Applications of Microbial Organophosphate-Degrading Enzymes to Detoxification of Organophosphorous Compounds for Medical Countermeasures against Poisoning and Environmental Remediation
Abstract
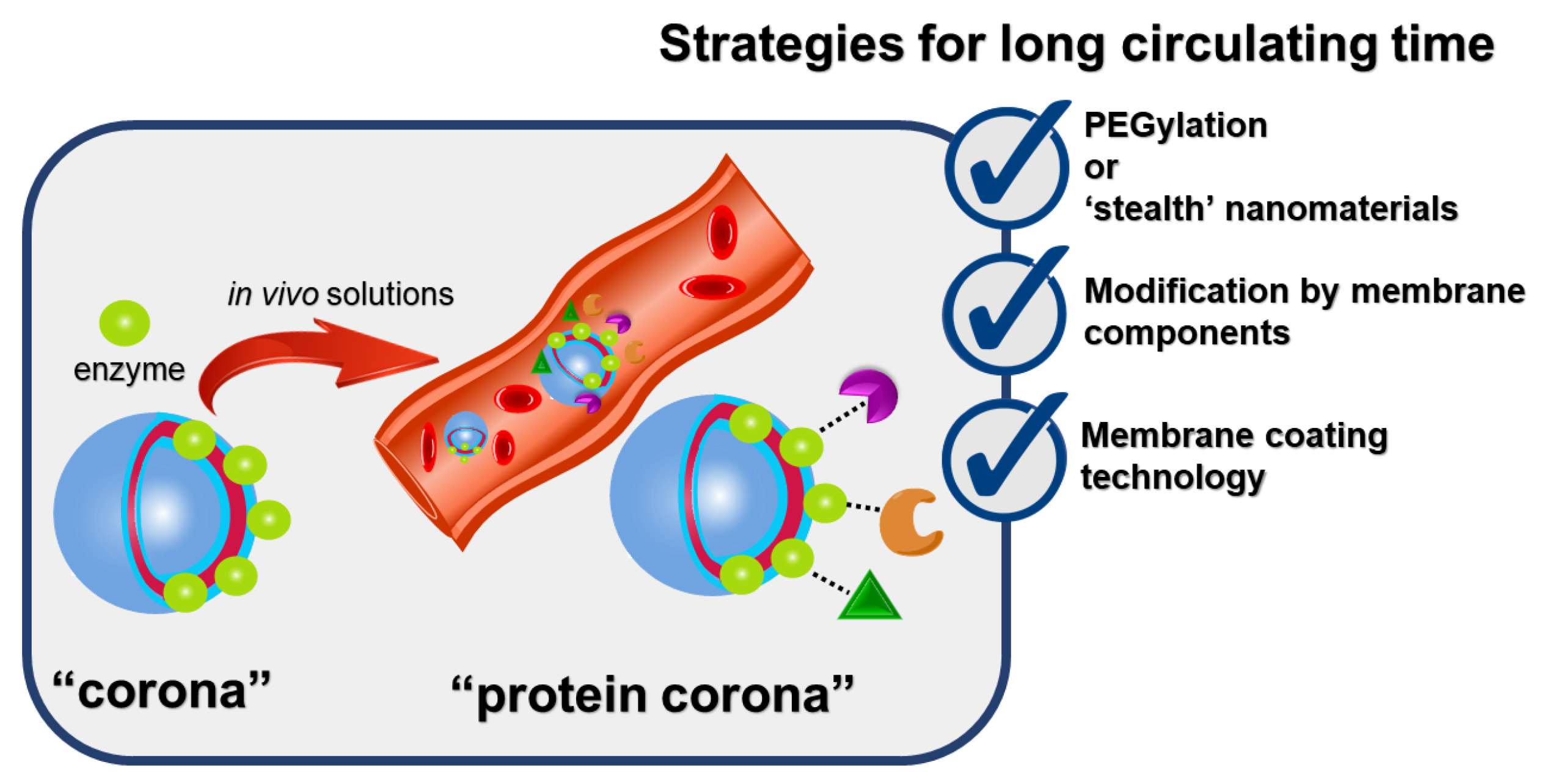
:1. Introduction
2. Toxicity of OPs
3. Sources of Organophosphate-Degrading Enzymes (Fungal, Bacterial and Archaeal Sources, Engineered Enzymes)
3.1. Organophosphate Degradation by Microbial Enzymes, Types of Enzymes and Mechanisms
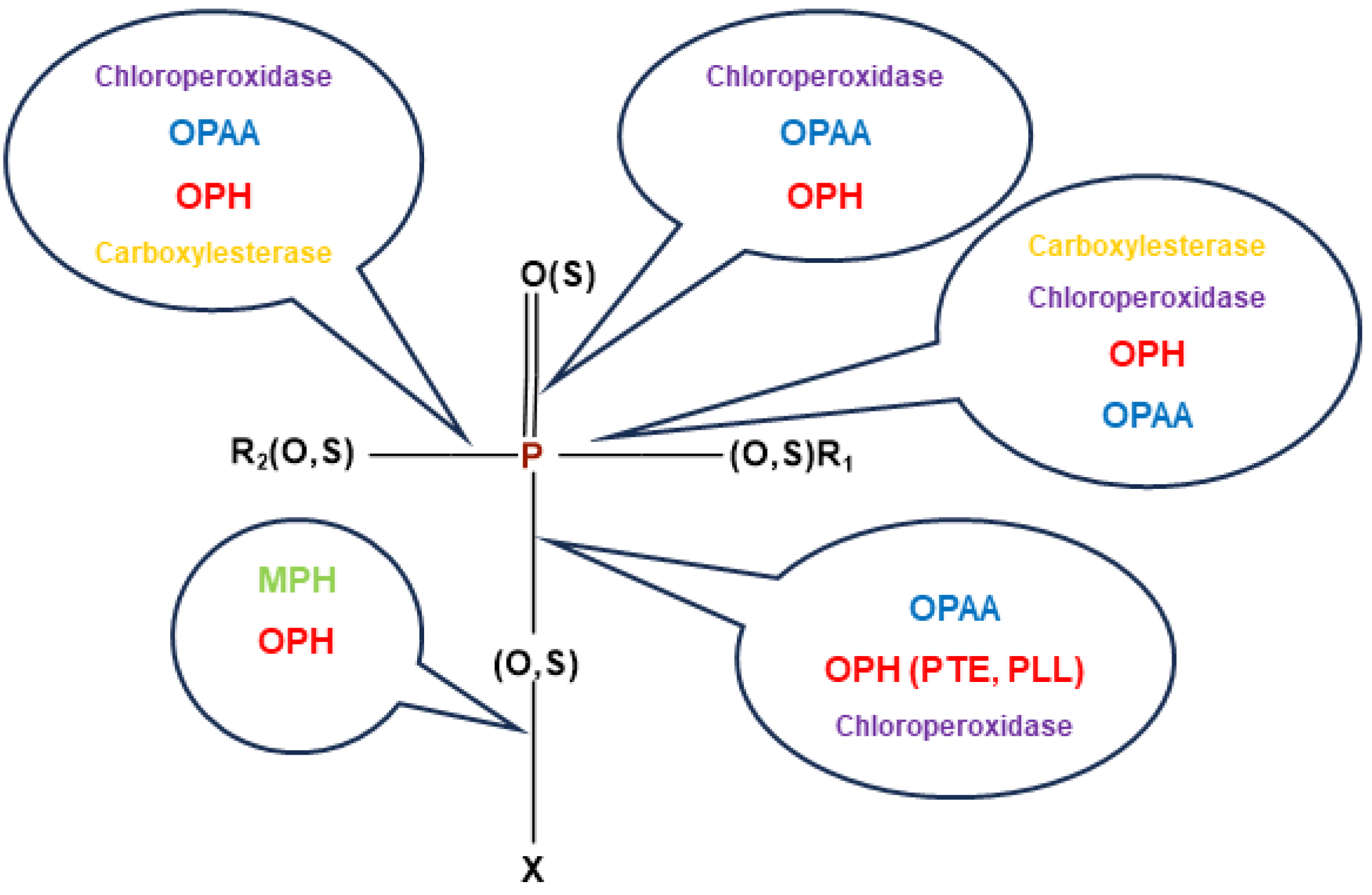
3.1.1. Phosphotriesterases
3.1.2. Phosphotriesterase-like Lactonase (PLL)
3.1.3. Other Bacterial Enzymes Reacting with OPs
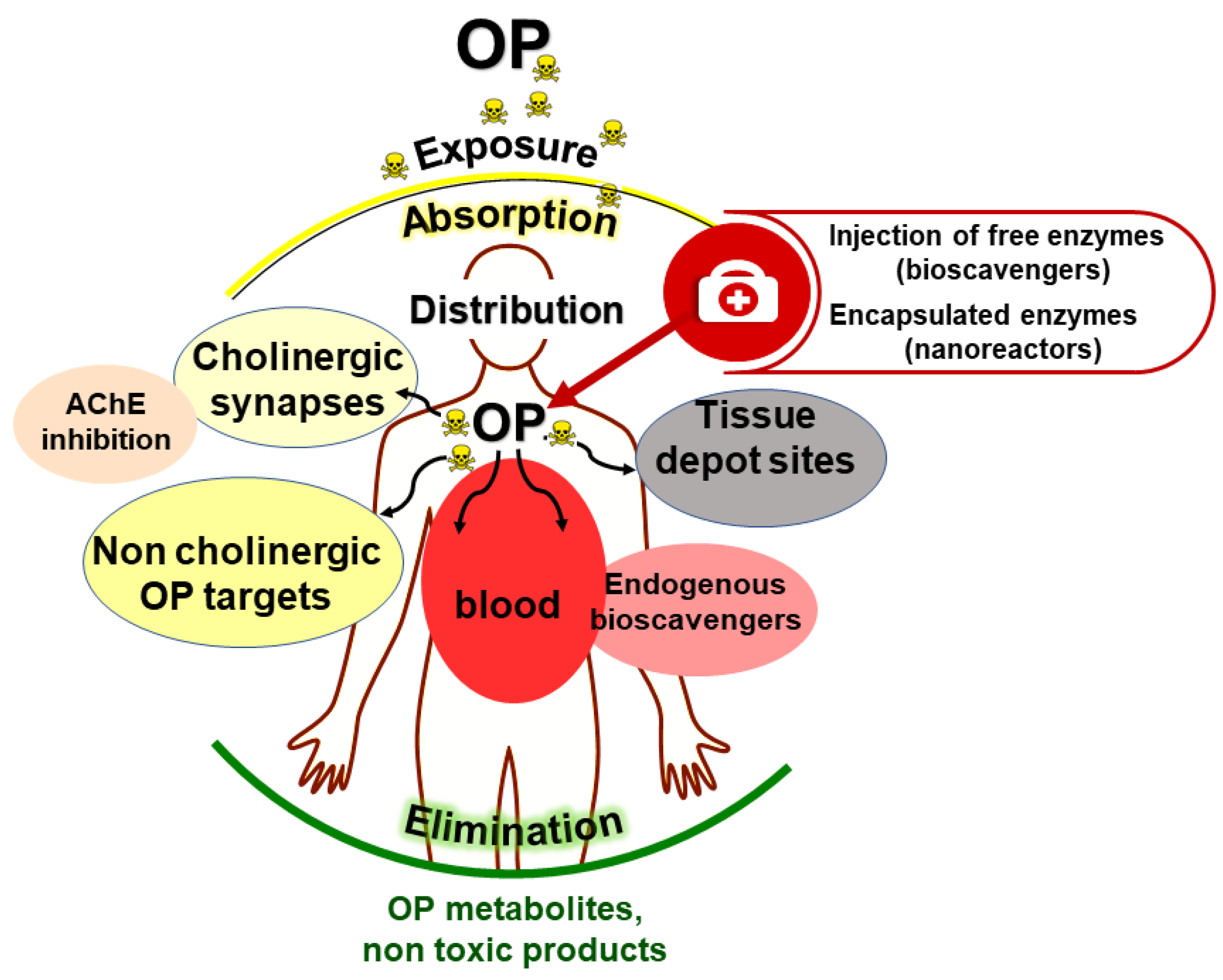
4. The Medical Bioscavenger Concept
4.1. Stoichiometric Bioscavengers
4.2. Pseudocatalytic Bioscavengers
4.3. Catalytic Bioscavengers
5. Requirements for Efficacy of Procaryotic Enzymes to Detoxify OPs
5.1. Requirements in Medicine for Efficacy and Safety of Injected Bacterial Enzymes
5.2. Requirements for Environmental Applications
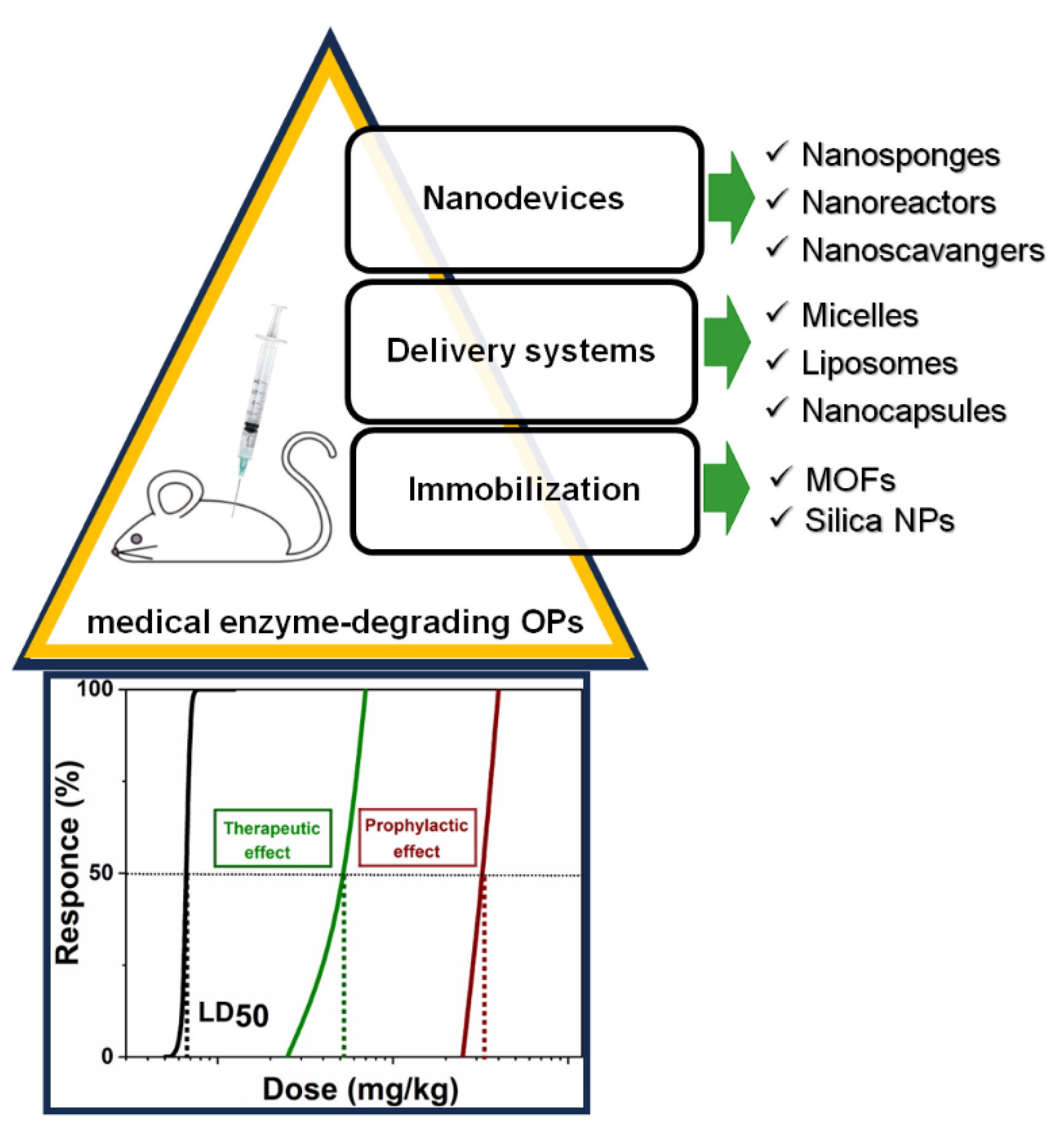
6. Role of Nanoparticles in Bioremediation
7. Combination of Nanoparticles and OP-Degrading Enzymes
| Enzyme/Source | OP | Carriers Used in the Immobilization Process | Efficiency | Ref |
|---|---|---|---|---|
| OP hydrolase | Methyl paraoxon | Poly-β-cyclodextrin microparticles | 100% | [247] |
| PTE | Methyl parathion | Cu2+-based enzyme hybrid nanoflowers | 62.5% | [248] |
| OP hydrolase from Flavobacterium sp. ATCC 27551 | Ethyl parathion | Two types of cellulose microfibers produced by using chemical coupling agents (1,4-butanediol diglycidyl ether and 1,1′-Carbonyldiimidazole) | 68% and 73% | [249] |
| OP hydrolase | Methyl parathion | Yolk-shell structured Co/C@SiO2@Ni/C nano-composites based MOFs ZIF-67 coated with PDA-Ni2+ shell | 100% | [250] |
| PTE | Paraoxon | DNA cage-semiconductor quantum dot nanocomposites | ~100% | [251] |
| Laccase from Bacillus sp. | Chlorpyrifos | Iron magnetic nanoparticles (Fe3O4) | [252] | |
| OP hydrolase from Flavobacterium sp. ATCC 27551 | Ethyl paraoxon | Magnetosomes of magnetite (Fe3O4) | 100% | [253] |
| OP hydrolase | Paraoxon | Mesoporous silica nanoparticles | 100% | [254] |
| Carboxylesterase from Spodoptera litura | Malathion | Mesoporous silica nanoparticles (SBA-15) | [255] |
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
References
- Singh, A.; Singh, A.; Singh, A.; Singh, P.; Singh, V.; Singh, Y.; Tuli, H.S.; Abdulabbas, H.S.; Chauhan, A. Chemistry, Metabolism and Neurotoxicity of Organophosphorus Insecticides: A Review. Nat. Environ. Pollut. Technol. 2023, 22, 1867–1880. [Google Scholar] [CrossRef]
- Masson, P.; Shaihutdinova, Z.; Lockridge, O. Drug and pro-drug substrates and pseudo-substrates of human butyrylcholinesterase. Biochem. Pharmacol. 2023, 218, 115910. [Google Scholar] [CrossRef] [PubMed]
- Hageman, G.; Mackenzie Ross, S.J.; Nihom, J.; van der Laan, G. Aerotoxic syndrome: A new occupational disease caused by contaminated cabin air? In Advances in Neurotoxicology; Academic Press: Cambridge, MA, USA, 2022; pp. 77–132. [Google Scholar]
- Opravil, J.; Pejchal, J.; Finger, V.; Korabecny, J.; Rozsypal, T.; Hrabinova, M.; Muckova, L.; Hepnarova, V.; Konecny, J.; Soukup, O.; et al. A-agents, misleadingly known as “Novichoks”: A narrative review. Arch. Toxicol. 2023, 97, 2587–2607. [Google Scholar] [CrossRef] [PubMed]
- Hrabinova, M.; Pejchal, J.; Hepnarova, V.; Muckova, L.; Junova, L.; Opravil, J.; Zdarova Karasova, J.; Rozsypal, T.; Dlabkova, A.; Rehulkova, H.; et al. A-series agent A-234: Initial in vitro and in vivo characterization. Arch. Toxicol. 2024, 98, 1135–1149. [Google Scholar] [CrossRef] [PubMed]
- FAOSTAT (Food and Agriculture Organization of the United Nations). Available online: https://www.fao.org/faostat/en/#data/RP (accessed on 11 February 2024).
- Mali, H.; Shah, C.; Raghunandan, B.H.; Prajapati, A.S.; Patel, D.H.; Trivedi, U.; Subramanian, R.B. Organophosphate pesticides an emerging environmental contaminant: Pollution, toxicity, bioremediation progress, and remaining challenges. J. Environ. Sci. 2023, 127, 234–250. [Google Scholar] [CrossRef] [PubMed]
- Tazdaït, D.; Salah-Tazdaït, R. Polycyclic Aromatic Hydrocarbons: Toxicity and Bioremediation Approaches. In Biotechnology for Sustainable Environment; Springer: Singapore, 2021; pp. 289–316. [Google Scholar]
- Tazdaït, D.; Salah, R.; Grib, H.; Abdi, N.; Mameri, N. Kinetic study on biodegradation of glyphosate with unacclimated activated sludge. Int. J. Environ. Health Res. 2018, 28, 448–459. [Google Scholar] [CrossRef] [PubMed]
- Salah-Tazdaït, R.; Tazdaït, D.; Berrahma, R.; Abdi, N.; Grib, H.; Mameri, N. Isolation and characterization of bacterial strains capable of growing on malathion and fenitrothion and the use of date syrup as an additional substrate. Int. J. Environ. Stud. 2018, 75, 466–483. [Google Scholar] [CrossRef]
- Hamad, M.T.M.H. Biodegradation of diazinon by fungal strain Apergillus niger MK640786 using response surface methodology. Environ. Technol. Innov. 2020, 18, 100691. [Google Scholar] [CrossRef]
- Nasiri, M.; Ahmadzadeh, H.; Amiri, A.H. Biodegradation and metabolic fate of organophosphorus pesticides in well water using Dunaliella salina. Int. J. Environ. Sci. Technol. 2023, 20, 981–992. [Google Scholar] [CrossRef]
- Fang, L.; Zhou, Y.; Chen, T.; Geng, Y.; Li, Z.; Zha, W.; Shi, T.; Hua, R. Efficient biodegradation characteristics and detoxification pathway of organophosphorus insecticide profenofos via Cupriavidus nantongensis X1T and enzyme OpdB. Sci. Total Environ. 2023, 862, 160782. [Google Scholar] [CrossRef]
- Chen, J.; Guo, Z.; Xin, Y.; Gu, Z.; Zhang, L.; Guo, X. Effective remediation and decontamination of organophosphorus compounds using enzymes: From rational design to potential applications. Sci. Total Environ. 2023, 867, 161510. [Google Scholar] [CrossRef]
- Huang, Y.; Xiao, L.; Li, F.; Xiao, M.; Lin, D.; Long, X.; Wu, Z. Microbial Degradation of Pesticide Residues and an Emphasis on the Degradation of Cypermethrin and 3-phenoxy Benzoic Acid: A Review. Molecules 2018, 23, 2313. [Google Scholar] [CrossRef]
- Guerrero Ramírez, J.R.; Ibarra Muñoz, L.A.; Balagurusamy, N.; Frías Ramírez, J.E.; Alfaro Hernández, L.; Carrillo Campos, J. Microbiology and Biochemistry of Pesticides Biodegradation. Int. J. Mol. Sci. 2023, 24, 15969. [Google Scholar] [CrossRef] [PubMed]
- Salah-Tazdaït, R.; Tazdaït, D. Use of microbial enzymes to degrade pesticide residues in agroecosystems-sustainable practices. In Biotechnology of Emerging Microbes; Elsevier: Amsterdam, The Netherlands, 2024; pp. 189–215. [Google Scholar]
- Masson, P.; Lushchekina, S.V. Catalytic bioscavengers: The second generation of bioscavenger-based medical countermeasures. In Handbook of Toxicology of Chemical Warfare Agents; Elsevier: Amsterdam, The Netherlands, 2020; Volume 2, pp. 1199–1229. ISBN 9780128190906. [Google Scholar]
- Matula, M.; Kucera, T.; Soukup, O.; Pejchal, J. Enzymatic Degradation of Organophosphorus Pesticides and Nerve Agents by EC: 3.1.8.2. Catalysts 2020, 10, 1365. [Google Scholar] [CrossRef]
- Paidi, M.K.; Satapute, P.; Haider, M.S.; Udikeri, S.S.; Ramachandra, Y.L.; Vo, D.-V.N.; Govarthanan, M.; Jogaiah, S. Mitigation of organophosphorus insecticides from environment: Residual detoxification by bioweapon catalytic scavengers. Environ. Res. 2021, 200, 111368. [Google Scholar] [CrossRef]
- Li, H.; Lu, C.; Liu, Z.; Fengshun, X.; Liu, B.; Wang, H.; Chang, J.; Li, P.; Chen, J. Advancements in bioscavenger mediated detoxification of organophosphorus poisoning. Toxicol. Res. 2024, 13, tfae089. [Google Scholar] [CrossRef]
- Jaiswal, S.; Singh, B.; Dhingra, I.; Joshi, A.; Kodgire, P. Bioremediation and bioscavenging for elimination of organophosphorus threats: An approach using enzymatic advancements. Environ. Res. 2024, 252, 118888. [Google Scholar] [CrossRef] [PubMed]
- Eddleston, M. Novel Clinical Toxicology and Pharmacology of Organophosphorus Insecticide Self-Poisoning. Annu. Rev. Pharmacol. Toxicol. 2019, 59, 341–360. [Google Scholar] [CrossRef] [PubMed]
- Worek, F.; Thiermann, H.; Wille, T. Organophosphorus compounds and oximes: A critical review. Arch. Toxicol. 2020, 94, 2275–2292. [Google Scholar] [CrossRef]
- Ratandeep; Ayushi; Garima; Devi, L.S.; Pooja. Role of Acetylcholinesterase (AChE) reactivators in the treatment of Organophosphorus poisoning: In vivo, in vitro, and in silico studies. Chem. Biol. Lett. 2023, 10, 538. [Google Scholar]
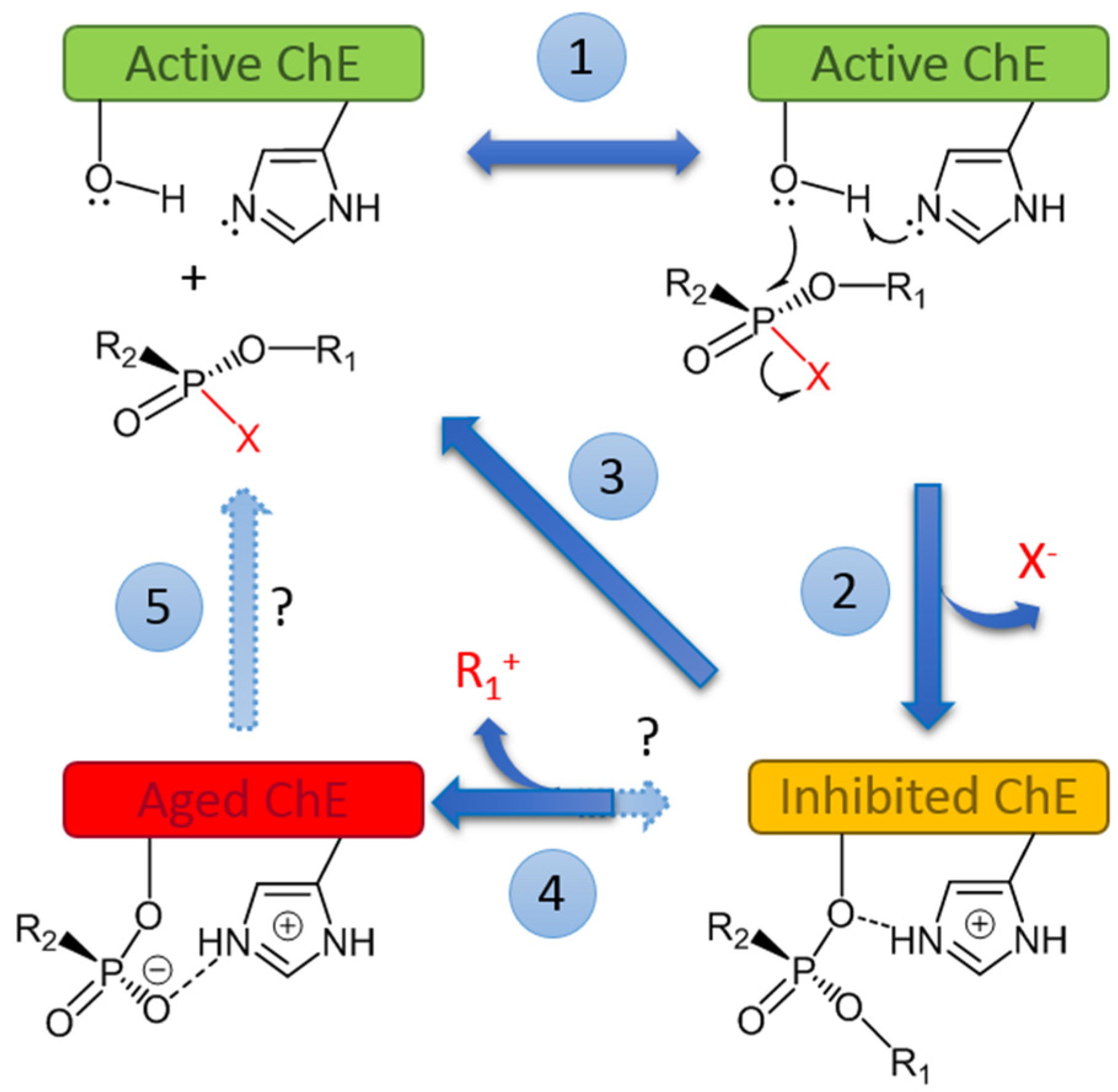
- Masson, P.; Nachon, F.; Lockridge, O. Structural approach to the aging of phosphylated cholinesterases. Chem. Biol. Interact. 2010, 187, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Q.; Franjesevic, A.J.; Corrigan, T.S.; Coldren, W.H.; Dicken, R.; Sillart, S.; DeYong, A.; Yoshino, N.; Smith, J.; Fabry, S.; et al. Demonstration of In Vitro Resurrection of Aged Acetylcholinesterase after Exposure to Organophosphorus Chemical Nerve Agents. J. Med. Chem. 2018, 61, 7034–7042. [Google Scholar] [CrossRef] [PubMed]
- Casida, J.E.; Quistad, G.B. Serine hydrolase targets of organophosphorus toxicants. Chem. Biol. Interact. 2005, 157–158, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Lockridge, O.; Schopfer, L.M. Review: Organophosphorus toxicants, in addition to inhibiting acetylcholinesterase activity, make covalent adducts on multiple proteins and promote protein crosslinking into high molecular weight aggregates. Chem. Biol. Interact. 2023, 376, 110460. [Google Scholar] [CrossRef] [PubMed]
- Willems, J.L.; De Bisschop, H.C.; Verstraete, A.G.; Declerck, C.; Christiaens, Y.; Vanscheeuwyck, P.; Buylaert, W.A.; Vogelaers, D.; Colardyn, F. Cholinesterase reactivation in organophosphorus poisoned patients depends on the plasma concentrations of the oxime pralidoxime methylsulphate and of the organophosphate. Arch. Toxicol. 1993, 67, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Thiermann, H.; Szinicz, L.; Eyer, P.; Felgenhauer, N.; Zilker, T.; Worek, F. Lessons to be learnt from organophosphorus pesticide poisoning for the treatment of nerve agent poisoning. Toxicology 2007, 233, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Eddleston, M.; Buckley, N.A.; Eyer, P.; Dawson, A.H. Management of acute organophosphorus pesticide poisoning. Lancet 2008, 371, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Masson, P. Novel approaches in prophylaxis/pretreatment and treatment of organophosphorus poisoning. Phosphorus Sulfur Silicon Relat. Elem. 2016, 191, 1433–1443. [Google Scholar] [CrossRef]
- Gorecki, L.; Soukup, O.; Korabecny, J. Countermeasures in organophosphorus intoxication: Pitfalls and prospects. Trends Pharmacol. Sci. 2022, 43, 593–606. [Google Scholar] [CrossRef]
- Prchalova, E.; Kohoutova, Z.; Knittelova, K.; Malinak, D.; Musilek, K. Strategies for enhanced bioavailability of oxime reactivators in the central nervous system. Arch. Toxicol. 2023, 97, 2839–2860. [Google Scholar] [CrossRef]
- Pulkrabkova, L.; Svobodova, B.; Konecny, J.; Kobrlova, T.; Muckova, L.; Janousek, J.; Pejchal, J.; Korabecny, J.; Soukup, O. Neurotoxicity evoked by organophosphates and available countermeasures. Arch. Toxicol. 2023, 97, 39–72. [Google Scholar] [CrossRef] [PubMed]
- Schallreuter, K.U.; Gibbons, N.C.J.; Elwary, S.M.; Parkin, S.M.; Wood, J.M. Calcium-activated butyrylcholinesterase in human skin protects acetylcholinesterase against suicide inhibition by neurotoxic organophosphates. Biochem. Biophys. Res. Commun. 2007, 355, 1069–1074. [Google Scholar] [CrossRef] [PubMed]
- Casida, J.E.; Durkin, K.A. Anticholinesterase insecticide retrospective. Chem. Biol. Interact. 2013, 203, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Redinbo, M.R.; Potter, P.M. Keynote review: Mammalian carboxylesterases: From drug targets to protein therapeutics. Drug Discov. Today 2005, 10, 313–325. [Google Scholar] [CrossRef] [PubMed]
- Kaliste-Korhonen, E.; Tuovinen, K.; Hänninen, O. Interspecies differences in enzymes reacting with organophosphates and their inhibition by paraoxon in vitro. Hum. Exp. Toxicol. 1996, 15, 972–978. [Google Scholar] [CrossRef]
- Shih, D.M.; Gu, L.; Xia, Y.-R.; Navab, M.; Li, W.-F.; Hama, S.; Castellani, L.W.; Furlong, C.E.; Costa, L.G.; Fogelman, A.M.; et al. Mice lacking serum paraoxonase are susceptible to organophosphate toxicity and atherosclerosis. Nature 1998, 394, 284–287. [Google Scholar] [CrossRef]
- Li, B.; Sedlacek, M.; Manoharan, I.; Boopathy, R.; Duysen, E.G.; Masson, P.; Lockridge, O. Butyrylcholinesterase, paraoxonase, and albumin esterase, but not carboxylesterase, are present in human plasma. Biochem. Pharmacol. 2005, 70, 1673–1684. [Google Scholar] [CrossRef]
- Napon, G.; Dafferner, A.J.; Saxena, A.; Lockridge, O. Identification of Carboxylesterase, Butyrylcholinesterase, Acetylcholinesterase, Paraoxonase, and Albumin Pseudoesterase in Guinea Pig Plasma through Nondenaturing Gel Electrophoresis. Comp. Med. 2018, 68, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Sogorb, M.A.; Álvarez-Escalante, C.; Carrera, V.; Vilanova, E. An in vitro approach for demonstrating the critical role of serum albumin in the detoxication of the carbamate carbaryl at in vivo toxicologically relevant concentrations. Arch. Toxicol. 2007, 81, 113–119. [Google Scholar] [CrossRef]
- Li, B.; Nachon, F.; Froment, M.T.; Verdier, L.; Debouzy, J.C.; Brasme, B.; Gillon, E.; Schopfer, L.M.; Lockridge, O.; Masson, P. Binding and hydrolysis of soman by human serum albumin. Chem. Res. Toxicol. 2008, 21, 421–431. [Google Scholar] [CrossRef]
- Belinskaia, D.A.; Goncharov, N.V. Theoretical and Practical Aspects of Albumin Esterase Activity. Russ. J. Bioorg. Chem. 2020, 46, 287–298. [Google Scholar] [CrossRef]
- Nomura, D.K.; Fujioka, K.; Issa, R.S.; Ward, A.M.; Cravatt, B.F.; Casida, J.E. Dual roles of brain serine hydrolase KIAA1363 in ether lipid metabolism and organophosphate detoxification. Toxicol. Appl. Pharmacol. 2008, 228, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Satvik Iyengar, A.R.; Tripathy, R.K.; Bajaj, P.; Pande, A.H. Improving storage stability of recombinant organophosphorus hydrolase. Protein Expr. Purif. 2015, 111, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Thakur, M.; Medintz, I.L.; Walper, S.A. Enzymatic Bioremediation of Organophosphate Compounds—Progress and Remaining Challenges. Front. Bioeng. Biotechnol. 2019, 7, 289. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sun, Y. Engineering organophosphate hydrolase for enhanced biocatalytic performance: A review. Biochem. Eng. J. 2021, 168, 107945. [Google Scholar] [CrossRef]
- Ambreen, S.; Yasmin, A.; Aziz, S. Isolation and characterization of organophosphorus phosphatases from Bacillus thuringiensis MB497 capable of degrading Chlorpyrifos, Triazophos and Dimethoate. Heliyon 2020, 6, e04221. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, P.; Zhou, X.; Huang, Y.; Zhang, W.; Chen, S. Characterization of the role of esterases in the biodegradation of organophosphate, carbamate, and pyrethroid pesticides. J. Hazard. Mater. 2021, 411, 125026. [Google Scholar] [CrossRef] [PubMed]
- Mali, H.; Shah, C.; Patel, D.H.; Trivedi, U.; Subramanian, R.B. Degradation insight of organophosphate pesticide chlorpyrifos through novel intermediate 2,6-dihydroxypyridine by Arthrobacter sp. HM01. Bioresour. Bioprocess. 2022, 9, 31. [Google Scholar] [CrossRef] [PubMed]
- Karbelkar, A.A.; Reynolds, E.E.; Ahlmark, R.; Furst, A.L. A Microbial Electrochemical Technology to Detect and Degrade Organophosphate Pesticides. ACS Cent. Sci. 2021, 7, 1718–1727. [Google Scholar] [CrossRef]
- Poirier, L.; Brun, L.; Jacquet, P.; Lepolard, C.; Armstrong, N.; Torre, C.; Daudé, D.; Ghigo, E.; Chabrière, E. Enzymatic degradation of organophosphorus insecticides decreases toxicity in planarians and enhances survival. Sci. Rep. 2017, 7, 15194. [Google Scholar] [CrossRef]
- Lin, Z.; Pang, S.; Zhang, W.; Mishra, S.; Bhatt, P.; Chen, S. Degradation of Acephate and Its Intermediate Methamidophos: Mechanisms and Biochemical Pathways. Front. Microbiol. 2020, 11, 2045. [Google Scholar] [CrossRef] [PubMed]
- Alejo-González, K.; Hanson-Viana, E.; Vazquez-Duhalt, R. Enzymatic detoxification of organophosphorus pesticides and related toxicants. J. Pestic. Sci. 2018, 43, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Leskovac, A.; Petrović, S. Pesticide Use and Degradation Strategies: Food Safety, Challenges and Perspectives. Foods 2023, 12, 2709. [Google Scholar] [CrossRef] [PubMed]
- Firozjaei, S.A.A.; Latifi, A.M.; Khodi, S.; Abolmaali, S.; Choopani, A. A review on biodegradation of toxic organophosphate compounds. J. Appl. Biotechnol. Rep. 2015, 2, 215–224. [Google Scholar]
- Kumar, S.S.; Ghosh, P.; Malyan, S.K.; Sharma, J.; Kumar, V. A comprehensive review on enzymatic degradation of the organophosphate pesticide malathion in the environment. J. Environ. Sci. Health C 2019, 37, 288–329. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lee, H.Y.; Cho, D.Y.; Kim, M.J.; Jung, J.G.; Jeong, E.H.; Haque, M.A.; Cho, K.M. Biodegradable properties of organophosphorus insecticides by the potential probiotic Lactobacillus plantarum WCP931 with a degrading gene (opdC). Appl. Biol. Chem. 2021, 64, 62. [Google Scholar] [CrossRef]
- Ghanem, E.; Raushel, F.M. Detoxification of organophosphate nerve agents by bacterial phosphotriesterase. Toxicol. Appl. Pharmacol. 2005, 207, 459–470. [Google Scholar] [CrossRef]
- Theriot, C.M.; Grunden, A.M. Hydrolysis of organophosphorus compounds by microbial enzymes. Appl. Microbiol. Biotechnol. 2011, 89, 35–43. [Google Scholar] [CrossRef]
- Bigley, A.N.; Raushel, F.M. Catalytic mechanisms for phosphotriesterases. Biochim. Biophys. Acta Proteins Proteom. 2013, 1834, 443–453. [Google Scholar] [CrossRef]
- Dickschat, J.S. Quorum sensing and bacterial biofilms. Nat. Prod. Rep. 2010, 27, 343. [Google Scholar] [CrossRef]
- Marone, M.; Porzio, E.; Lampitella, E.A.; Manco, G. A mesophilic phosphotriesterase-like lactonase shows high stability and proficiency as quorum quenching enzyme. Chem. Biol. Interact. 2023, 383, 110657. [Google Scholar] [CrossRef] [PubMed]
- Amara, N.; Krom, B.P.; Kaufmann, G.F.; Meijler, M.M. Macromolecular Inhibition of Quorum Sensing: Enzymes, Antibodies, and Beyond. Chem. Rev. 2011, 111, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Afriat, L.; Roodveldt, C.; Manco, G.; Tawfik, D.S. The Latent Promiscuity of Newly Identified Microbial Lactonases Is Linked to a Recently Diverged Phosphotriesterase. Biochemistry 2006, 45, 13677–13686. [Google Scholar] [CrossRef]
- Elias, M.; Dupuy, J.; Merone, L.; Mandrich, L.; Porzio, E.; Moniot, S.; Rochu, D.; Lecomte, C.; Rossi, M.; Masson, P.; et al. Structural Basis for Natural Lactonase and Promiscuous Phosphotriesterase Activities. J. Mol. Biol. 2008, 379, 1017–1028. [Google Scholar] [CrossRef] [PubMed]
- Afriat-Jurnou, L.; Jackson, C.J.; Tawfik, D.S. Reconstructing a Missing Link in the Evolution of a Recently Diverged Phosphotriesterase by Active-Site Loop Remodeling. Biochemistry 2012, 51, 6047–6055. [Google Scholar] [CrossRef] [PubMed]
- Hiblot, J.; Gotthard, G.; Chabriere, E.; Elias, M. Structural and Enzymatic characterization of the lactonase SisLac from Sulfolobus islandicus. PLoS ONE 2012, 7, e47028. [Google Scholar] [CrossRef] [PubMed]
- Jacquet, P.; Billot, R.; Shimon, A.; Hoekstra, N.; Bergonzi, C.; Jenks, A.; Chabrière, E.; Daudé, D.; Elias, M.H. Changes in Active Site Loop Conformation Relate to the Transition toward a Novel Enzymatic Activity. JACS Au 2024, 4, 1941–1953. [Google Scholar] [CrossRef] [PubMed]
- Horne, I.; Sutherland, T.D.; Harcourt, R.L.; Russell, R.J.; Oakeshott, J.G. Identification of an opd (Organophosphate Degradation) Gene in an Agrobacterium Isolate. Appl. Environ. Microbiol. 2002, 68, 3371–3376. [Google Scholar] [CrossRef] [PubMed]
- Gotthard, G.; Hiblot, J.; Gonzalez, D.; Elias, M.; Chabriere, E. Structural and Enzymatic Characterization of the Phosphotriesterase OPHC2 from Pseudomonas pseudoalcaligenes. PLoS ONE 2013, 8, e77995. [Google Scholar] [CrossRef]
- Merone, L.; Mandrich, L.; Rossi, M.; Manco, G. A thermostable phosphotriesterase from the archaeon Sulfolobus solfataricus: Cloning, overexpression and properties. Extremophiles 2005, 9, 297–305. [Google Scholar] [CrossRef]
- de Castro, A.A.; Prandi, I.G.; Kuca, K.; Ramalho, T.C. Enzimas degradantes de organofosforados: Base molecular e perspectivas para biorremediação enzimática de agroquímicos. Ciênc. Agrotecnol. 2017, 41, 471–482. [Google Scholar] [CrossRef]
- Carletti, E.; Jacquamet, L.; Loiodice, M.; Rochu, D.; Masson, P.; Nachon, F. Update on biochemical properties of recombinant Pseudomonas diminuta phosphotriesterase. J. Enzym. Inhib. Med. Chem. 2009, 24, 1045–1055. [Google Scholar] [CrossRef]
- Benning, M.M.; Kuo, J.M.; Raushel, F.M.; Holden, H.M. Three-Dimensional Structure of Phosphotriesterase: An Enzyme Capable of Detoxifying Organophosphate Nerve Agents. Biochemistry 1994, 33, 15001–15007. [Google Scholar] [CrossRef]
- Samples, C.R.; Howard, T.; Raushel, F.M.; DeRose, V.J. Protonation of the Binuclear Metal Center within the Active Site of Phosphotriesterase. Biochemistry 2005, 44, 11005–11013. [Google Scholar] [CrossRef] [PubMed]
- Aubert, S.D.; Li, Y.; Raushel, F.M. Mechanism for the Hydrolysis of Organophosphates by the Bacterial Phosphotriesterase. Biochemistry 2004, 43, 5707–5715. [Google Scholar] [CrossRef]
- Jackson, C.J.; Carr, P.D.; Kim, H.-K.; Liu, J.-W.; Herrald, P.; Mitić, N.; Schenk, G.; Smith, C.A.; Ollis, D.L. Anomalous scattering analysis of Agrobacterium radiobacter phosphotriesterase: The prominent role of iron in the heterobinuclear active site. Biochem. J. 2006, 397, 501–508. [Google Scholar] [CrossRef]
- Chen, S.-L.; Fang, W.-H.; Himo, F. Theoretical Study of the Phosphotriesterase Reaction Mechanism. J. Phys. Chem. B 2007, 111, 1253–1255. [Google Scholar] [CrossRef] [PubMed]
- Samples, C.R.; Raushel, F.M.; DeRose, V.J. Activation of the Binuclear Metal Center through Formation of Phosphotriesterase−Inhibitor Complexes. Biochemistry 2007, 46, 3435–3442. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.-Y.; Gao, J. The Reaction Mechanism of Paraoxon Hydrolysis by Phosphotriesterase from Combined QM/MM Simulations. Biochemistry 2007, 46, 13352–13369. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.J.; Foo, J.-L.; Kim, H.-K.; Carr, P.D.; Liu, J.-W.; Salem, G.; Ollis, D.L. In Crystallo Capture of a Michaelis Complex and Product-binding Modes of a Bacterial Phosphotriesterase. J. Mol. Biol. 2008, 375, 1189–1196. [Google Scholar] [CrossRef]
- Benning, M.M.; Hong, S.-B.; Raushel, F.M.; Holden, H.M. The Binding of Substrate Analogs to Phosphotriesterase. J. Biol. Chem. 2000, 275, 30556–30560. [Google Scholar] [CrossRef] [PubMed]
- Bigley, A.N.; Raushel, F.M. The evolution of phosphotriesterase for decontamination and detoxification of organophosphorus chemical warfare agents. Chem. Biol. Interact. 2019, 308, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Tsai, P.-C.; Chen, S.-L.; Himo, F.; Almo, S.C.; Raushel, F.M. Structure of Diethyl Phosphate Bound to the Binuclear Metal Center of Phosphotriesterase. Biochemistry 2008, 47, 9497–9504. [Google Scholar] [CrossRef] [PubMed]
- Bigley, A.N.; Xiang, D.F.; Ren, Z.; Xue, H.; Hull, K.G.; Romo, D.; Raushel, F.M. Chemical Mechanism of the Phosphotriesterase from Sphingobium sp. Strain TCM1, an Enzyme Capable of Hydrolyzing Organophosphate Flame Retardants. J. Am. Chem. Soc. 2016, 138, 2921–2924. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, R.; Song, L.; Lin, Y.; Lin, M.; Cao, Z.; Wu, W.; Mo, Y. Molecular dynamics simulations of the detoxification of paraoxon catalyzed by phosphotriesterase. J. Comput. Chem. 2009, 30, 2388–2401. [Google Scholar] [CrossRef] [PubMed]
- Blum, M.-M.; Löhr, F.; Richardt, A.; Rüterjans, H.; Chen, J.C.-H. Binding of a Designed Substrate Analogue to Diisopropyl Fluorophosphatase: Implications for the Phosphotriesterase Mechanism. J. Am. Chem. Soc. 2006, 128, 12750–12757. [Google Scholar] [CrossRef] [PubMed]
- Elias, M.; Liebschner, D.; Koepke, J.; Lecomte, C.; Guillot, B.; Jelsch, C.; Chabriere, E. Hydrogen atoms in protein structures: High-resolution X-ray diffraction structure of the DFPase. BMC Res. Notes 2013, 6, 308. [Google Scholar] [CrossRef] [PubMed]
- Ben-David, M.; Wieczorek, G.; Elias, M.; Silman, I.; Sussman, J.L.; Tawfik, D.S. Catalytic Metal Ion Rearrangements Underline Promiscuity and Evolvability of a Metalloenzyme. J. Mol. Biol. 2013, 425, 1028–1038. [Google Scholar] [CrossRef] [PubMed]
- Grunkemeyer, T.J.; Mata, D.G.; Doddapaneni, K.; Murali, S.; Magliery, T.J. Insights into the mechanism of paraoxonase-1: Comparing the reactivity of the six-bladed β-propeller hydrolases. Biochemistry 2018, acs.biochem.8b01115. [Google Scholar] [CrossRef]
- Kuo, J.M.; Chae, M.Y.; Raushel, F.M. Perturbations to the Active Site of Phosphotriesterase. Biochemistry 1997, 36, 1982–1988. [Google Scholar] [CrossRef]
- Job, L.; Köhler, A.; Testanera, M.; Escher, B.; Worek, F.; Skerra, A. Engineering of a phosphotriesterase with improved stability and enhanced activity for detoxification of the pesticide metabolite malaoxon. Protein Eng. Des. Sel. 2023, 36, gzad020. [Google Scholar] [CrossRef] [PubMed]
- Dumas, D.P.; Durst, H.D.; Landis, W.G.; Raushel, F.M.; Wild, J.R. Inactivation of organophosphorus nerve agents by the phosphotriesterase from Pseudomonas diminuta. Arch. Biochem. Biophys. 1990, 277, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.M.; Li, W.-S.; Thoden, J.B.; Holden, H.M.; Raushel, F.M. Enhanced Degradation of Chemical Warfare Agents through Molecular Engineering of the Phosphotriesterase Active Site. J. Am. Chem. Soc. 2003, 125, 8990–8991. [Google Scholar] [CrossRef] [PubMed]
- Tsai, P.-C.; Bigley, A.; Li, Y.; Ghanem, E.; Cadieux, C.L.; Kasten, S.A.; Reeves, T.E.; Cerasoli, D.M.; Raushel, F.M. Stereoselective Hydrolysis of Organophosphate Nerve Agents by the Bacterial Phosphotriesterase. Biochemistry 2010, 49, 7978–7987. [Google Scholar] [CrossRef] [PubMed]
- Tsai, P.-C.; Fox, N.; Bigley, A.N.; Harvey, S.P.; Barondeau, D.P.; Raushel, F.M. Enzymes for the Homeland Defense: Optimizing Phosphotriesterase for the Hydrolysis of Organophosphate Nerve Agents. Biochemistry 2012, 51, 6463–6475. [Google Scholar] [CrossRef]
- Bigley, A.N.; Desormeaux, E.; Xiang, D.F.; Bae, S.Y.; Harvey, S.P.; Raushel, F.M. Overcoming the Challenges of Enzyme Evolution to Adapt Phosphotriesterase for V-Agent Decontamination. Biochemistry 2019, 58, 2039–2053. [Google Scholar] [CrossRef] [PubMed]
- Stigler, L.; Köhler, A.; Koller, M.; Job, L.; Escher, B.; Potschka, H.; Thiermann, H.; Skerra, A.; Worek, F.; Wille, T. Post-VX exposure treatment of rats with engineered phosphotriesterases. Arch. Toxicol. 2022, 96, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Lyagin, I.; Efremenko, E. Theoretical evaluation of suspected enzymatic hydrolysis of Novichok agents. Catal. Commun. 2019, 120, 91–94. [Google Scholar] [CrossRef]
- Jacquet, P.; Rémy, B.; Bross, R.P.T.; van Grol, M.; Gaucher, F.; Chabrière, E.; de Koning, M.C.; Daudé, D. Enzymatic Decontamination of G-Type, V-Type and Novichok Nerve Agents. Int. J. Mol. Sci. 2021, 22, 8152. [Google Scholar] [CrossRef]
- LeJeune, K.E.; Russell, A.J. Biocatalytic nerve agent detoxification in fire fighting foams. Biotechnol. Bioeng. 1999, 62, 659–665. [Google Scholar] [CrossRef]
- Gill, I.; Ballesteros, A. Degradation of organophosphorous nerve agents by enzyme-polymer nanocomposites: Efficient biocatalytic materials for personal protection and large-scale detoxification. Biotechnol. Bioeng. 2000, 70, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Doctor, B.P.; Saxena, A. Bioscavengers for the protection of humans against organophosphate toxicity. Chem. Biol. Interact. 2005, 157–158, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Jun, D.; Kuca, K.; Bajgar, J.; Hruby, M.; Kucka, J.; Renault, F.; Masson, P. Phosphotriesterase modified by poly[N-(2-hydroxypropyl)methacrylamide]. Toxicology 2007, 233, 235. [Google Scholar] [CrossRef]
- Pei, L.; Omburo, G.; Mcguinn, W.D.; Petrikovics, I.; Dave, K.; Raushel, F.M.; Wild, J.R.; Deloach, J.R.; Way, J.L. Encapsulation of Phosphotriesterase within Murine Erythrocytes. Toxicol. Appl. Pharmacol. 1994, 124, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Petrikovics, I.; Papahadjopoulos, D.; Hong, K.; Cheng, T.C.; Baskin, S.I.; Jiang, J.; Jaszberenyi, J.C.; Logue, B.A.; Szilasi, M.; McGuinn, W.D.; et al. Comparing therapeutic and prophylactic protection against the lethal effect of paraoxon. Toxicol. Sci. 2004, 77, 258–262. [Google Scholar] [CrossRef] [PubMed]
- Masson, P.; Josse, D.; Lockridge, O.; Viguié, N.; Taupin, C.; Buhler, C. Enzymes hydrolyzing organophosphates as potential catalytic scavengers against organophosphate poisoning. J. Physiol. 1998, 92, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Pashirova, T.N.; Bogdanov, A.; Masson, P. Therapeutic nanoreactors for detoxification of xenobiotics: Concepts, challenges and biotechnological trends with special emphasis to organophosphate bioscavenging. Chem. Biol. Interact. 2021, 346, 109577. [Google Scholar] [CrossRef] [PubMed]
- Parsa, R.; Green, H. Destruction of DFP by organophosphorus acid anhydrase covalently coupled to the cornified layer of human epidermis. In Proceedings of the International Symposium on Applications of Enzymes in Chemical and Biological Defense, Orlando, FL, USA, 13–18 May 2001. [Google Scholar]
- McDaniel, C.S.; McDaniel, J.; Wales, M.E.; Wild, J.R. Enzyme-based additives for paints and coatings. Prog. Org. Coat. 2006, 55, 182–188. [Google Scholar] [CrossRef]
- Wang, X.; Wu, N.; Guo, J.; Chu, X.; Tian, J.; Yao, B.; Fan, Y. Phytodegradation of organophosphorus compounds by transgenic plants expressing a bacterial organophosphorus hydrolase. Biochem. Biophys. Res. Commun. 2008, 365, 453–458. [Google Scholar] [CrossRef]
- Efremenko, E.; Lyagin, I.; Votchitseva, Y.; Sirotkina, M.; Varfolomeyev, S. Polyhistidine-containing organophosphorus hydrolase with outstanding properties. Biocatal. Biotransform. 2007, 25, 103–108. [Google Scholar] [CrossRef]
- Mandrich, L.; Merone, L.; Manco, G. Hyperthermophilic phosphotriesterases/lactonases for the environment and human health. Environ. Technol. 2010, 31, 1115–1127. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Gupta, R.D. Organophosphorus Nerve Agents: Types, Toxicity, and Treatments. J. Toxicol. 2020, 2020, 3007984. [Google Scholar] [CrossRef]
- Zhang, Y.; An, J.; Ye, W.; Yang, G.; Qian, Z.-G.; Chen, H.-F.; Cui, L.; Feng, Y. Enhancing the Promiscuous Phosphotriesterase Activity of a Thermostable Lactonase (GkaP) for the Efficient Degradation of Organophosphate Pesticides. Appl. Environ. Microbiol. 2012, 78, 6647–6655. [Google Scholar] [CrossRef]
- Hiblot, J.; Gotthard, G.; Elias, M.; Chabriere, E. Differential Active Site Loop Conformations Mediate Promiscuous Activities in the Lactonase SsoPox. PLoS ONE 2013, 8, e75272. [Google Scholar] [CrossRef]
- Hiblot, J.; Gotthard, G.; Champion, C.; Chabriere, E.; Elias, M. Crystallization and preliminary X-ray diffraction analysis of the lactonase Vmo Lac from Vulcanisaeta moutnovskia. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013, 69, 1235–1238. [Google Scholar] [CrossRef]
- Porzio, E.; Di Gennaro, S.; Palma, A.; Manco, G. Mn2+ modulates the kinetic properties of an archaeal member of the PLL family. Chem. Biol. Interact. 2013, 203, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Merone, L.; Mandrich, L.; Porzio, E.; Rossi, M.; Müller, S.; Reiter, G.; Worek, F.; Manco, G. Improving the promiscuous nerve agent hydrolase activity of a thermostable archaeal lactonase. Bioresour. Technol. 2010, 101, 9204–9212. [Google Scholar] [CrossRef]
- Jacquet, P.; Hiblot, J.; Daudé, D.; Bergonzi, C.; Gotthard, G.; Armstrong, N.; Chabrière, E.; Elias, M. Rational engineering of a native hyperthermostable lactonase into a broad spectrum phosphotriesterase. Sci. Rep. 2017, 7, 16745. [Google Scholar] [CrossRef] [PubMed]
- Restaino, O.F.; Borzacchiello, M.G.; Scognamiglio, I.; Fedele, L.; Alfano, A.; Porzio, E.; Manco, G.; De Rosa, M.; Schiraldi, C. High yield production and purification of two recombinant thermostable phosphotriesterase-like lactonases from Sulfolobus acidocaldarius and Sulfolobus solfataricus useful as bioremediation tools and bioscavengers. BMC Biotechnol. 2018, 18, 18. [Google Scholar] [CrossRef]
- Singh, B.K. Organophosphorus-degrading bacteria: Ecology and industrial applications. Nat. Rev. Microbiol. 2009, 7, 156–164. [Google Scholar] [CrossRef]
- Ng, T.-K.; Gahan, L.R.; Schenk, G.; Ollis, D.L. Altering the substrate specificity of methyl parathion hydrolase with directed evolution. Arch. Biochem. Biophys. 2015, 573, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Hawwa, R.; Larsen, S.D.; Ratia, K.; Mesecar, A.D. Structure-Based and Random Mutagenesis Approaches Increase the Organophosphate-Degrading Activity of a Phosphotriesterase Homologue from Deinococcus radiodurans. J. Mol. Biol. 2009, 393, 36–57. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.J.; Weir, K.; Herlt, A.; Khurana, J.; Sutherland, T.D.; Horne, I.; Easton, C.; Russell, R.J.; Scott, C.; Oakeshott, J.G. Structure-Based Rational Design of a Phosphotriesterase. Appl. Environ. Microbiol. 2009, 75, 5153–5156. [Google Scholar] [CrossRef] [PubMed]
- Ely, F.; Hadler, K.S.; Gahan, L.R.; Guddat, L.W.; Ollis, D.L.; Schenk, G. The organophosphate-degrading enzyme from Agrobacterium radiobacter displays mechanistic flexibility for catalysis. Biochem. J. 2010, 432, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.Y.; Myslinski, J.M.; McMahon, L.R.; Height, J.J.; Bigley, A.N.; Raushel, F.M.; Harvey, S.P. An OPAA enzyme mutant with increased catalytic efficiency on the nerve agents sarin, soman, and GP. Enzym. Microb. Technol. 2018, 112, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Bzdrenga, J.; Trenet, E.; Chantegreil, F.; Bernal, K.; Nachon, F.; Brazzolotto, X. A Thermophilic Bacterial Esterase for Scavenging Nerve Agents: A Kinetic, Biophysical and Structural Study. Molecules 2021, 26, 657. [Google Scholar] [CrossRef] [PubMed]
- Santillan, J.Y.; Dettorre, L.A.; Lewkowicz, E.S.; Iribarren, A.M. New and highly active microbial phosphotriesterase sources. FEMS Microbiol. Lett. 2016, 363, fnw276. [Google Scholar] [CrossRef] [PubMed]
- Theriot, C.M.; Tove, S.R.; Grunden, A.M. Chapter 3—Biotechnological Applications of Recombinant Microbial Prolidases. In Advances in Applied Microbiology; Elsevier: Amsterdam, The Netherlands, 2009; pp. 99–132. [Google Scholar]
- Vyas, N.K.; Nickitenko, A.; Rastogi, V.K.; Shah, S.S.; Quiocho, F.A. Structural Insights into the Dual Activities of the Nerve Agent Degrading Organophosphate Anhydrolase/Prolidase. Biochemistry 2010, 49, 547–559. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.C.; Harvey, S.P.; Chen, G.L. Cloning and expression of a gene encoding a bacterial enzyme for decontamination of organophosphorus nerve agents and nucleotide sequence of the enzyme. Appl. Environ. Microbiol. 1996, 62, 1636–1641. [Google Scholar] [CrossRef]
- Cheng, T.; DeFrank, J.J.; Rastogi, V.K. Alteromonas prolidase for organophosphorus G-agent decontamination. Chem. Biol. Interact. 1999, 119–120, 455–462. [Google Scholar] [CrossRef]
- Theriot, C.M.; Du, X.; Tove, S.R.; Grunden, A.M. Improving the catalytic activity of hyperthermophilic Pyrococcus prolidases for detoxification of organophosphorus nerve agents over a broad range of temperatures. Appl. Microbiol. Biotechnol. 2010, 87, 1715–1726. [Google Scholar] [CrossRef] [PubMed]
- Theriot, C.M.; Semcer, R.L.; Shah, S.S.; Grunden, A.M. Improving the Catalytic Activity of Hyperthermophilic Pyrococcus horikoshii Prolidase for Detoxification of Organophosphorus Nerve Agents over a Broad Range of Temperatures. Archaea 2011, 2011, 565127. [Google Scholar] [CrossRef] [PubMed]
- Tani, Y.; Nagasawa, T.; Oda, H.; Ogata, K. Distribution and Some Properties of Bacterial Cholinesterase. Agric. Biol. Chem. 1975, 39, 105–111. [Google Scholar] [CrossRef]
- Rochu, D.; Rothlisberger, C.; Taupin, C.; Renault, F.; Gagnon, J.; Masson, P. Purification, molecular characterization and catalytic properties of a Pseudomonas fluorescens enzyme having cholinesterase-like activity. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1998, 1385, 126–138. [Google Scholar] [CrossRef] [PubMed]
- Pham, V.D.; To, T.A.; Gagné-Thivierge, C.; Couture, M.; Lagüe, P.; Yao, D.; Picard, M.-È.; Lortie, L.-A.; Attéré, S.A.; Zhu, X.; et al. Structural insights into the putative bacterial acetylcholinesterase ChoE and its substrate inhibition mechanism. J. Biol. Chem. 2020, 295, 8708–8724. [Google Scholar] [CrossRef] [PubMed]
- Silman, I. The multiple biological roles of the cholinesterases. Prog. Biophys. Mol. Biol. 2021, 162, 41–56. [Google Scholar] [CrossRef] [PubMed]
- Lushchekina, S.V.; Schopfer, L.M.; Grigorenko, B.L.; Nemukhin, A.V.; Varfolomeev, S.D.; Lockridge, O.; Masson, P. Optimization of Cholinesterase-Based Catalytic Bioscavengers against Organophosphorus Agents. Front. Pharmacol. 2018, 9, 211. [Google Scholar] [CrossRef] [PubMed]
- Lushchekina, S.; Masson, P. Catalytic bioscavengers against organophosphorus agents: Mechanistic issues of self-reactivating cholinesterases. Toxicology 2018, 409, 91–102. [Google Scholar] [CrossRef]
- Chauhan, P.S.; Jha, B. Pilot scale production of extracellular thermo-alkali stable laccase from Pseudomonas sp. S2 using agro waste and its application in organophosphorous pesticides degradation. J. Chem. Technol. Biotechnol. 2018, 93, 1022–1030. [Google Scholar] [CrossRef]
- Agarwal, N.; Solanki, V.S.; Gacem, A.; Hasan, M.A.; Pare, B.; Srivastava, A.; Singh, A.; Yadav, V.K.; Yadav, K.K.; Lee, C.; et al. Bacterial Laccases as Biocatalysts for the Remediation of Environmental Toxic Pollutants: A Green and Eco-Friendly Approach—A Review. Water 2022, 14, 4068. [Google Scholar] [CrossRef]
- Russell, A.J.; Berberich, J.A.; Drevon, G.F.; Koepsel, R.R. Biomaterials for mediation of chemical and biological warfare agents. Annu. Rev. Biomed. Eng. 2003, 5, 1–27. [Google Scholar] [CrossRef]
- Raj, A.; Kumar, A.; Khare, P.K. The looming threat of profenofos organophosphate and microbes in action for their sustainable degradation. Environ. Sci. Pollut. Res. 2024, 31, 14367–14387. [Google Scholar] [CrossRef]
- Hiblot, J.; Bzdrenga, J.; Champion, C.; Chabriere, E.; Elias, M. Crystal structure of VmoLac, a tentative quorum quenching lactonase from the extremophilic crenarchaeon Vulcanisaeta moutnovskia. Sci. Rep. 2015, 5, 8372. [Google Scholar] [CrossRef] [PubMed]
- Masson, P.; Lockridge, O. Butyrylcholinesterase for protection from organophosphorus poisons: Catalytic complexities and hysteretic behavior. Arch. Biochem. Biophys. 2010, 494, 107–120. [Google Scholar] [CrossRef]
- Masson, P.; Nachon, F. Cholinesterase reactivators and bioscavengers for pre- and post-exposure treatments of organophosphorus poisoning. J. Neurochem. 2017, 142, 26–40. [Google Scholar] [CrossRef]
- Lyagin, I.; Efremenko, E. Enzymes, Reacting with Organophosphorus Compounds as Detoxifiers: Diversity and Functions. Int. J. Mol. Sci. 2021, 22, 1761. [Google Scholar] [CrossRef]
- Allard, J.L.; Shields, K.A.; Munro, T.P.; Lua, L.H.L. Strategies for developing a recombinant butyrylcholinesterase medical countermeasure for Organophosphorus poisoning. Chem. Biol. Interact. 2022, 363, 109996. [Google Scholar] [CrossRef]
- Ashani, Y.; Pistinner, S. Estimation of the Upper Limit of Human Butyrylcholinesterase Dose Required for Protection against Organophosphates Toxicity: A Mathematically Based Toxicokinetic Model. Toxicol. Sci. 2004, 77, 358–367. [Google Scholar] [CrossRef] [PubMed]
- Masson, P.; Lushchekina, S.V. Emergence of catalytic bioscavengers against organophosphorus agents. Chem. Biol. Interact. 2016, 259, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Lockridge, O. Review of human butyrylcholinesterase structure, function, genetic variants, history of use in the clinic, and potential therapeutic uses. Pharmacol. Ther. 2015, 148, 34–46. [Google Scholar] [CrossRef]
- Xing, S.; Li, Q.; Xiong, B.; Chen, Y.; Feng, F.; Liu, W.; Sun, H. Structure and therapeutic uses of butyrylcholinesterase: Application in detoxification, Alzheimer’s disease, and fat metabolism. Med. Res. Rev. 2021, 41, 858–901. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, Y.; Saxena, A. Acetylcholinesterase inhibition resulting from exposure to inhaled OP can be prevented by pretreatment with BChE in both macaques and minipigs. Neuropharmacology 2020, 174, 108150. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.-J.; Carr, J.; Carlson, J.E.; Tong, L.; Xue, W.; Li, Y.; Schopfer, L.M.; Li, B.; Nachon, F.; Asojo, O.; et al. Five Tyrosines and Two Serines in Human Albumin Are Labeled by the Organophosphorus Agent FP-Biotin. Chem. Res. Toxicol. 2008, 21, 1787–1794. [Google Scholar] [CrossRef]
- Cochran, R.; Kalisiak, J.; Küçükkılınç, T.; Radić, Z.; Garcia, E.; Zhang, L.; Ho, K.-Y.; Amitai, G.; Kovarik, Z.; Fokin, V.V.; et al. Oxime-assisted Acetylcholinesterase Catalytic Scavengers of Organophosphates That Resist Aging. J. Biol. Chem. 2011, 286, 29718–29724. [Google Scholar] [CrossRef] [PubMed]
- Kovarik, Z.; Maček Hrvat, N. Efficient detoxification of nerve agents by oxime-assisted reactivation of acetylcholinesterase mutants. Neuropharmacology 2020, 171, 108111. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Murata, H.; Amitai, G.; Smith, P.N.; Matyjaszewski, K.; Russell, A.J. Catalytic Detoxification of Organophosphorus Nerve Agents by Butyrylcholinesterase-Polymer-Oxime Bioscavengers. Biomacromolecules 2020, 21, 3867–3877. [Google Scholar] [CrossRef] [PubMed]
- Shajhutdinova, Z.; Pashirova, T.; Masson, P. Kinetic Processes in Enzymatic Nanoreactors for In Vivo Detoxification. Biomedicines 2022, 10, 784. [Google Scholar] [CrossRef]
- Worek, F.; Thiermann, H.; Wille, T. Catalytic bioscavengers in nerve agent poisoning: A promising approach? Toxicol. Lett. 2016, 244, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Masson, P.; Lushchekina, S.V. Slow-binding inhibition of cholinesterases, pharmacological and toxicological relevance. Arch. Biochem. Biophys. 2016, 593, 60–68. [Google Scholar] [CrossRef]
- Goldsmith, M.; Ashani, Y. Catalytic bioscavengers as countermeasures against organophosphate nerve agents. Chem. Biol. Interact. 2018, 292, 50–64. [Google Scholar] [CrossRef]
- Ashani, Y.; Leader, H.; Rothschild, N.; Dosoretz, C. Combined Effect of Organophosphorus Hydrolase and Oxime on the Reactivation Rate of Diethylphosphoryl-Acetylcholinesterase Conjugates. Biochem. Pharmacol. 1998, 55, 159–168. [Google Scholar] [CrossRef]
- Saxena, A.; Sun, W.; Luo, C.; Myers, T.M.; Koplovitz, I.; Lenz, D.E.; Doctor, B.P. Bioscavenger for Protection from Toxicity of Organophosphorus Compounds. J. Mol. Neurosci. 2006, 30, 145–148. [Google Scholar] [CrossRef]
- Katyal, P.; Chu, S.; Montclare, J.K. Enhancing organophosphate hydrolase efficacy via protein engineering and immobilization strategies. Ann. N. Y. Acad. Sci. 2020, 1480, 54–72. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.; Arad, A.; Margalit, R. Lipsome-formulated enzymes for organophosphate scavenging: Butyrylcholinesterase and Demeton-S. Arch. Biochem. Biophys. 2005, 434, 108–115. [Google Scholar] [CrossRef]
- Chen, W.; Mulchandani, A. The use of live biocatalysts for pesticide detoxification. Trends Biotechnol. 1998, 16, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Moon, S.Y.; Guelta, M.A.; Lin, L.; Gómez-Gualdrón, D.A.; Snurr, R.Q.; Harvey, S.P.; Hupp, J.T.; Farha, O.K. Nanosizing a Metal-Organic Framework Enzyme Carrier for Accelerating Nerve Agent Hydrolysis. ACS Nano 2016, 10, 9174–9182. [Google Scholar] [CrossRef]
- Li, P.; Moon, S.-Y.; Guelta, M.A.; Harvey, S.P.; Hupp, J.T.; Farha, O.K. Encapsulation of a Nerve Agent Detoxifying Enzyme by a Mesoporous Zirconium Metal–Organic Framework Engenders Thermal and Long-Term Stability. J. Am. Chem. Soc. 2016, 138, 8052–8055. [Google Scholar] [CrossRef]
- Li, J.; Jin, X.; Liu, Y.; Li, F.; Zhang, L.; Zhu, X.; Lu, Y. Robust enzyme–silica composites made from enzyme nanocapsules. Chem. Commun. 2015, 51, 9628–9631. [Google Scholar] [CrossRef] [PubMed]
- Pei, L.; Petrikovics, I.; Way, J.L. Antagonism of the Lethal Effects of Paraoxon by Carrier Erythrocytes Containing Phosphotriesterase. Toxicol. Sci. 1995, 28, 209–214. [Google Scholar] [CrossRef]
- Petrikovics, I.; Hong, K.; Omburo, G.; Hu, Q.Z.; Pei, L.; McGuinn, W.D.; Sylvester, D.; Tamulinas, C.; Papahadjopoulos, D.; Jaszberenyi, J.C.; et al. Antagonism of paraoxon intoxication by recombinant phosphotriesterase encapsulated within sterically stabilized liposames. Toxicol. Appl. Pharmacol. 1999, 156, 56–63. [Google Scholar] [CrossRef]
- Petrikovics, I.; McGuinn, W.D.; Sylvester, D.; Yuzapavik, P.; Jiang, J.; Way, J.L.; Papahadjopoulos, D.; Hong, K.; Yin, R.; Cheng, T.C.; et al. In vitro studies on sterically stabilized liposomes (SL) as enzyme carriers in organophosphorus (OP) antagonism. Drug Deliv. J. Deliv. Target. Ther. Agents 2000, 7, 83–89. [Google Scholar] [CrossRef]
- Petrikovics, I. Long Circulating Liposomes Encapsulating Organophosphorus Acid Anhydrolase in Diisopropylfluorophosphate Antagonism. Toxicol. Sci. 2000, 57, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.K.; Liu, Z.N.; Yuan, L.; Zhang, P.S.; Zhao, M. Preparation of paraoxonase-1 liposomes and studies on their in vivo pharmacokinetics in rats. Clin. Exp. Pharmacol. Physiol. 2014, 41, 825–829. [Google Scholar] [CrossRef] [PubMed]
- Petrikovics, I.; Wales, M.; Budai, M.; Yu, J.C.C.; Szilasi, M. Nano-intercalated organophosphorus-hydrolyzing enzymes in organophosphorus antagonism. AAPS PharmSciTech 2012, 13, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Du, J.; Li, J.; Yan, M.; Zhu, Q.; Jin, X.; Zhu, X.; Hu, Z.; Tang, Y.; Lu, Y. Construction of Robust Enzyme Nanocapsules for Effective Organophosphate Decontamination, Detoxification, and Protection. Adv. Mater. 2013, 25, 2212–2218. [Google Scholar] [CrossRef] [PubMed]
- Efremenko, E.N.; Lyagin, I.V.; Klyachko, N.L.; Bronich, T.; Zavyalova, N.V.; Jiang, Y.; Kabanov, A.V. A simple and highly effective catalytic nanozyme scavenger for organophosphorus neurotoxins. J. Control. Release 2017, 247, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Wang, X.; Song, W.; Cheng, H.; Zhang, X. Metal-Organic Framework Mediated Multifunctional Nanoplatforms for Cancer Therapy. Adv. Ther. 2019, 2, 1800100. [Google Scholar] [CrossRef]
- Zhu, H.; Li, B.; Yu Chan, C.; Low Qian Ling, B.; Tor, J.; Yi Oh, X.; Jiang, W.; Ye, E.; Li, Z.; Jun Loh, X. Advances in Single-component inorganic nanostructures for photoacoustic imaging guided photothermal therapy. Adv. Drug Deliv. Rev. 2023, 192, 114644. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Zhao, Y.L.; Luo, X.J.; Xu, D.S.; Cao, X.; Xu, J.H.; Dai, Q.; Zhang, X.Y.; Ge, J.; Bai, Y.P. Cross-linked enzyme-polymer conjugates with excellent stability and detergent-enhanced activity for efficient organophosphate degradation. Bioresour. Bioprocess. 2018, 5, 49. [Google Scholar] [CrossRef]
- Sun, Z.; Hou, Y. Micro/Nanorobots as Active Delivery Systems for Biomedicine: From Self-Propulsion to Controllable Navigation. Adv. Ther. 2022, 5, 2100228. [Google Scholar] [CrossRef]
- Li, J.; Esteban-Fernández de Ávila, B.; Gao, W.; Zhang, L.; Wang, J. Micro/nanorobots for biomedicine: Delivery, surgery, sensing, and detoxification. Sci. Robot. 2017, 2, eaam6431. [Google Scholar] [CrossRef] [PubMed]
- Pashirova, T.N.; Shaihutdinova, Z.M.; Mironov, V.F.; Masson, P. Biomedical Nanosystems for in vivo Detoxification: From Passive Delivery Systems to Functional Nanodevices and Nanorobots. Acta Naturae 2023, 15, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Forster, V.; Leroux, J.-C. Nano-antidotes for drug overdose and poisoning. Sci. Transl. Med. 2015, 7, 290ps14. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, H.; Zou, H.; Ding, J. Polymer Nanoantidotes. Chem. Eur. J. 2023, 29, e202301107. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.-M.J.; Fang, R.H.; Copp, J.; Luk, B.T.; Zhang, L. A biomimetic nanosponge that absorbs pore-forming toxins. Nat. Nanotechnol. 2013, 8, 336–340. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Anton, N.; Niko, Y.; Klymchenko, A.S. Controlled Release and Capture of Aldehydes by Dynamic Imine Chemistry in Nanoemulsions: From Delivery to Detoxification. ACS Appl. Bio Mater. 2023, 6, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, D.; Kai, M.; Shen, W.-T.; Sun, L.; Gao, W.; Zhang, L. Design Strategies for Cellular Nanosponges as Medical Countermeasures. BME Front. 2023, 4, 0018. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Liu, E.J.; Tsao, C.; Kasten, S.A.; Boeri, M.V.; Dao, T.L.; DeBus, S.J.; Cadieux, C.L.; Baker, C.A.; Otto, T.C.; et al. Nanoscavenger provides long-term prophylactic protection against nerve agents in rodents. Sci. Transl. Med. 2019, 11, eaau7091. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.; Wang, B.; Wang, Q.; Liu, G.; Song, J.; Zhang, F.; Li, J.; Wang, F.; He, Q.; Zhu, Y.; et al. Dual-Modal Nanoscavenger for Detoxification of Organophosphorus Compounds. ACS Appl. Mater. Interfaces 2022, 14, 42454–42467. [Google Scholar] [CrossRef]
- Zou, S.; Wang, Q.; Song, J.; Liu, G.; Zhang, F.; Li, J.; Wang, F.; Hu, Y.; Lv, Y.; Zhou, D.; et al. Top-down Nanoscavengers for the protection of organophosphate-challenged cells. Giant 2024, 17, 100213. [Google Scholar] [CrossRef]
- Pashirova, T.; Shaihutdinova, Z.; Mansurova, M.; Kazakova, R.; Shambazova, D.; Bogdanov, A.; Tatarinov, D.; Daudé, D.; Jacquet, P.; Chabrière, E.; et al. Enzyme Nanoreactor for In Vivo Detoxification of Organophosphates. ACS Appl. Mater. Interfaces 2022, 14, 19241–19252. [Google Scholar] [CrossRef]
- Pashirova, T.; Shaihutdinova, Z.; Tatarinov, D.; Mansurova, M.; Kazakova, R.; Bogdanov, A.; Chabrière, E.; Jacquet, P.; Daudé, D.; Akhunzianov, A.A.; et al. Tuning the Envelope Structure of Enzyme Nanoreactors for In Vivo Detoxification of Organophosphates. Int. J. Mol. Sci. 2023, 24, 15756. [Google Scholar] [CrossRef] [PubMed]
- Gaur, D.; Dubey, N.C.; Tripathi, B.P. Biocatalytic self-assembled synthetic vesicles and coacervates: From single compartment to artificial cells. Adv. Colloid Interface Sci. 2022, 299, 102566. [Google Scholar] [CrossRef] [PubMed]
- Maffeis, V.; Heuberger, L.; Nikoletić, A.; Schoenenberger, C.; Palivan, C.G. Synthetic Cells Revisited: Artificial Cells Construction Using Polymeric Building Blocks. Adv. Sci. 2024, 11, 2305837. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, Y.; Zhuang, J.; Lee, J.H.; Wang, L.; Fang, R.H.; Gao, W.; Zhang, L. Cell-membrane-cloaked oil nanosponges enable dual-modal detoxification. ACS Nano 2019, 13, 7209–7215. [Google Scholar] [CrossRef]
- Zou, S.; Wang, Q.; He, Q.; Liu, G.; Song, J.; Li, J.; Wang, F.; Huang, Y.; Hu, Y.; Zhou, D.; et al. Brain-targeted nanoreactors prevent the development of organophosphate-induced delayed neurological damage. J. Nanobiotechnol. 2023, 21, 256. [Google Scholar] [CrossRef] [PubMed]
- Kazakova, R.R.; Masson, P. Quantitative Measurements of Pharmacological and Toxicological Activity of Molecules. Chemistry 2022, 4, 1466–1474. [Google Scholar] [CrossRef]
- Polhuijs, M.; Langenberg, J.P.; Benschop, H.P. New Method for Retrospective Detection of Exposure to Organophosphorus Anticholinesterases: Application to Alleged Sarin Victims of Japanese Terrorists. Toxicol. Appl. Pharmacol. 1997, 146, 156–161. [Google Scholar] [CrossRef]
- Griffiths, A.D. Directed evolution of an extremely fast phosphotriesterase by in vitro compartmentalization. EMBO J. 2003, 22, 24–35. [Google Scholar] [CrossRef]
- Goldsmith, M.; Aggarwal, N.; Ashani, Y.; Jubran, H.; Greisen, P.J.; Ovchinnikov, S.; Leader, H.; Baker, D.; Sussman, J.L.; Goldenzweig, A.; et al. Overcoming an optimization plateau in the directed evolution of highly efficient nerve agent bioscavengers. Protein Eng. Des. Sel. 2017, 30, 333–345. [Google Scholar] [CrossRef]
- Goldsmith, M.; Tawfik, D.S. Enzyme engineering: Reaching the maximal catalytic efficiency peak. Curr. Opin. Struct. Biol. 2017, 47, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.J.; Scott, C.; Carville, A.; Mansfield, K.; Ollis, D.L.; Bird, S.B. Pharmacokinetics of OpdA, an organophosphorus hydrolase, in the African green monkey. Biochem. Pharmacol. 2010, 80, 1075–1079. [Google Scholar] [CrossRef]
- Liu, Y.; Li, J.; Lu, Y. Enzyme therapeutics for systemic detoxification. Adv. Drug Deliv. Rev. 2015, 90, 24–39. [Google Scholar] [CrossRef] [PubMed]
- Schadauer, F.; Geiss, A.F.; Srajer, J.; Siebenhofer, B.; Frank, P.; Reiner-Rozman, C.; Ludwig, B.; Richter, O.-M.H.; Nowak, C.; Naumann, R.L.C. Silica Nanoparticles for the Oriented Encapsulation of Membrane Proteins into Artificial Bilayer Lipid Membranes. Langmuir 2015, 31, 2511–2516. [Google Scholar] [CrossRef]
- Godoy, S.; Violot, S.; Boullanger, P.; Bouchu, M.; Leca-Bouvier, B.D.; Blum, L.J.; Girard-Egrot, A.P. Kinetics Study of Bungarus fasciatus Venom Acetylcholinesterase Immobilised on a Langmuir–Blodgett Proteo-Glycolipidic Bilayer. ChemBioChem 2005, 6, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Felsztyna, I.; Perillo, M.A.; Clop, E.M. Nanoarchitectonic approaches for measuring the catalytic behavior of a membrane anchored enzyme. From Langmuir-Blodgett to a novel Langmuir-Schaefer based nanofilm building device. Biochim. Biophys. Acta Biomembr. 2023, 1865, 184177. [Google Scholar] [CrossRef] [PubMed]
- Girard-Egrot, A.P.; Godoy, S.; Blum, L.J. Enzyme association with lipidic Langmuir–Blodgett films: Interests and applications in nanobioscience. Adv. Colloid Interface Sci. 2005, 116, 205–225. [Google Scholar] [CrossRef] [PubMed]
- Marques, C.; Hajipour, M.J.; Marets, C.; Oudot, A.; Safavi-sohi, R.; Guillemin, M.; Borchard, G.; Jordan, O.; Saviot, L.; Maurizi, L. Identification of the Proteins Determining the Blood Circulation Time of Nanoparticles. ACS Nano 2023, 17, 12458–12470. [Google Scholar] [CrossRef] [PubMed]
- Berrecoso, G.; Crecente-Campo, J.; Alonso, M.J. Unveiling the pitfalls of the protein corona of polymeric drug nanocarriers. Drug Deliv. Transl. Res. 2020, 10, 730–750. [Google Scholar] [CrossRef]
- Marques, C.; Borchard, G.; Jordan, O. Unveiling the challenges of engineered protein corona from the proteins’ perspective. Int. J. Pharm. 2024, 654, 123987. [Google Scholar] [CrossRef]
- Rodriguez, P.L.; Harada, T.; Christian, D.A.; Pantano, D.A.; Tsai, R.K.; Discher, D.E. Minimal “Self” Peptides That Inhibit Phagocytic Clearance and Enhance Delivery of Nanoparticles. Science 2013, 339, 971–975. [Google Scholar] [CrossRef]
- Pinals, R.L.; Yang, D.; Rosenberg, D.J.; Chaudhary, T.; Crothers, A.R.; Iavarone, A.T.; Hammel, M.; Landry, M.P. Quantitative Protein Corona Composition and Dynamics on Carbon Nanotubes in Biological Environments. Angew. Chem. Int. Ed. 2020, 59, 23668–23677. [Google Scholar] [CrossRef] [PubMed]
- Vincent, M.P.; Bobbala, S.; Karabin, N.B.; Frey, M.; Liu, Y.; Navidzadeh, J.O.; Stack, T.; Scott, E.A. Surface chemistry-mediated modulation of adsorbed albumin folding state specifies nanocarrier clearance by distinct macrophage subsets. Nat. Commun. 2021, 12, 648. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.H.D.; Jayasinghe, M.K.; Le, A.H.; Peng, B.; Le, M.T.N. Advances in Drug Delivery Systems Based on Red Blood Cells and Their Membrane-Derived Nanoparticles. ACS Nano 2023, 17, 5187–5210. [Google Scholar] [CrossRef]
- Smith, P.N.; Mao, L.; Sinha, K.; Russell, A.J. Organophosphate detoxification by membrane-engineered red blood cells. Acta Biomater. 2021, 124, 270–281. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, M. The need for improved methodology in protein corona analysis. Nat. Commun. 2022, 13, 49. [Google Scholar] [CrossRef]
- Sun, W.; Luo, C.; Naik, R.S.; Doctor, B.P.; Saxena, A. Pharmacokinetics and immunologic consequences of repeated administrations of purified heterologous and homologous butyrylcholinesterase in mice. Life Sci. 2009, 85, 657–661. [Google Scholar] [CrossRef] [PubMed]
- Novikov, B.N.; Grimsley, J.K.; Kern, R.J.; Wild, J.R.; Wales, M.E. Improved pharmacokinetics and immunogenicity profile of organophosphorus hydrolase by chemical modification with polyethylene glycol. J. Control. Release 2010, 146, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Trovaslet-Leroy, M.; Musilova, L.; Renault, F.; Brazzolotto, X.; Misik, J.; Novotny, L.; Froment, M.-T.; Gillon, E.; Loiodice, M.; Verdier, L.; et al. Organophosphate hydrolases as catalytic bioscavengers of organophosphorus nerve agents. Toxicol. Lett. 2011, 206, 14–23. [Google Scholar] [CrossRef]
- Katre, N.V.; Asherman, J.; Schaefer, H.; Hora, M. Multivesicular Liposome (DepoFoam) Technology for the Sustained Delivery of Insulin-Like Growth Factor-I (IGF-I). J. Pharm. Sci. 1998, 87, 1341–1346. [Google Scholar] [CrossRef]
- Ye, Q.; Asherman, J.; Stevenson, M.; Brownson, E.; Katre, N.V. DepoFoam™ technology: A vehicle for controlled delivery of protein and peptide drugs. J. Control. Release 2000, 64, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Shum, H.C.; Kim, J.W.; Cho, J.C.; Weitz, D.A. Multiple polymersomes for programmed release of multiple components. J. Am. Chem. Soc. 2011, 133, 15165–15171. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Shen, M.; Möhwald, H. Polyelectrolyte multilayer nanoreactors toward the synthesis of diverse nanostructured materials. Prog. Polym. Sci. 2004, 29, 987–1019. [Google Scholar] [CrossRef]
- Vranckx, C.; Lambricht, L.; Préat, V.; Cornu, O.; Dupont-Gillain, C.; vander Straeten, A. Layer-by-Layer Nanoarchitectonics Using Protein–Polyelectrolyte Complexes toward a Generalizable Tool for Protein Surface Immobilization. Langmuir 2022, 38, 5579–5589. [Google Scholar] [CrossRef] [PubMed]
- Borges, J.; Zeng, J.; Liu, X.Q.; Chang, H.; Monge, C.; Garot, C.; Ren, K.; Machillot, P.; Vrana, N.E.; Lavalle, P.; et al. Recent Developments in Layer-by-Layer Assembly for Drug Delivery and Tissue Engineering Applications. Adv. Healthc. Mater. 2024, 13, 2302713. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.D.; Langer, R. Immobilized enzymes in clinical medicine: An emerging approach to new drug therapies. Trends Biotechnol. 1986, 4, 179–186. [Google Scholar] [CrossRef]
- Haque, A.M.; Hwang, C.E.; Kim, S.C.; Cho, D.Y.; Lee, H.Y.; Cho, K.M.; Lee, J.H. Biodegradation of organophosphorus insecticides by two organophosphorus hydrolase genes (opdA and opdE) from isolated Leuconostoc mesenteroides WCP307 of kimchi origin. Process Biochem. 2020, 94, 340–348. [Google Scholar] [CrossRef]
- Santillan, J.Y.; Rojas, N.L.; Lewkowicz, E.S.; Iribarren, A.M. Novel fungal organophosphorus hydrolases in acidic media: An application to apples decontamination. Environ. Sci. Pollut. Res. 2022, 30, 10803–10811. [Google Scholar] [CrossRef]
- Santillan, J.Y.; Muzlera, A.; Molina, M.; Lewkowicz, E.S.; Iribarren, A.M. Microbial degradation of organophosphorus pesticides using whole cells and enzyme extracts. Biodegradation 2020, 31, 423–433. [Google Scholar] [CrossRef]
- Parthipan, P.; Prakash, C.; Perumal, D.; Elumalai, P.; Rajasekar, A.; Cheng, L. Biogenic Nanoparticles and Strategies of Nano-bioremediation to Remediate PAHs for a Sustainable Future. In Biotechnology for Sustainable Environment; Springer: Singapore, 2021; pp. 317–337. [Google Scholar]
- Chauhan, P.; Imam, A.; Kanaujia, P.K.; Suman, S.K. Nano-bioremediation: An eco-friendly and effective step towards petroleum hydrocarbon removal from environment. Environ. Res. 2023, 231, 116224. [Google Scholar] [CrossRef]
- Feng, J.-R.; Deng, Q.-X.; Han, S.-K.; Ni, H.-G. Use of nanoparticle-coated bacteria for the bioremediation of organic pollution: A mini review. Chemosphere 2023, 313, 137391. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, P.; Ahamad, L.; Chaudhary, A.; Kumar, G.; Chen, W.-J.; Chen, S. Nanoparticle-mediated bioremediation as a powerful weapon in the removal of environmental pollutants. J. Environ. Chem. Eng. 2023, 11, 109591. [Google Scholar] [CrossRef]
- Karthik, V.; Senthil Kumar, P.; Vo, D.-V.N.; Selvakumar, P.; Gokulakrishnan, M.; Keerthana, P.; Audilakshmi, V.; Jeyanthi, J. Enzyme-loaded nanoparticles for the degradation of wastewater contaminants: A review. Environ. Chem. Lett. 2021, 19, 2331–2350. [Google Scholar] [CrossRef]
- Zhu, H.; Prince, E.; Narayanan, P.; Liu, K.; Nie, Z.; Kumacheva, E. Colloidal stability of nanoparticles stabilized with mixed ligands in solvents with varying polarity. Chem. Commun. 2020, 56, 8131–8134. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Z.; Yu, L.; Shi, Q.; Dong, X.; Sun, Y. Zwitterionic polymer-mediated immobilization of organophosphorus hydrolase enhances hydrolysis of methyl parathion by substrate enrichment. Biochem. Eng. J. 2022, 184, 108491. [Google Scholar] [CrossRef]
- Wang, Z.; Gao, J.; Shi, Q.; Dong, X.; Sun, Y. Facile purification and immobilization of organophosphorus hydrolase on protein-inorganic hybrid phosphate nanosheets. Chin. J. Chem. Eng. 2023, 56, 119–125. [Google Scholar] [CrossRef]
- Chen, J.; Guo, Z.; Zhang, H.; Xin, Y.; Shi, Y.; Gu, Z.; Zhang, L.; Zhong, J.; Guo, X.; Li, Y.; et al. Development of a multimetal-based phosphotriesterase hybrid nanoflowers for decontamination of environmental organophosphorus compounds pollution. Chem. Eng. J. 2022, 446, 136933. [Google Scholar] [CrossRef]
- Das, A.; Jaswal, V.; Yogalakshmi, K.N. Degradation of chlorpyrifos in soil using laccase immobilized iron oxide nanoparticles and their competent role in deterring the mobility of chlorpyrifos. Chemosphere 2020, 246, 125676. [Google Scholar] [CrossRef] [PubMed]
- Moon, Y.; Jafry, A.T.; Bang Kang, S.; Young Seo, J.; Baek, K.-Y.; Kim, E.-J.; Pan, J.-G.; Choi, J.-Y.; Kim, H.-J.; Han Lee, K.; et al. Organophosphorus hydrolase-poly-β-cyclodextrin as a stable self-decontaminating bio-catalytic material for sorption and degradation of organophosphate pesticide. J. Hazard. Mater. 2019, 365, 261–269. [Google Scholar] [CrossRef]
- Chen, J.; Guo, Z.; Xin, Y.; Shi, Y.; Li, Y.; Gu, Z.; Zhong, J.; Guo, X.; Zhang, L. Preparation of efficient, stable, and reusable copper-phosphotriesterase hybrid nanoflowers for biodegradation of organophosphorus pesticides. Enzym. Microb. Technol. 2021, 146, 109766. [Google Scholar] [CrossRef]
- Sharifi, M.; Robatjazi, S.-M.; Sadri, M.; Mosaabadi, J.M. Immobilization of organophosphorus hydrolase enzyme by covalent attachment on modified cellulose microfibers using different chemical activation strategies: Characterization and stability studies. Chin. J. Chem. Eng. 2019, 27, 191–199. [Google Scholar] [CrossRef]
- Li, Y.; Luan, P.; Zhou, L.; Xue, S.; Liu, Y.; Liu, Y.; Jiang, Y.; Gao, J. Purification and immobilization of His-tagged organophosphohydrolase on yolk–shell Co/C@SiO2@Ni/C nanoparticles for cascade degradation and detection of organophosphates. Biochem. Eng. J. 2021, 167, 107895. [Google Scholar] [CrossRef]
- Samanta, A.; Breger, J.C.; Susumu, K.; Oh, E.; Walper, S.A.; Bassim, N.; Medintz, I.L. DNA–Nanoparticle Composites Synergistically Enhance Organophosphate Hydrolase Enzymatic Activity. ACS Appl. Nano Mater. 2018, 1, 3091–3097. [Google Scholar] [CrossRef]
- Srinivasan, P.; Selvankumar, T.; Paray, B.A.; Rehman, M.U.; Kamala-Kannan, S.; Govarthanan, M.; Kim, W.; Selvam, K. Chlorpyrifos degradation efficiency of Bacillus sp. laccase immobilized on iron magnetic nanoparticles. 3 Biotech 2020, 10, 366. [Google Scholar] [CrossRef] [PubMed]
- Ginet, N.; Pardoux, R.; Adryanczyk, G.; Garcia, D.; Brutesco, C.; Pignol, D. Single-Step Production of a Recyclable Nanobiocatalyst for Organophosphate Pesticides Biodegradation Using Functionalized Bacterial Magnetosomes. PLoS ONE 2011, 6, e21442. [Google Scholar] [CrossRef] [PubMed]
- El-Boubbou, K.; Schofield, D.A.; Landry, C.C. Enhanced Enzymatic Activity of OPH in Ammonium-Functionalized Mesoporous Silica: Surface Modification and Pore Effects. J. Phys. Chem. C 2012, 116, 17501–17506. [Google Scholar] [CrossRef]
- Diao, J.; Zhao, G.; Li, Y.; Huang, J.; Sun, Y. Carboxylesterase from Spodoptera Litura: Immobilization and use for the Degradation of Pesticides. Procedia Environ. Sci. 2013, 18, 610–619. [Google Scholar] [CrossRef]
- Despotović, D.; Aharon, E.; Dubovetskyi, A.; Leader, H.; Ashani, Y.; Tawfik, D.S. A mixture of three engineered phosphotriesterases enables rapid detoxification of the entire spectrum of known threat nerve agents. Protein Eng. Des. Sel. 2019, 32, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Otto, T.C.; Scott, J.R.; Kauffman, M.A.; Hodgins, S.M.; DiTargiani, R.C.; Hughes, J.H.; Sarricks, E.P.; Saturday, G.A.; Hamilton, T.A.; Cerasoli, D.M. Identification and characterization of novel catalytic bioscavengers of organophosphorus nerve agents. Chem. Biol. Interact. 2013, 203, 186–190. [Google Scholar] [CrossRef]
- Ferrer, M.; Golyshina, O.; Beloqui, A.; Golyshin, P.N. Mining enzymes from extreme environments. Curr. Opin. Microbiol. 2007, 10, 207–214. [Google Scholar] [CrossRef]
- Jacob, R.B.; Michaels, K.C.; Anderson, C.J.; Fay, J.M.; Dokholyan, N.V. Harnessing Nature’s Diversity: Discovering organophosphate bioscavenger characteristics among low molecular weight proteins. Sci. Rep. 2016, 6, 37175. [Google Scholar] [CrossRef] [PubMed]
- Bershtein, S.; Tawfik, D.S. Advances in laboratory evolution of enzymes. Curr. Opin. Chem. Biol. 2008, 12, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Bigley, A.N.; Xu, C.; Henderson, T.J.; Harvey, S.P.; Raushel, F.M. Enzymatic Neutralization of the Chemical Warfare Agent VX: Evolution of Phosphotriesterase for Phosphorothiolate Hydrolysis. J. Am. Chem. Soc. 2013, 135, 10426–10432. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.H.; Zhi, Q.W.; Sun, M.J. Dual activities of human prolidase. Toxicol. In Vitro 2006, 20, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Farnoosh, G.; Khajeh, K.; Mohammadi, M.; Hassanpour, K.; Latifi, A.M.; Aghamollaei, H. Catalytic and structural effects of flexible loop deletion in organophosphorus hydrolase enzyme: A thermostability improvement mechanism. J. Biosci. 2020, 45, 54. [Google Scholar] [CrossRef]
- Shi, C.; Liu, S.; Du, G. Improving Catalytic Activity and Thermal Stability of Methyl-Parathion Hydrolase for Degrading the Pesticide of Methyl-Parathion. Int. J. Chem. Eng. 2022, 2022, 7355170. [Google Scholar] [CrossRef]

| Type of Organism | Enzyme | Ref. | |
|---|---|---|---|
| Bacteria | Agrobacterium radiobacter | OpdA (organophosphate hydrolase) | [50] |
| Bacillus thuringiensis MB497 | OPP (organophosphorus phosphatase) | [51] | |
| Alteromonas sp., Alteromonas haloplanktis, Alteromonas undin | OPAA (organophosphorus acid anhydrolase) | [19] | |
| Flavobacterium sp. | OPD (organophosphate hydrolase) | [52] | |
| Achromobacter xylosoxidans GH9OP, Arthrobacter sp. HM01, Brevundimonas diminuta. | OPH/OpdH (organophosphosphate hydrolase)/PTE (phospho-triesterase)/aryl-dialkyl-phosphatase | [49,52,53] | |
| Cronobacter muytjensii GH10, Pseudaminobacter sp. mp-1, Pseudomonas aeruginosa GH2NO8, Brevundimonas diminuta MG (formely Pseudomonas diminuta MG), Pseudomonas monteilii C11 | |||
| Pseudomonas sp. WBC-3, Plesiomonas sp. M6 | MPH (methyl parathion hydrolase) | [50,52] | |
| Engineered | Engineered Escherichia coli | OPH-E (organophosphosphate hydrolase)/parathion hydrolase | [54] |
| Archaebacteria | Sulfolobus solfataricus Vulcanisaeta moutnovskia | Ssopox Phosphotriesterase-like lactonase | [55] [56] |
| Fungi | Caldariomyces fumago | Chloroperoxidase | [57] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pashirova, T.; Salah-Tazdaït, R.; Tazdaït, D.; Masson, P. Applications of Microbial Organophosphate-Degrading Enzymes to Detoxification of Organophosphorous Compounds for Medical Countermeasures against Poisoning and Environmental Remediation. Int. J. Mol. Sci. 2024, 25, 7822. https://doi.org/10.3390/ijms25147822
Pashirova T, Salah-Tazdaït R, Tazdaït D, Masson P. Applications of Microbial Organophosphate-Degrading Enzymes to Detoxification of Organophosphorous Compounds for Medical Countermeasures against Poisoning and Environmental Remediation. International Journal of Molecular Sciences. 2024; 25(14):7822. https://doi.org/10.3390/ijms25147822
Chicago/Turabian StylePashirova, Tatiana, Rym Salah-Tazdaït, Djaber Tazdaït, and Patrick Masson. 2024. "Applications of Microbial Organophosphate-Degrading Enzymes to Detoxification of Organophosphorous Compounds for Medical Countermeasures against Poisoning and Environmental Remediation" International Journal of Molecular Sciences 25, no. 14: 7822. https://doi.org/10.3390/ijms25147822







