MEG3-Mediated Oral Squamous-Cell-Carcinoma-Derived Exosomal miR-421 Activates Angiogenesis by Targeting HS2ST1 in Vascular Endothelial Cells
Abstract
1. Introduction
2. Results
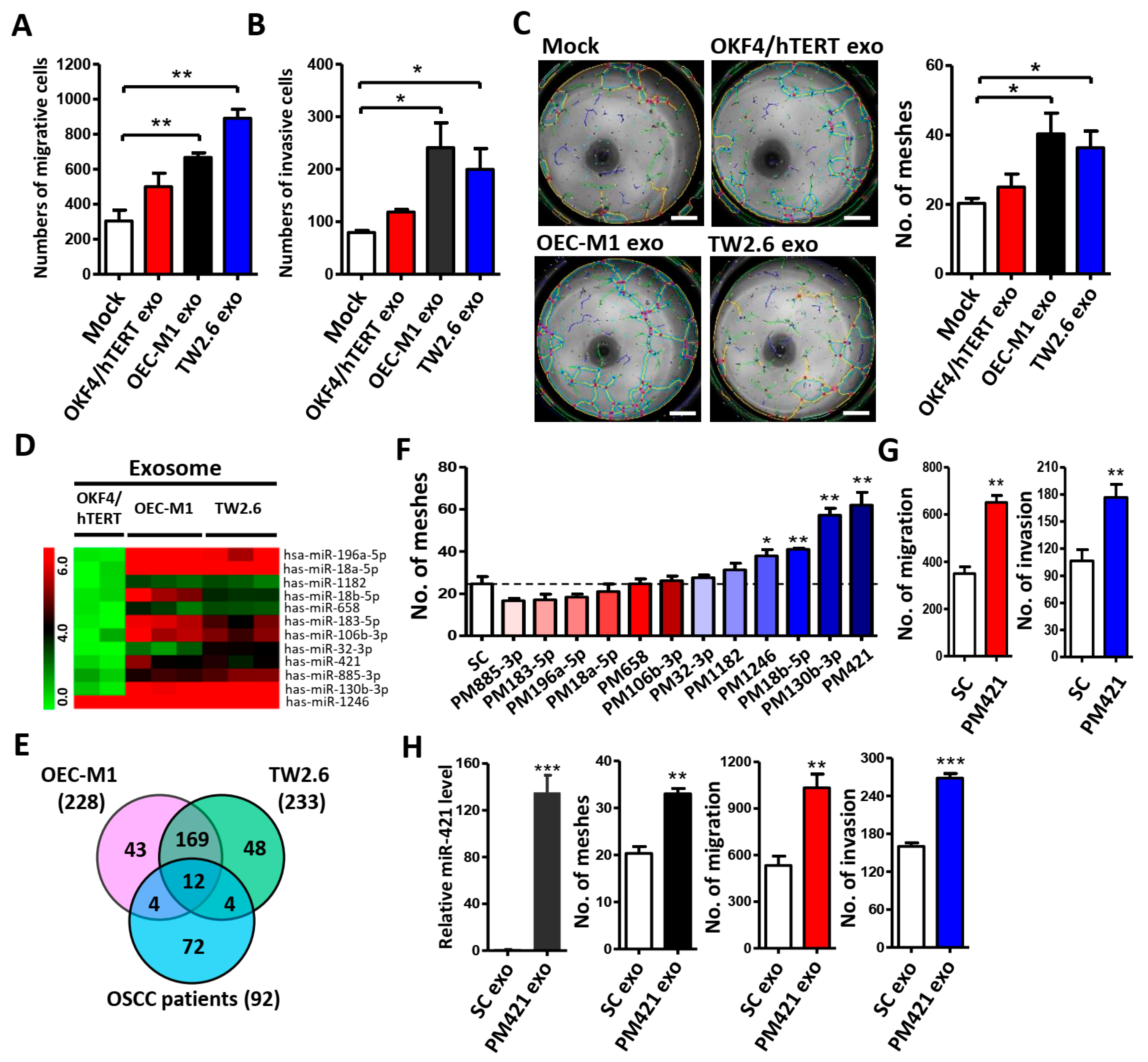
2.1. OSCC Cell-Derived Exosomal miR-421 Promotes HUVEC Tube Formation, Migration, and Invasion
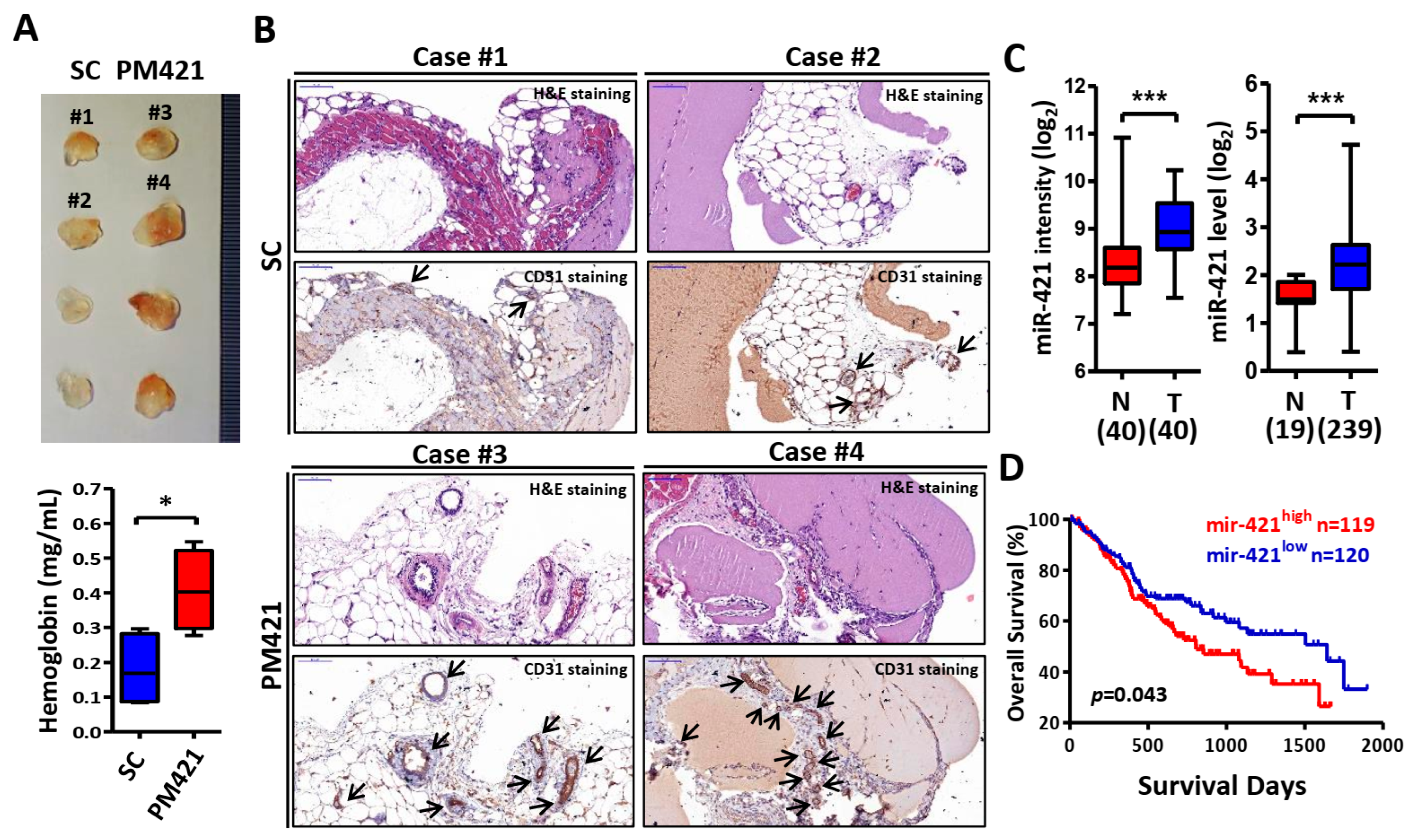
2.2. miR-421 Induces Neovascularization In Vivo
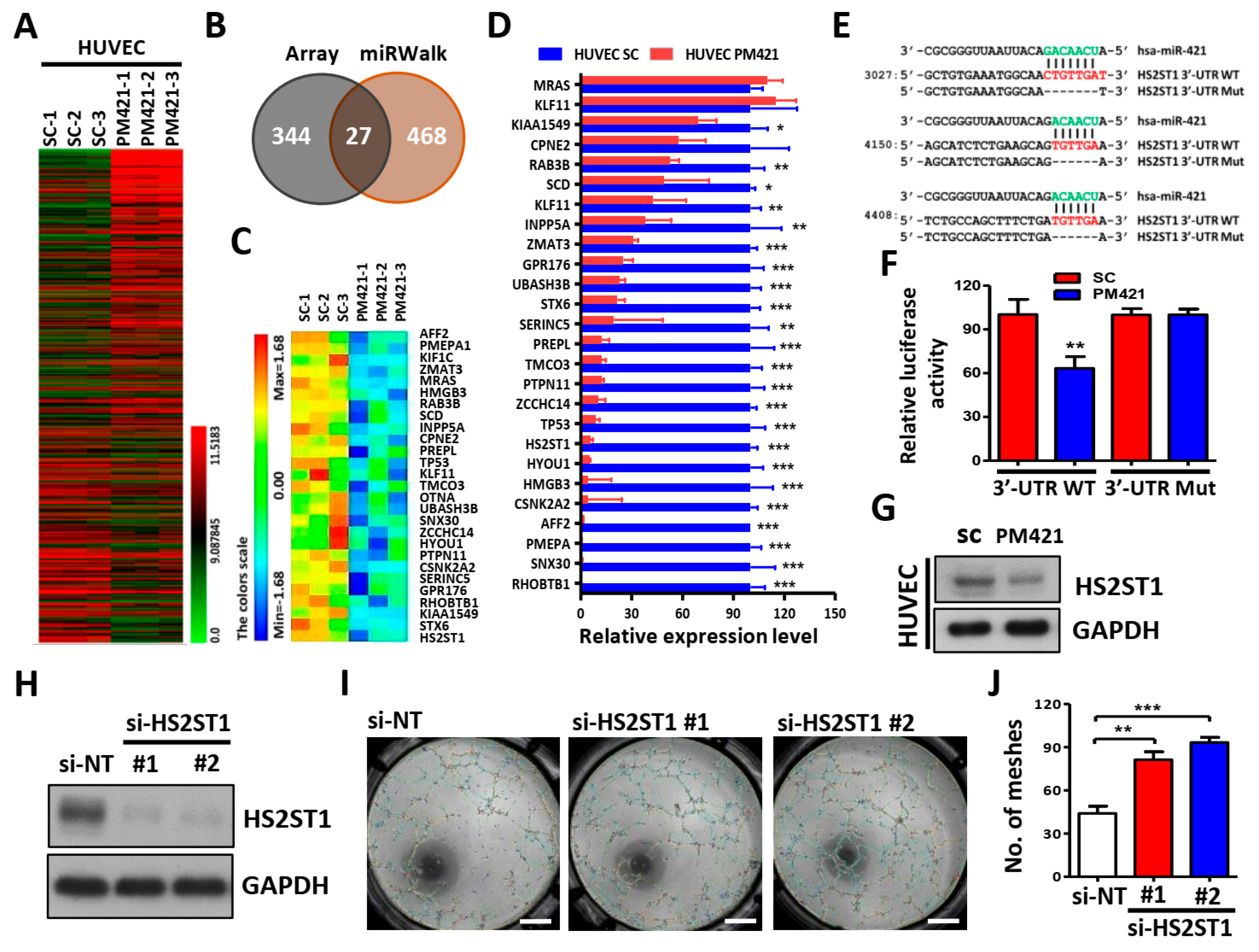
2.3. HS2ST1 Is a Target of miR-421 in HUVECs
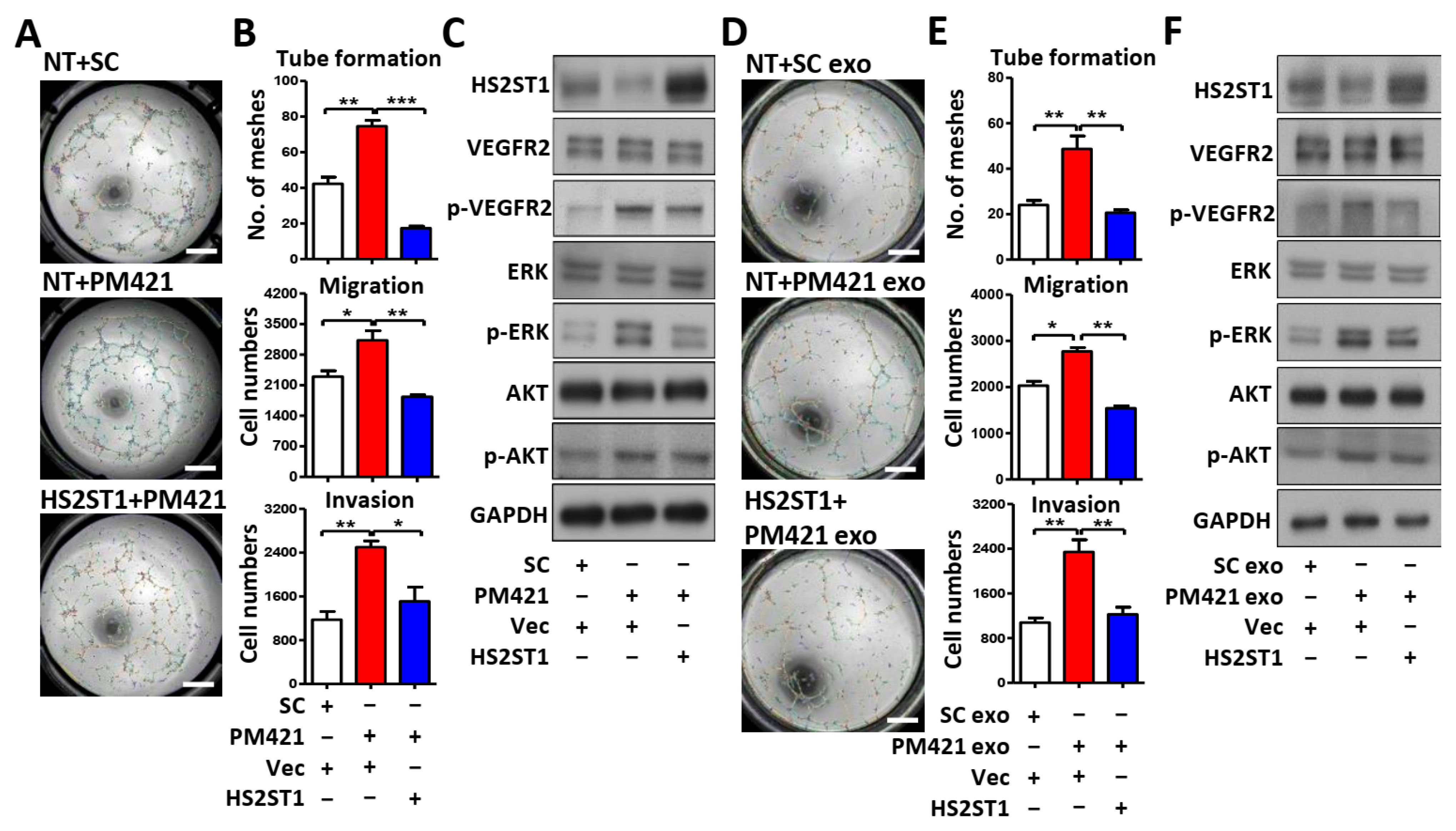
2.4. HS2ST1 Modulates VEGF-Mediated Activation of ERK and AKT in HUVECs
2.5. Exosomal miR-421 Promotes Tube Formation by Targeting HS2ST1
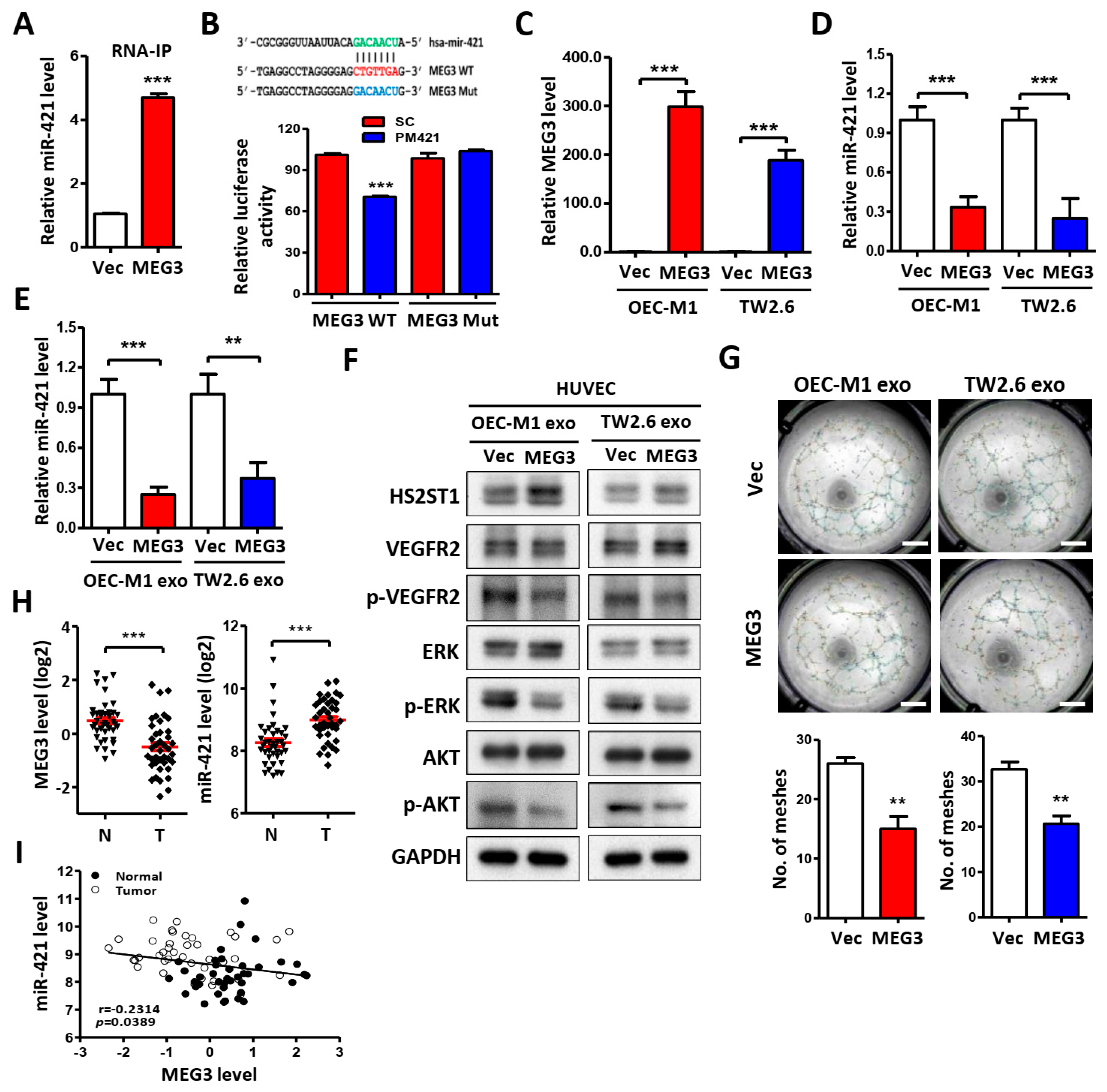
2.6. LncRNA MEG3 Inhibits Tube Formation via Targeting miR-421
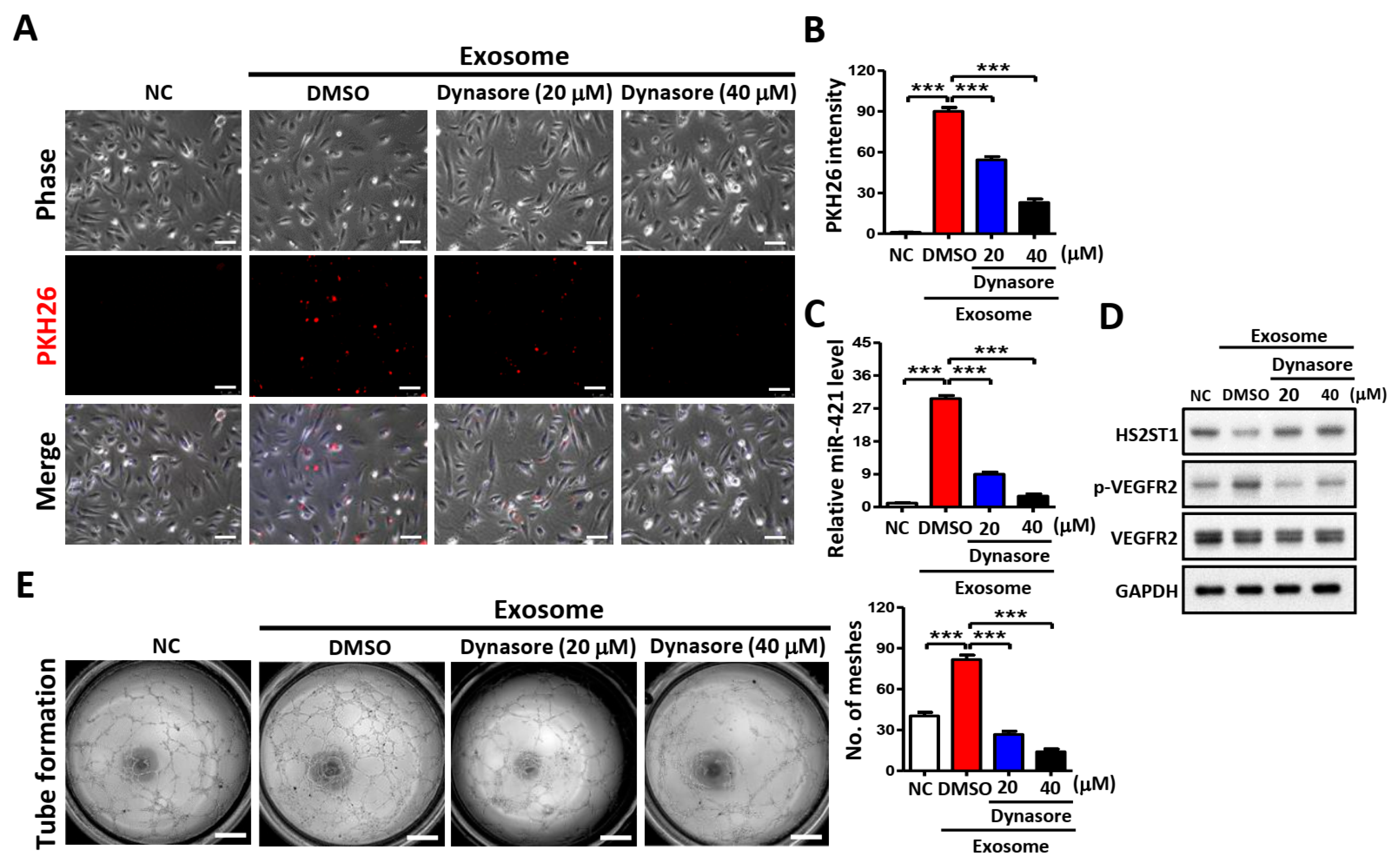
2.7. Blocking Exosome Uptake Inhibits Tube Formation in HUVECs
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Exosome Purification, Characterization, and Quantification
4.3. Tube Formation Assay
4.4. Cell Migration and Invasion Assays
4.5. RNA Extraction, Reverse Transcription, and Quantitative PCR (q-PCR)
4.6. Exosomal miRNA Profiling
4.7. In Vivo Matrigel Plug Assay
4.8. Clinical Samples and Microarray Profiling
4.9. Plasmids, Transfection, and Virus Infection Assays
4.10. Luciferase Reporter Assay
4.11. Protein Extraction and Western Blotting Analysis
4.12. RNA Immunoprecipitation (RNA-IP)
4.13. Fluorescence Analysis and Exosome Uptake Assay
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Panarese, I.; Aquino, G.; Ronchi, A.; Longo, F.; Montella, M.; Cozzolino, I.; Roccuzzo, G.; Colella, G.; Caraglia, M.; Franco, R. Oral and Oropharyngeal squamous cell carcinoma: Prognostic and predictive parameters in the etiopathogenetic route. Expert. Rev. Anticancer Ther. 2019, 19, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Primers 2020, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Roi, A.; Roi, C.I.; Negrutiu, M.L.; Rivis, M.; Sinescu, C.; Rusu, L.C. The Challenges of OSCC Diagnosis: Salivary Cytokines as Potential Biomarkers. J. Clin. Med. 2020, 9, 2866. [Google Scholar] [CrossRef] [PubMed]
- Macluskey, M.; Chandrachud, L.M.; Pazouki, S.; Green, M.; Chisholm, D.M.; Ogden, G.R.; Schor, S.L.; Schor, A.M. Apoptosis, proliferation, and angiogenesis in oral tissues. Possible relevance to tumour progression. J. Pathol. 2000, 191, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Koontongkaew, S. The tumor microenvironment contribution to development, growth, invasion and metastasis of head and neck squamous cell carcinomas. J. Cancer 2013, 4, 66–83. [Google Scholar] [CrossRef] [PubMed]
- Sarrazin, S.; Lamanna, W.C.; Esko, J.D. Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 2011, 3, a004952. [Google Scholar] [CrossRef] [PubMed]
- Bishop, J.R.; Schuksz, M.; Esko, J.D. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 2007, 446, 1030–1037. [Google Scholar] [CrossRef] [PubMed]
- Espinoza-Sanchez, N.A.; Gotte, M. Role of cell surface proteoglycans in cancer immunotherapy. Semin. Cancer Biol. 2020, 62, 48–67. [Google Scholar] [CrossRef] [PubMed]
- Denys, A.; Allain, F. The Emerging Roles of Heparan Sulfate 3-O-Sulfotransferases in Cancer. Front. Oncol. 2019, 9, 507. [Google Scholar] [CrossRef] [PubMed]
- Karamanos, N.K.; Piperigkou, Z.; Theocharis, A.D.; Watanabe, H.; Franchi, M.; Baud, S.; Brezillon, S.; Gotte, M.; Passi, A.; Vigetti, D.; et al. Proteoglycan Chemical Diversity Drives Multifunctional Cell Regulation and Therapeutics. Chem. Rev. 2018, 118, 9152–9232. [Google Scholar] [CrossRef]
- Hull, E.E.; Montgomery, M.R.; Leyva, K.J. Epigenetic Regulation of the Biosynthesis & Enzymatic Modification of Heparan Sulfate Proteoglycans: Implications for Tumorigenesis and Cancer Biomarkers. Int. J. Mol. Sci. 2017, 18, 1361. [Google Scholar] [CrossRef]
- Witt, R.M.; Hecht, M.L.; Pazyra-Murphy, M.F.; Cohen, S.M.; Noti, C.; van Kuppevelt, T.H.; Fuller, M.; Chan, J.A.; Hopwood, J.J.; Seeberger, P.H.; et al. Heparan sulfate proteoglycans containing a glypican 5 core and 2-O-sulfo-iduronic acid function as Sonic Hedgehog co-receptors to promote proliferation. J. Biol. Chem. 2013, 288, 26275–26288. [Google Scholar] [CrossRef] [PubMed]
- Esko, J.D.; Selleck, S.B. Order out of chaos: Assembly of ligand binding sites in heparan sulfate. Annu. Rev. Biochem. 2002, 71, 435–471. [Google Scholar] [CrossRef] [PubMed]
- Vijaya Kumar, A.; Brezillon, S.; Untereiner, V.; Sockalingum, G.D.; Kumar Katakam, S.; Mohamed, H.T.; Kemper, B.; Greve, B.; Mohr, B.; Ibrahim, S.A.; et al. HS2ST1-dependent signaling pathways determine breast cancer cell viability, matrix interactions, and invasive behavior. Cancer Sci. 2020, 111, 2907–2922. [Google Scholar] [CrossRef] [PubMed]
- Bonuccelli, G.; Casimiro, M.C.; Sotgia, F.; Wang, C.; Liu, M.; Katiyar, S.; Zhou, J.; Dew, E.; Capozza, F.; Daumer, K.M.; et al. Caveolin-1 (P132L), a common breast cancer mutation, confers mammary cell invasiveness and defines a novel stem cell/metastasis-associated gene signature. Am. J. Pathol. 2009, 174, 1650–1662. [Google Scholar] [CrossRef] [PubMed]
- Constantinescu, A.A.; Vink, H.; Spaan, J.A. Endothelial cell glycocalyx modulates immobilization of leukocytes at the endothelial surface. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 1541–1547. [Google Scholar] [CrossRef]
- Pretorius, D.; Richter, R.P.; Anand, T.; Cardenas, J.C.; Richter, J.R. Alterations in heparan sulfate proteoglycan synthesis and sulfation and the impact on vascular endothelial function. Matrix Biol. Plus 2022, 16, 100121. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Zheng, S.; Luo, Y.; Wang, B. Exosome Theranostics: Biology and Translational Medicine. Theranostics 2018, 8, 237–255. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Peng, F.; Chen, J. The Role of Exosomal MicroRNAs in the Tumor Microenvironment of Breast Cancer. Int. J. Mol. Sci. 2019, 20, 3884. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.B.S.; Anjali, K. Tumour generated exosomal miRNAs: A major player in tumour angiogenesis. Biochim. Biophys. Acta Mol. Basis Dis. 2022, 1868, 166383. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Zhang, W.; Li, X.; Wang, J.; Chen, X.; Chen, Y.; Lai, R. Oral squamous cell carcinoma (OSCC)-derived exosomal MiR-221 targets and regulates phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1) to promote human umbilical vein endothelial cells migration and tube formation. Bioengineered 2021, 12, 2164–2174. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, L.; Zhou, X.; Luo, X.; Liu, K.; Jiang, E.; Chen, Y.; Shao, Z.; Shang, Z. OSCC Exosomes Regulate miR-210-3p Targeting EFNA3 to Promote Oral Cancer Angiogenesis through the PI3K/AKT Pathway. Biomed. Res. Int. 2020, 2020, 2125656. [Google Scholar] [CrossRef] [PubMed]
- Hao, C.; Wangzhou, K.; Liang, Z.; Liu, C.; Wang, L.; Gong, L.; Tan, Y.; Li, C.; Lai, Z.; Hu, G. Circular RNA ITCH Suppresses Cell Proliferation but Induces Apoptosis in Oral Squamous Cell Carcinoma by Regulating miR-421/PDCD4 Axis. Cancer Manag. Res. 2020, 12, 5651–5658. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lin, Z.; Gao, Y.; Yao, T. Downregulation of long noncoding RNA MEG3 is associated with poor prognosis and promoter hypermethylation in cervical cancer. J. Exp. Clin. Cancer Res. 2017, 36, 5. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Gejman, R.; Mahta, A.; Zhong, Y.; Rice, K.A.; Zhou, Y.; Cheunsuchon, P.; Louis, D.N.; Klibanski, A. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res. 2010, 70, 2350–2358. [Google Scholar] [CrossRef] [PubMed]
- Shiah, S.G.; Hsiao, J.R.; Chang, W.M.; Chen, Y.W.; Jin, Y.T.; Wong, T.Y.; Huang, J.S.; Tsai, S.T.; Hsu, Y.M.; Chou, S.T.; et al. Downregulated miR329 and miR410 promote the proliferation and invasion of oral squamous cell carcinoma by targeting Wnt-7b. Cancer Res. 2014, 74, 7560–7572. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Wang, Y.; Chen, Y.; Guo, Y.; Li, Q.; Wei, X. Exosomal miR-130b-3p Promotes Progression and Tubular Formation Through Targeting PTEN in Oral Squamous Cell Carcinoma. Front. Cell Dev. Biol. 2021, 9, 616306. [Google Scholar] [CrossRef] [PubMed]
- Li, J.P.; Kusche-Gullberg, M. Heparan Sulfate: Biosynthesis, Structure, and Function. Int. Rev. Cell Mol. Biol. 2016, 325, 215–273. [Google Scholar] [CrossRef]
- Sasisekharan, R.; Shriver, Z.; Venkataraman, G.; Narayanasami, U. Roles of heparan-sulphate glycosaminoglycans in cancer. Nat. Rev. Cancer 2002, 2, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Beckouche, N.; Bignon, M.; Lelarge, V.; Mathivet, T.; Pichol-Thievend, C.; Berndt, S.; Hardouin, J.; Garand, M.; Ardidie-Robouant, C.; Barret, A.; et al. The interaction of heparan sulfate proteoglycans with endothelial transglutaminase-2 limits VEGF165-induced angiogenesis. Sci. Signal. 2015, 8, ra70. [Google Scholar] [CrossRef] [PubMed]
- Ashikari-Hada, S.; Habuchi, H.; Sugaya, N.; Kobayashi, T.; Kimata, K. Specific inhibition of FGF-2 signaling with 2-O-sulfated octasaccharides of heparan sulfate. Glycobiology 2009, 19, 644–654. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Zhang, A.; Liu, S.; Lu, F.; Guo, Y.; Zhang, G.; Xu, F.; Shi, Y.; Shen, S.; Liang, J.; et al. Aberrant Methylation-Mediated Silencing of lncRNA MEG3 Functions as a ceRNA in Esophageal Cancer. Mol. Cancer Res. 2017, 15, 800–810. [Google Scholar] [CrossRef]
- Zheng, Y.; Tu, C.; Zhang, J.; Wang, J. Inhibition of multiple myeloma-derived exosomes uptake suppresses the functional response in bone marrow stromal cell. Int. J. Oncol. 2019, 54, 1061–1070. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, X.; Klibanski, A. MEG3 noncoding RNA: A tumor suppressor. J. Mol. Endocrinol. 2012, 48, R45–R53. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Luo, Y.; Liang, B.; Ye, L.; Lu, G.; He, W. Potential applications of MEG3 in cancer diagnosis and prognosis. Oncotarget 2017, 8, 73282–73295. [Google Scholar] [CrossRef] [PubMed]
- Deng, D.; Liang, H. Silencing MEG3 protects PC12 cells from hypoxic injury by targeting miR-21. Artif. Cells Nanomed. Biotechnol. 2020, 48, 610–619. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Ji, W.W.; Wei, W.; Zhan, L.X.; Huang, X. Long noncoding RNA Meg3 sponges miR-708 to inhibit intestinal tumorigenesis via SOCS3-repressed cancer stem cells growth. Cell Death Dis. 2021, 13, 25. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Feng, G.; Hou, Y.; Yu, Y.; Wang, R.; Yuan, H. Long noncoding RNA MEG3 decreases the growth of head and neck squamous cell carcinoma by regulating the expression of miR-421 and E-cadherin. Cancer Med. 2020, 9, 3954–3963. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Ni, Z.; Yicheng, S.; Pan, H.; Huang, Y.; Xiong, Y.; Liu, T. Anisomycin inhibits angiogenesis in ovarian cancer by attenuating the molecular sponge effect of the lncRNA-Meg3/miR-421/PDGFRA axis. Int. J. Oncol. 2019, 55, 1296–1312. [Google Scholar] [CrossRef] [PubMed]
- Rezaie, J.; Feghhi, M.; Etemadi, T. A review on exosomes application in clinical trials: Perspective, questions, and challenges. Cell Commun. Signal. 2022, 20, 145. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Bi, J.; Huang, J.; Tang, Y.; Du, S.; Li, P. Exosome: A Review of Its Classification, Isolation Techniques, Storage, Diagnostic and Targeted Therapy Applications. Int. J. Nanomed. 2020, 15, 6917–6934. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, C.; Liu, Z.; Guo, H.; Lu, W.; Hu, W.; Lin, Z. PABPC1-induced stabilization of IFI27 mRNA promotes angiogenesis and malignant progression in esophageal squamous cell carcinoma through exosomal miRNA-21-5p. J. Exp. Clin. Cancer Res. 2022, 41, 111. [Google Scholar] [CrossRef] [PubMed]
- Catalano, M.; O’Driscoll, L. Inhibiting extracellular vesicles formation and release: A review of EV inhibitors. J. Extracell. Vesicles 2020, 9, 1703244. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Lee, C.H.; Baek, M.C. Dissecting exosome inhibitors: Therapeutic insights into small-molecule chemicals against cancer. Exp. Mol. Med. 2022, 54, 1833–1843. [Google Scholar] [CrossRef] [PubMed]
- Escudier, B.; Dorval, T.; Chaput, N.; Andre, F.; Caby, M.P.; Novault, S.; Flament, C.; Leboulaire, C.; Borg, C.; Amigorena, S.; et al. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: Results of thefirst phase I clinical trial. J. Transl. Med. 2005, 3, 10. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yu, F.; Ding, H.; Wang, Y.; Li, P.; Wang, K. Emerging Function and Clinical Values of Exosomal MicroRNAs in Cancer. Mol. Ther. Nucleic Acids 2019, 16, 791–804. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, F.; Vijaya Kumar, A.; Kumar Katakam, S.; Cocola, C.; Pelucchi, P.; Graf, M.; Kiesel, L.; Reinbold, R.; Pavao, M.S.G.; Greve, B.; et al. The Heparan Sulfate Sulfotransferases HS2ST1 and HS3ST2 Are Novel Regulators of Breast Cancer Stem-Cell Properties. Front. Cell Dev. Biol. 2020, 8, 559554. [Google Scholar] [CrossRef] [PubMed]
- Chou, S.T.; Peng, H.Y.; Mo, K.C.; Hsu, Y.M.; Wu, G.H.; Hsiao, J.R.; Lin, S.F.; Wang, H.D.; Shiah, S.G. MicroRNA-486-3p functions as a tumor suppressor in oral cancer by targeting DDR1. J. Exp. Clin. Cancer Res. 2019, 38, 281. [Google Scholar] [CrossRef] [PubMed]
- Shiah, S.G.; Hsiao, J.R.; Chang, H.J.; Hsu, Y.M.; Wu, G.H.; Peng, H.Y.; Chou, S.T.; Kuo, C.C.; Chang, J.Y. MiR-30a and miR-379 modulate retinoic acid pathway by targeting DNA methyltransferase 3B in oral cancer. J. Biomed. Sci. 2020, 27, 46. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.Y.; Jiang, S.S.; Hsiao, J.R.; Hsiao, M.; Hsu, Y.M.; Wu, G.H.; Chang, W.M.; Chang, J.Y.; Jin, S.L.; Shiah, S.G. IL-8 induces miR-424-5p expression and modulates SOCS2/STAT5 signaling pathway in oral squamous cell carcinoma. Mol. Oncol. 2016, 10, 895–909. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhu, J.; Jiang, T.; Zhong, Y.; Tie, Y.; Wu, Y.; Zheng, X.; Jin, Y.; Fu, H. Identification of lncRNA MEG3 Binding Protein Using MS2-Tagged RNA Affinity Purification and Mass Spectrometry. Appl. Biochem. Biotechnol. 2015, 176, 1834–1845. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, C.-Y.; Chou, S.-T.; Hsu, Y.-M.; Chao, W.-J.; Wu, G.-H.; Hsiao, J.-R.; Wang, H.-D.; Shiah, S.-G. MEG3-Mediated Oral Squamous-Cell-Carcinoma-Derived Exosomal miR-421 Activates Angiogenesis by Targeting HS2ST1 in Vascular Endothelial Cells. Int. J. Mol. Sci. 2024, 25, 7576. https://doi.org/10.3390/ijms25147576
Huang C-Y, Chou S-T, Hsu Y-M, Chao W-J, Wu G-H, Hsiao J-R, Wang H-D, Shiah S-G. MEG3-Mediated Oral Squamous-Cell-Carcinoma-Derived Exosomal miR-421 Activates Angiogenesis by Targeting HS2ST1 in Vascular Endothelial Cells. International Journal of Molecular Sciences. 2024; 25(14):7576. https://doi.org/10.3390/ijms25147576
Chicago/Turabian StyleHuang, Chia-Yun, Sung-Tau Chou, Yuan-Ming Hsu, Wan-Ju Chao, Guan-Hsun Wu, Jenn-Ren Hsiao, Horng-Dar Wang, and Shine-Gwo Shiah. 2024. "MEG3-Mediated Oral Squamous-Cell-Carcinoma-Derived Exosomal miR-421 Activates Angiogenesis by Targeting HS2ST1 in Vascular Endothelial Cells" International Journal of Molecular Sciences 25, no. 14: 7576. https://doi.org/10.3390/ijms25147576
APA StyleHuang, C.-Y., Chou, S.-T., Hsu, Y.-M., Chao, W.-J., Wu, G.-H., Hsiao, J.-R., Wang, H.-D., & Shiah, S.-G. (2024). MEG3-Mediated Oral Squamous-Cell-Carcinoma-Derived Exosomal miR-421 Activates Angiogenesis by Targeting HS2ST1 in Vascular Endothelial Cells. International Journal of Molecular Sciences, 25(14), 7576. https://doi.org/10.3390/ijms25147576





