/ijms25147526
Chicago/Turabian StyleBecerra-Cervera, Adriana, Anna D. Argoty-Pantoja, Diana I. Aparicio-Bautista, Priscilla López-Montoya, Berenice Rivera-Paredez, Alberto Hidalgo-Bravo, and Rafael Velázquez-Cruz. 2024. "Proteomic Biomarkers Associated with Low Bone Mineral Density: A Systematic Review" International Journal of Molecular Sciences 25, no. 14: 7526. https://doi.org/10.3390/ijms25147526
APA StyleBecerra-Cervera, A., Argoty-Pantoja, A. D., Aparicio-Bautista, D. I., López-Montoya, P., Rivera-Paredez, B., Hidalgo-Bravo, A., & Velázquez-Cruz, R. (2024). Proteomic Biomarkers Associated with Low Bone Mineral Density: A Systematic Review. International Journal of Molecular Sciences, 25(14), 7526. https://doi.org/10.3390/ijms25147526








