Effect of Arthrospira maxima Phycobiliproteins, Rosiglitazone, and 17β-Estradiol on Lipogenic and Inflammatory Gene Expression during 3T3-L1 Preadipocyte Cell Differentiation
Abstract
:1. Introduction
2. Results
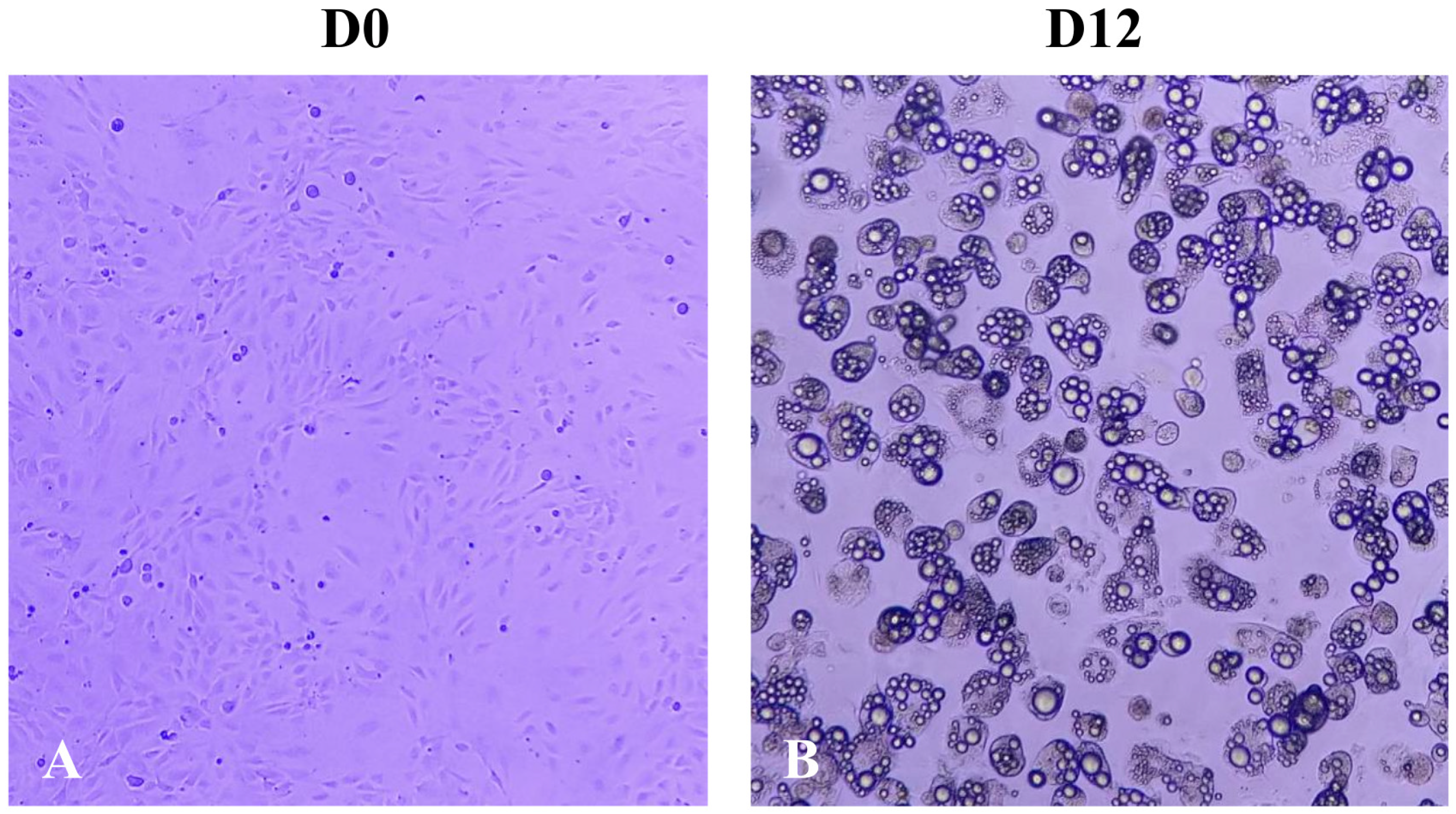
2.1. Effect of A. maxima Phycobiliproteins (PBPs) on Cell Proliferation of 3T3-L1 Preadipocytes
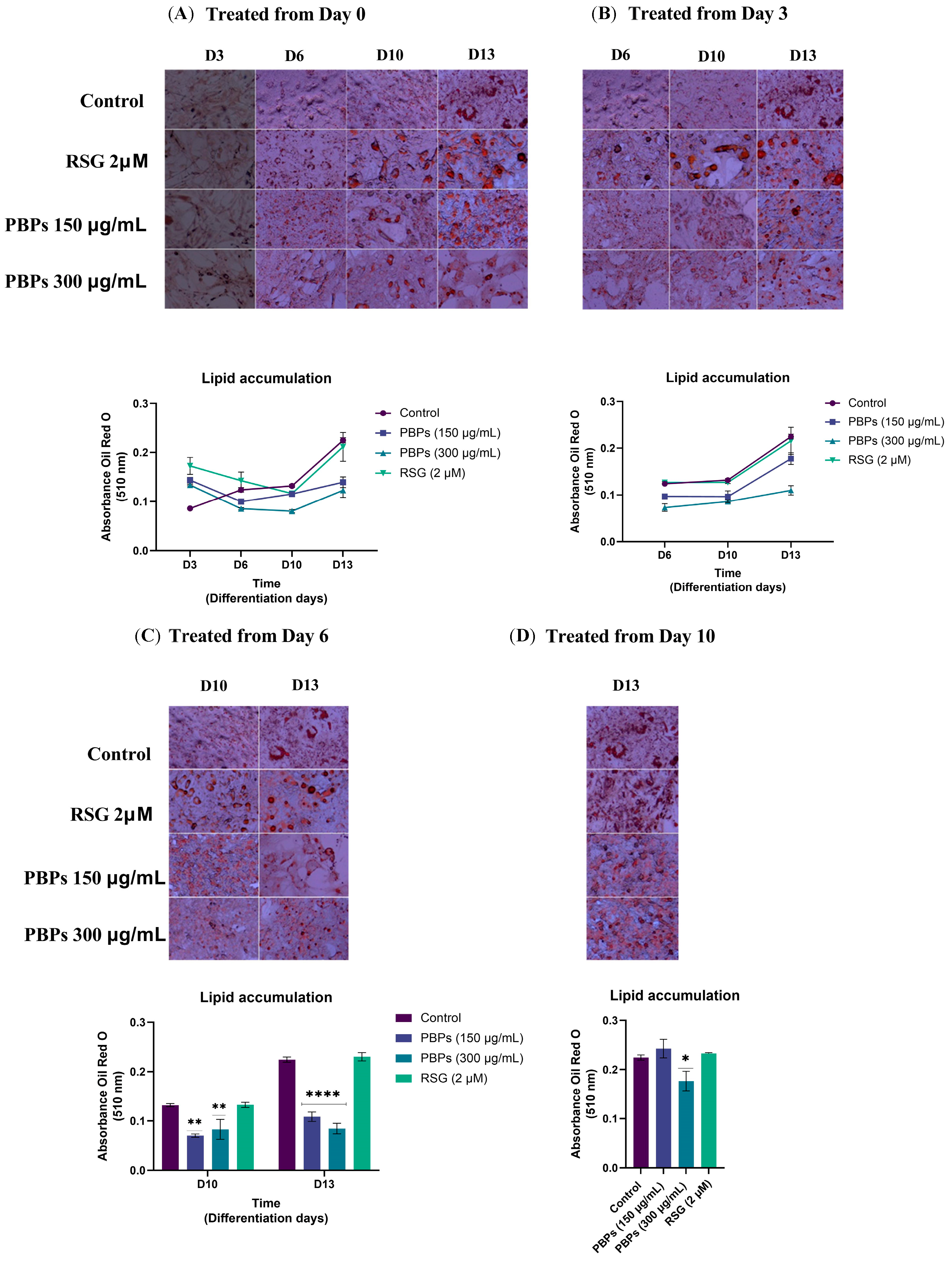
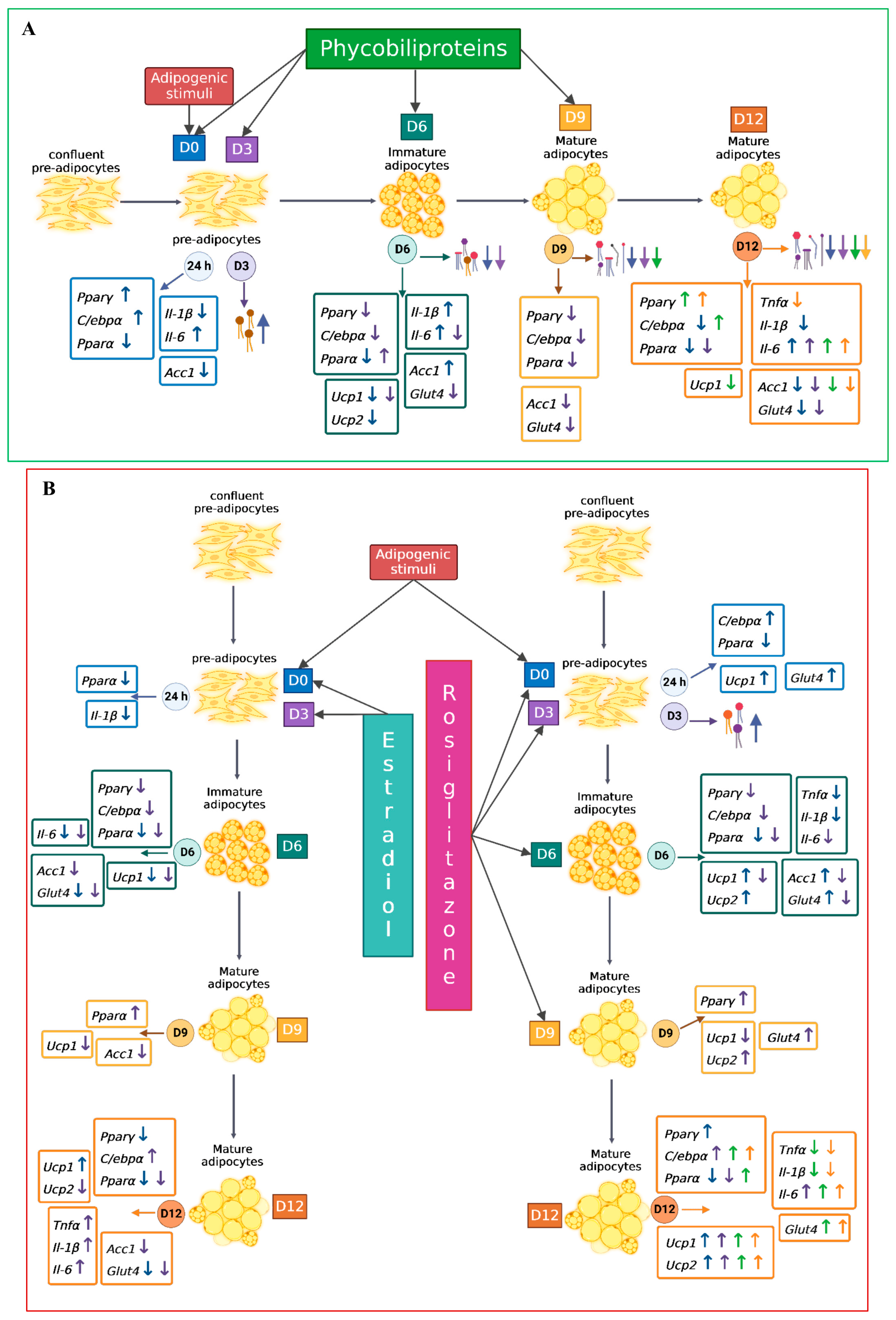
2.2. Effect of Treatment with A. maxima Phycobiliproteins (PBPs) and Rosiglitazone (RSG) on Adipogenesis in 3T3-L1 Cells
2.3. Effect of A. maxima Phycobiliproteins (PBPs) and Rosiglitazone (RSG) on Adipogenesis in 3T3-L1 Cells after Treatment at Different Time Points during 12 Days of Differentiation
2.4. Effect of A. maxima Phycobiliproteins (PBPs) on the Relative mRNA Expression of Genes Involved in Lipid Metabolism, Glucose Metabolism, Thermogenesis, and Antioxidant Defence in the Adipogenic Process in 3T3-L1 Cells
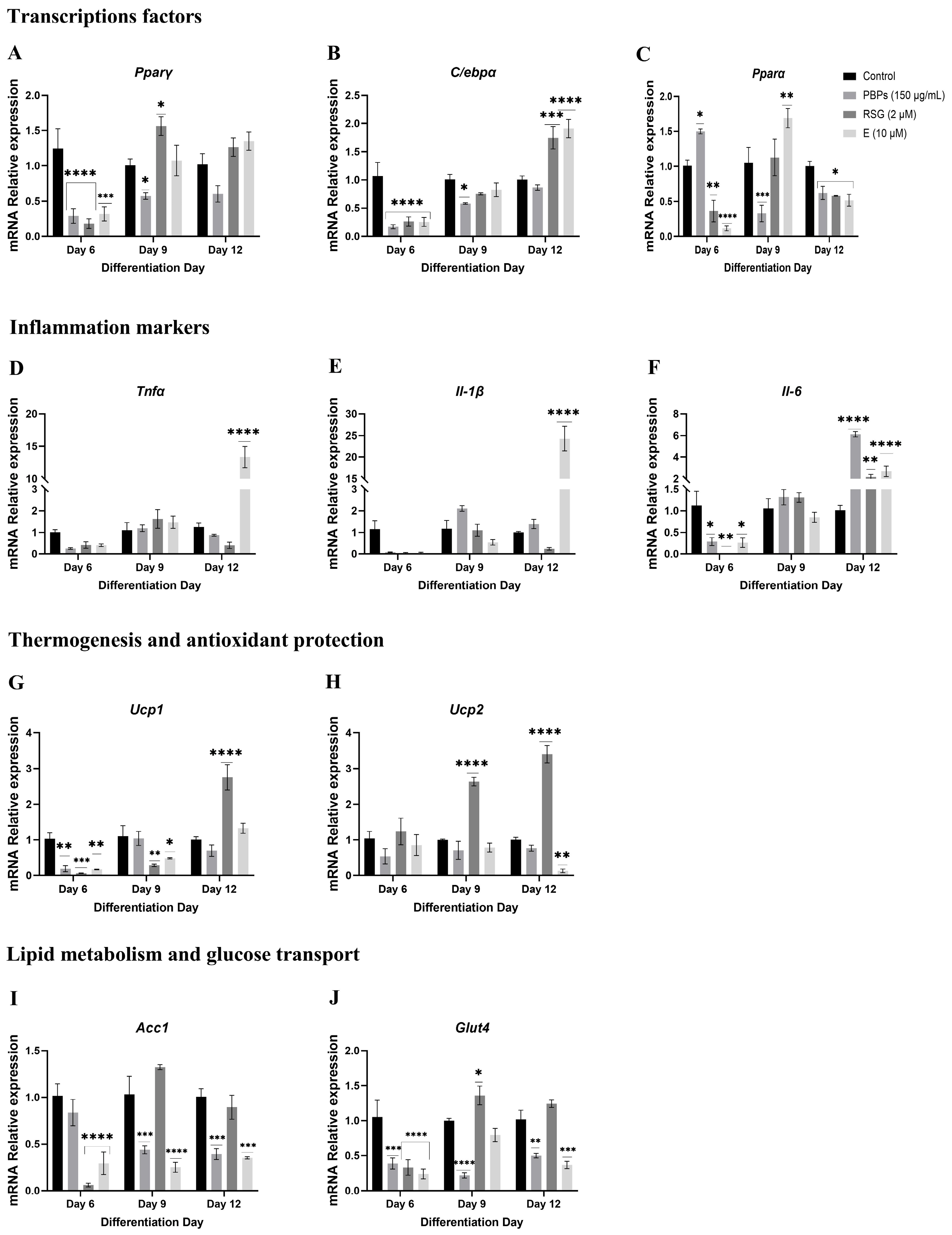
2.4.1. Treatment Initiation from Day 0 of Differentiation
mRNA Expression of Markers at 24 h of the Adipogenic Process
mRNA Expression of Markers at D6 of the Adipogenic Process
Expression of mRNA of Markers at Day 12 of the Adipogenic Process
2.4.2. Treatment Initiation from Day 3 of Differentiation
mRNA Expression of Markers at Day 6 of the Adipogenic Process
mRNA Expression of Markers at Day 9 of the Adipogenic Process
mRNA Expression of Markers at Day 12 of the Adipogenic Process
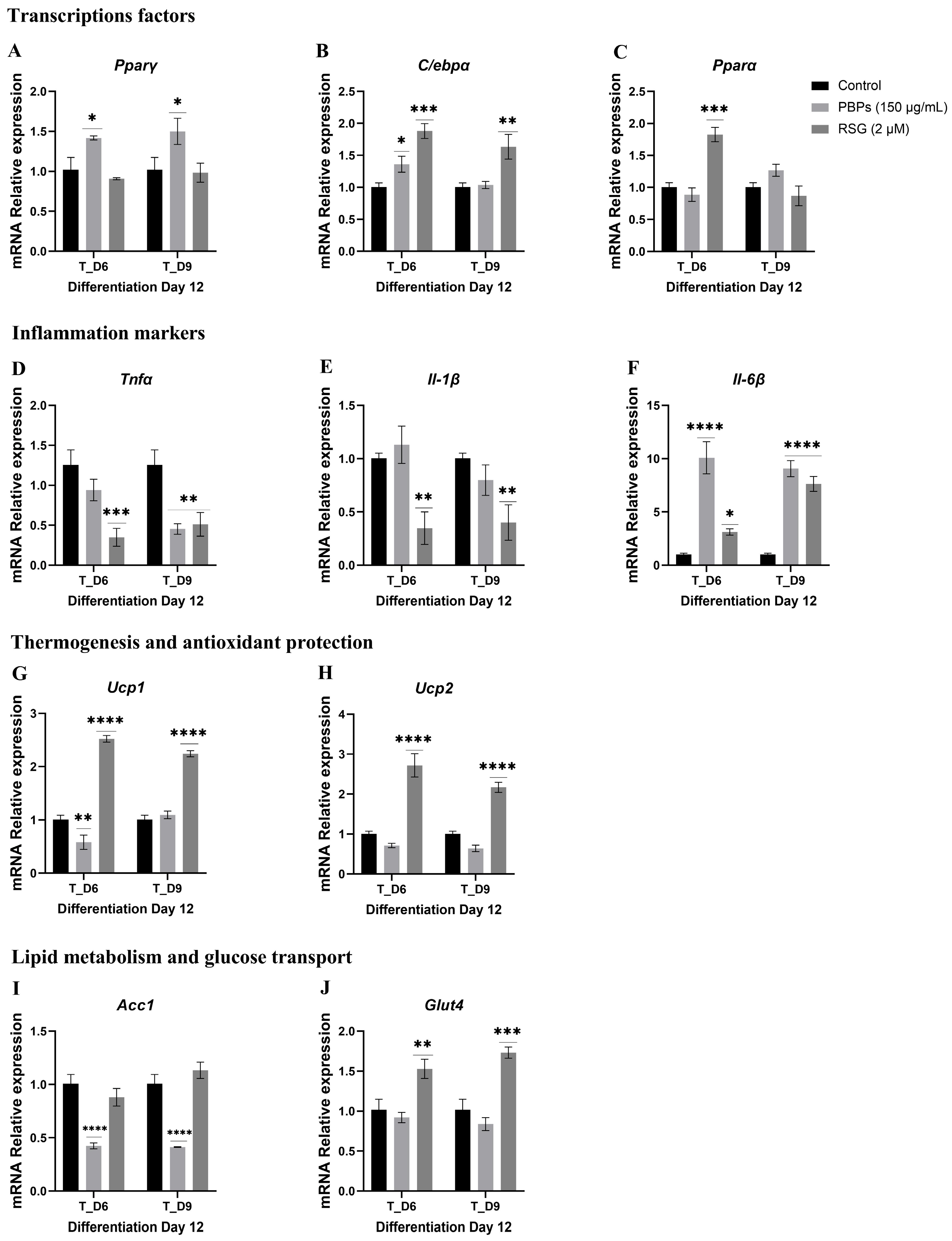
2.4.3. Treatment Initiation from Day 6 (T_D6) and Day 9 (T_D9) of Differentiation
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Cell Proliferation Assay
4.3. Differentiation of Preadipocytes into Adipocytes
4.4. Real-Time Quantitative Reverse Transcription-PCR (qRT-PCR)
4.5. Statistical Analyses
4.5.1. Cell Proliferation
4.5.2. Lipid Accumulation and Gene Expression
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Bernlohr, D.A.; Jenkins, A.E.; Bennaars, A.A. Adipose Tissue and Lipid Metabolism. In Biochemistry of Lipids, Lipoproteins and Membranes, 4th ed.; Vance, D.E., Vance, J.E., Eds.; Elsevier Science B.V.: Amsterdam, The Netherlands, 2002; pp. 263–288. [Google Scholar]
- Arsenijevic, T.; Grégoire, F.; Delforge, V.; Delporte, C.; Perret, J. Murine 3T3-L1 Adipocyte cell differentiation model: Validated reference genes for qPCR gene expression analysis. PLoS ONE 2012, 7, e37517. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Qiu, T.; Li, L.; Yu, R.; Chen, X.; Li, C.; Proud, C.G.; Jiang, T. Pathophysiology of obesity and its associated diseases. Acta Pharm. Sin. B 2023, 13, 2403–2424. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Sun, T.; Bookout, A.L.; Downes, M.; Yu, R.T.; Evans, R.M.; Mangelsdorf, D.J. A Nuclear receptor atlas: 3T3-L1 adipogenesis. Mol. Endocrinol. 2005, 19, 2437–2450. [Google Scholar] [CrossRef] [PubMed]
- Clavijo, M.A.; Gómez Camargo, D.; Gómez Alegría, C. In vitro Adipogenesis of 3T3-L1 Cells. Rev. Fac. Med. 2007, 15, 170–176. [Google Scholar]
- Balfour, J.A.; Plosker, G.L. Rosiglitazone. Drugs 1999, 57, 921–932. [Google Scholar] [CrossRef] [PubMed]
- Aydemir, D.; Sarayloo, E.; Ulusu, N.N. Rosiglitazone-induced changes in the oxidative stress metabolism and fatty acid composition in relation with trace element status in the primary adipocytes. J. Med. Biochem. 2020, 39, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, F.; Kamble, P.G.; Hetty, S.; Fanni, G.; Vranic, M.; Sarsenbayeva, A.; Kristófi, R.; Almby, K.; Svensson, M.K.; Pereira, M.J.; et al. Role of Estrogen and Its Receptors in Adipose Tissue Glucose Metabolism in Pre- and Postmenopausal Women. J. Clin. Endocrinol. Metab. 2022, 107, e1879–e1889. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.; Yoon, M. 17β-Estradiol inhibition of Pparγ-induced adipogenesis and adipocyte-specific gene expression. Acta Pharmacol. Sin. 2011, 32, 230–238. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Romero, S.; Torrella, J.R.; Pagès, T.; Viscor, G.; Torres, J.L. Edible Microalgae and Their Bioactive Compounds in the Prevention and Treatment of Metabolic Alterations. Nutrients 2021, 13, 563. [Google Scholar] [CrossRef]
- Daliri, E.B.; Oh, D.H.; Lee, B.H. Bioactive Peptides. Foods 2017, 6, 32. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.S. Peptides and Peptidomimetics as Potential Antiobesity Agents: Overview of Current Status. Front. Nutr. 2019, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Freitas, S.; Silva, N.G.; Sousa, M.L.; Ribeiro, T.; Rosa, F.; Leão, P.N.; Vasconcelos, V.; Reis, M.A.; Urbatzka, R. Chlorophyll derivatives from marine cyanobacteria with lipid-reducing activities. Mar. Drugs 2019, 17, 229. [Google Scholar] [CrossRef]
- Khan, M.I.; Khan, M.Z.; Shin, J.H.; Shin, T.S.; Lee, Y.B.; Kim, M.Y.; Kim, J.D. Pharmacological Approaches to Attenuate Inflammation and Obesity with Natural Products Formulations by Regulating the Associated Promoting Molecular Signaling Pathways. BioMed Res. Int. 2021, 2021, 2521273. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, Y.; Chen, W.; Ding, L.; Li, P.; Zhao, X.; Wang, X.; Li, A.; Bao, Q. Identification of differentially expressed proteins of Arthrospira (Spirulina) plantensis-YZ under salt-stress conditions by proteomics and qRT-PCR analysis. Proteome Sci. 2013, 11, 6. [Google Scholar] [CrossRef] [PubMed]
- Furmaniak, M.A.; Misztak, A.E.; Franczuk, M.D.; Wilmotte, A.; Waleron, M.; Waleron, K.F. Edible cyanobacterial genus Arthrospira: Actual state of the art in cultivation methods, genetics, and application in medicine. Front. Microbiol. 2017, 8, 2541. [Google Scholar] [CrossRef]
- Chen, H.; Qi, H.; Xiong, P. Phycobiliproteins-A Family of Algae-Derived Biliproteins: Productions, Characterization and Pharmaceutical Potentials. Mar. Drugs 2022, 20, 450. [Google Scholar] [CrossRef] [PubMed]
- Pagels, F.; Guedes, A.C.; Amaro, H.M.; Kijjoa, A.; Vasconcelos, V. Phycobiliproteins from cyanobacteria: Chemistry and biotechnological applications. Biotechnol. Adv. 2019, 37, 422–443. [Google Scholar] [CrossRef] [PubMed]
- Marles, R.J.; Barrett, M.L.; Barnes, J.; Chavez, M.L.; Gardiner, P.; Ko, R.; Mahady, G.B.; Dog, T.L.; Sarma, N.D.; Giancaspro, G.I.; et al. United states pharmacopeia safety evaluation of Spirulina. Crit. Rev. Food Sci. 2011, 51, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.J.; Kim, K.J.; Choi, J.; Koh, E.J.; Lee, B.Y. Spirulina maxima extract reduces obesity through suppression of adipogenesis and activation of browning in 3T3-L1 cells and high-fat diet-induced obese mice. Nutrients 2018, 10, 712. [Google Scholar] [CrossRef] [PubMed]
- Esteve Ràfols, M. Tejido adiposo: Heterogeneidad celular y diversidad funcional. Endocrinol. Nutr. 2014, 61, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Qiang, X.; Guo, C.; Gu, W.; Song, Y.; Zhang, Y.; Gong, X.; Wang, L.; Wang, G. The Complex of Phycobiliproteins, Fucoxanthin, and Krill Oil Ameliorates Obesity through Modulation of Lipid Metabolism and Antioxidants in Obese Rats. Nutrients 2022, 14, 4815. [Google Scholar] [CrossRef] [PubMed]
- Corrales, P.; Vidal-Puig, A.; Medina-Gómez, G. PPARS and metabolic disorders associated with challenged adipose tissue plasticity. Int. J. Mol. Sci. 2018, 19, 2124. [Google Scholar] [CrossRef] [PubMed]
- Halama, A.; Horsch, M.; Kastenmüller, G.; Möller, G.; Kumar, P.; Prehn, C.; Laumen, H.; Hauner, H.; Hrabě De Angelis, M.; Beckers, J.; et al. Metabolic switch during adipogenesis: From branched chain amino acid catabolism to lipid synthesis. Arch. Biochem. Biophys. 2016, 589, 93–107. [Google Scholar] [CrossRef] [PubMed]
- Ros Pérez, M.; Medina-Gómez, G. Obesidad, adipogénesis y resistencia a la insulina. Endocrinol. Nutr. 2011, 58, 360–369. [Google Scholar] [CrossRef]
- Zebisch, K.; Voigt, V.; Wabitsch, M.; Brandsch, M. Protocol for effective differentiation of 3T3-L1 cells to adipocytes. Anal. Biochem. 2012, 425, 88–90. [Google Scholar] [CrossRef] [PubMed]
- Mallikarjuna, B.G.; Manjappara, U.V. Obestatin and Rosiglitazone Differentially Modulate Lipid Metabolism through Peroxisome Proliferator-activated Receptor-γ (Pparγ) in Pre-adipose and Mature 3T3-L1 Cells. Cell Biochem. Biophys. 2021, 79, 73–85. [Google Scholar]
- Konno, T.; Sasaki, K.; Kobayashi, K.; Murata, T. Indirubin promotes adipocyte differentiation and reduces lipid accumulation in 3T3–L1 cells via peroxisome proliferator–activated receptor γ activation. Mol. Med. Rep. 2020, 21, 1552–1560. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.D.; Spiegelman, B.M. Molecular Regulation of Adipogenesis. Annu. Rev. Cell Dev. Biol. 2000, 16, 145–171. [Google Scholar] [CrossRef] [PubMed]
- Miehle, F.; Möller, G.; Cecil, A.; Lintelmann, J.; Wabitsch, M.; Tokarz, J.; Adamski, J.; Haid, M. Lipidomic phenotyping reveals extensive lipid remodeling during adipogenesis in human adipocytes. Metabolites 2020, 10, 217. [Google Scholar] [CrossRef] [PubMed]
- Farmer, S.R. Transcriptional control of adipocyte formation. Cell Metab. 2006, 4, 263–273. [Google Scholar] [CrossRef]
- Hinds, T.D.; Kipp, Z.A.; Xu, M.; Yiannikouris, F.B.; Morris, A.J.; Stec, D.F.; Wahli, W.; Stec, D.E. Adipose-specific Pparα knockout mice have increased lipogenesis by pask–srebp1 signaling and a polarity shift to inflammatory macrophages in white adipose tissue. Cells 2022, 11, 4. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.X.; Park, Y.K.; Lee, J.Y. Anti-inflammatory effects of spirulina platensis extract via the modulation of histone deacetylases. Nutrients 2016, 8, 381. [Google Scholar] [CrossRef] [PubMed]
- Ku, C.S.; Pham, T.X.; Park, Y.; Kim, B.; Shin, M.; Kang, I.; Lee, J. Edible blue-green algae reduce the production of pro- inflammatory cytokines by inhibiting NF-κB pathway in macrophages and splenocytes. Bone 2012, 23, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Choy, E.H.; De Benedetti, F.; Takeuchi, T.; Hashizume, M.; John, M.R.; Kishimoto, T. Translating IL-6 biology into effective treatments. Nat. Rev. Rheumatol. 2020, 16, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Franckhauser, S.; Elias, I.; Sopasakis, V.R.; Ferré, T.; Nagaev, I.; Andersson, C.X.; Agudo, J.; Ruberte, J.; Bosch, F.; Smith, U.; et al. Overexpression of Il-6 leads to hyperinsulinaemia, liver inflammation and reduced body weight in mice Abbreviations AMPK AMP kinase CMV cytomegalovirus CoA coenzyme A 2-DG 2-deoxy-D-[1-3 H]glucose GFP green fluorescent protein GLUT glucose transporter PKB. Diabetologia 2008, 51, 1306–1316. [Google Scholar] [CrossRef] [PubMed]
- Ellingsgaard, H.; Hauselmann, I.; Schuler, B.; Habib, A.M.; Baggio, L.L.; Meier, D.T.; Eppler, E.; Bouzakri, K.; Wueest, S.; Muller, Y.D.; et al. Interleukin-6 enhances insulin secretion by increasing glucagon-like peptide-1 secretion from L cells and alpha cells. Nat. Med. 2011, 17, 1481–1489. [Google Scholar] [CrossRef]
- Behl, S.; Adem, A.; Hussain, A.; Singh, J.; Hussain, A. Effects of rilpivirine, 17β-estradiol and β-naphthoflavone on the inflammatory status of release of adipocytokines in 3T3-L1 adipocytes in vitro. Mol. Biol. Rep. 2019, 46, 2643–2655. [Google Scholar] [CrossRef] [PubMed]
- Mandujano-Lázaro, G.; Galaviz-Hernández, C.; Reyes-López, C.A.; Almanza-Pérez, J.C.; Giacoman-Martínez, A.; López-Camarillo, C.; Huang, F.; Marchat, L.A.; Sanchez, R.A. Short S-Equol Exposure Has a Long-Term Inhibitory Effect on Adipogenesis in Mouse 3T3-L1 Cells. Appl. Sci. 2021, 11, 9657. [Google Scholar] [CrossRef]
- Li, J.; Jiang, R.; Cong, X.; Zhao, Y.; Zhao, C.Y. UCP2 gene polymorphisms in obesity and diabetes, and the role of UCP2 in cancer. FEBS Lett. 2019, 593, 2525–2534. [Google Scholar] [CrossRef] [PubMed]
- Azevedo Machado, S.; Pasquarelli-do-Nascimento, G.; Santos da Silva, D.; Ribeiro Farias, G.; de Oliveira Santos, I.; Borges Baptista, L.; Magalhães, K.G. Browning of the white adipose tissue regulation: New insights into nutritional and metabolic relevance in health and diseases. Nutr. Metab. 2022, 19, 61. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, J.C.; Romero, R.; Muñoz, L.V.; Rivera, R.A. Adipose organ, a metabolic and endocrine regulating rainbow. Rev. Cuba. Endocrinol. 2016, 27, 105–119. [Google Scholar]
- Moradi, S.; Khaje-Bishak, Y.; Alipour, M.; Alivand, M.; Alipour, B. The review of the relationship between UCP2 and obesity: Focusing on inflammatory-obesity. New Insights Obes. Gene Beyond 2021, 5, 001–013. [Google Scholar]
- Guzmán-Gómez, O.; García-Rodríguez, R.V.; Quevedo-Corona, L.; Pérez-Pastén-Borja, R.; Rivero-Ramírez, N.L.; Ríos-Castro, E.; Pérez-Gutiérrez, S.; Pérez-Ramos, J.; Chamorro-Cevallos, G.A. Amelioration of ethanol-induced gastric ulcers in rats pretreated with phycobiliproteins of Arthrospira (Spirulina) maxima. Nutrients 2018, 10, 763. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-García, R.M.; Jaramillo-Flores, M.E. Effect of Arthrospira maxima Phycobiliproteins, Rosiglitazone, and 17β-Estradiol on Lipogenic and Inflammatory Gene Expression during 3T3-L1 Preadipocyte Cell Differentiation. Int. J. Mol. Sci. 2024, 25, 7566. https://doi.org/10.3390/ijms25147566
García-García RM, Jaramillo-Flores ME. Effect of Arthrospira maxima Phycobiliproteins, Rosiglitazone, and 17β-Estradiol on Lipogenic and Inflammatory Gene Expression during 3T3-L1 Preadipocyte Cell Differentiation. International Journal of Molecular Sciences. 2024; 25(14):7566. https://doi.org/10.3390/ijms25147566
Chicago/Turabian StyleGarcía-García, Ruth Marina, and María Eugenia Jaramillo-Flores. 2024. "Effect of Arthrospira maxima Phycobiliproteins, Rosiglitazone, and 17β-Estradiol on Lipogenic and Inflammatory Gene Expression during 3T3-L1 Preadipocyte Cell Differentiation" International Journal of Molecular Sciences 25, no. 14: 7566. https://doi.org/10.3390/ijms25147566







