Analysis of Carcinogenic Involvement of MicroRNA Pattern in Peripheral Non-Cancerous Tissues and Chronic Viral Liver Injury
Abstract
1. Introduction
2. Results
2.1. Clinical Characteristics of Each Disease Group by Age
2.2. Differences between Age Groups in the Same Disease
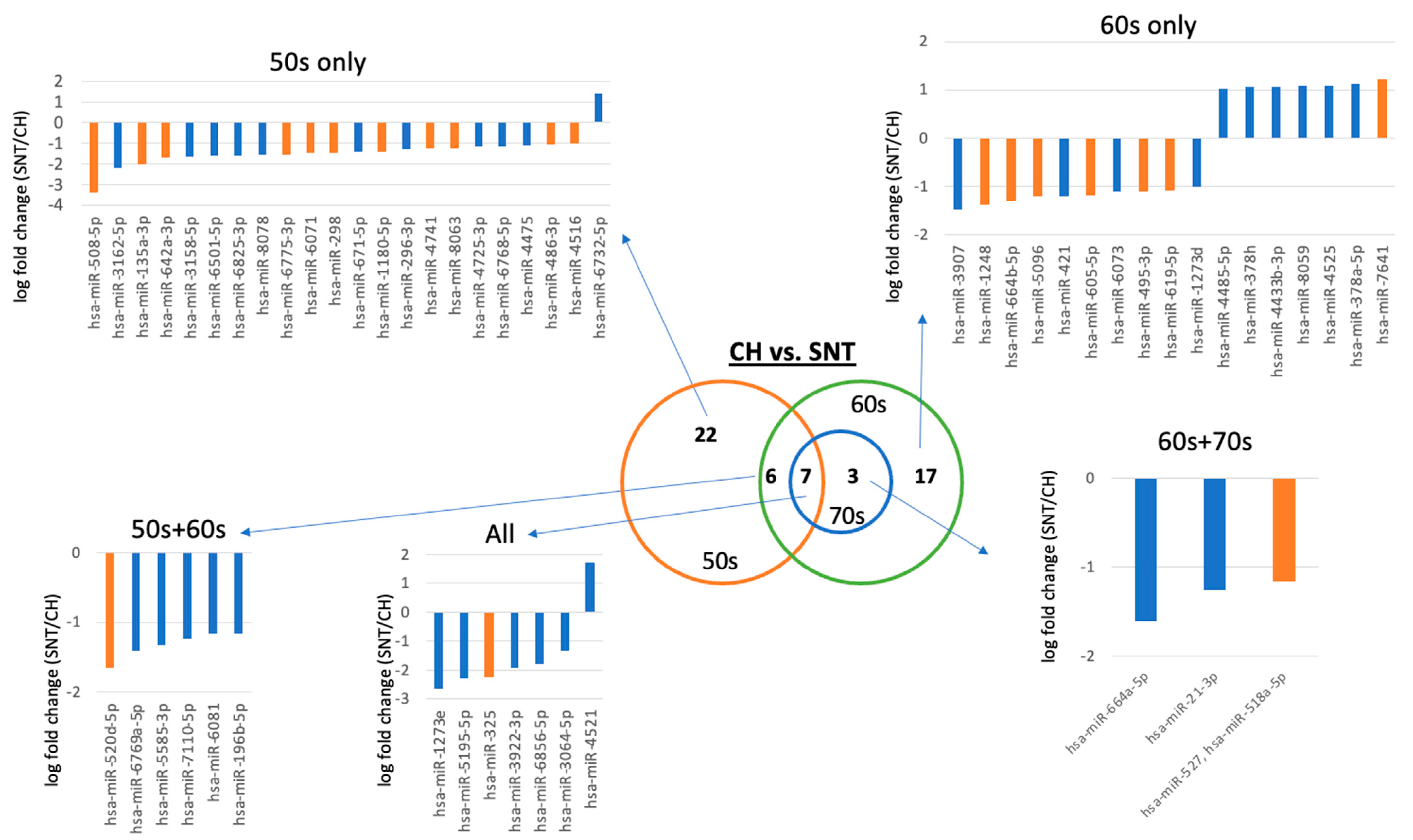
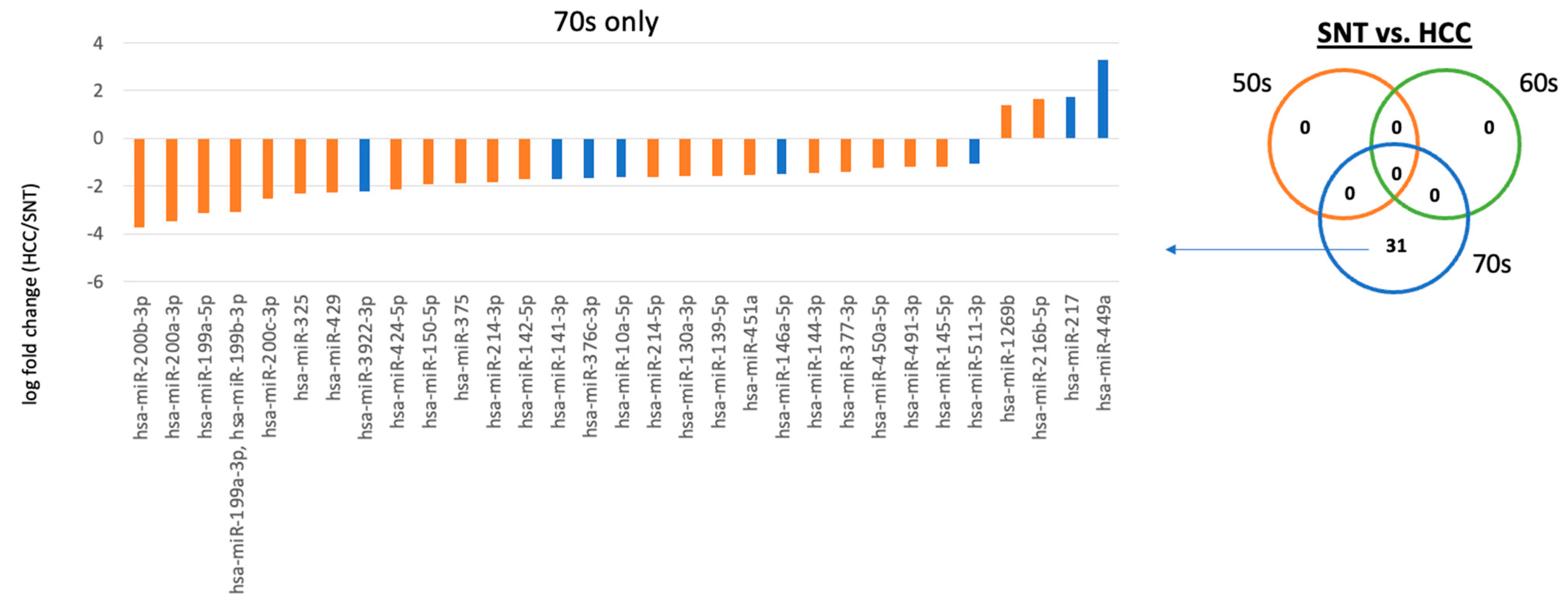
2.3. Differences between Diseases in the Same Age Group
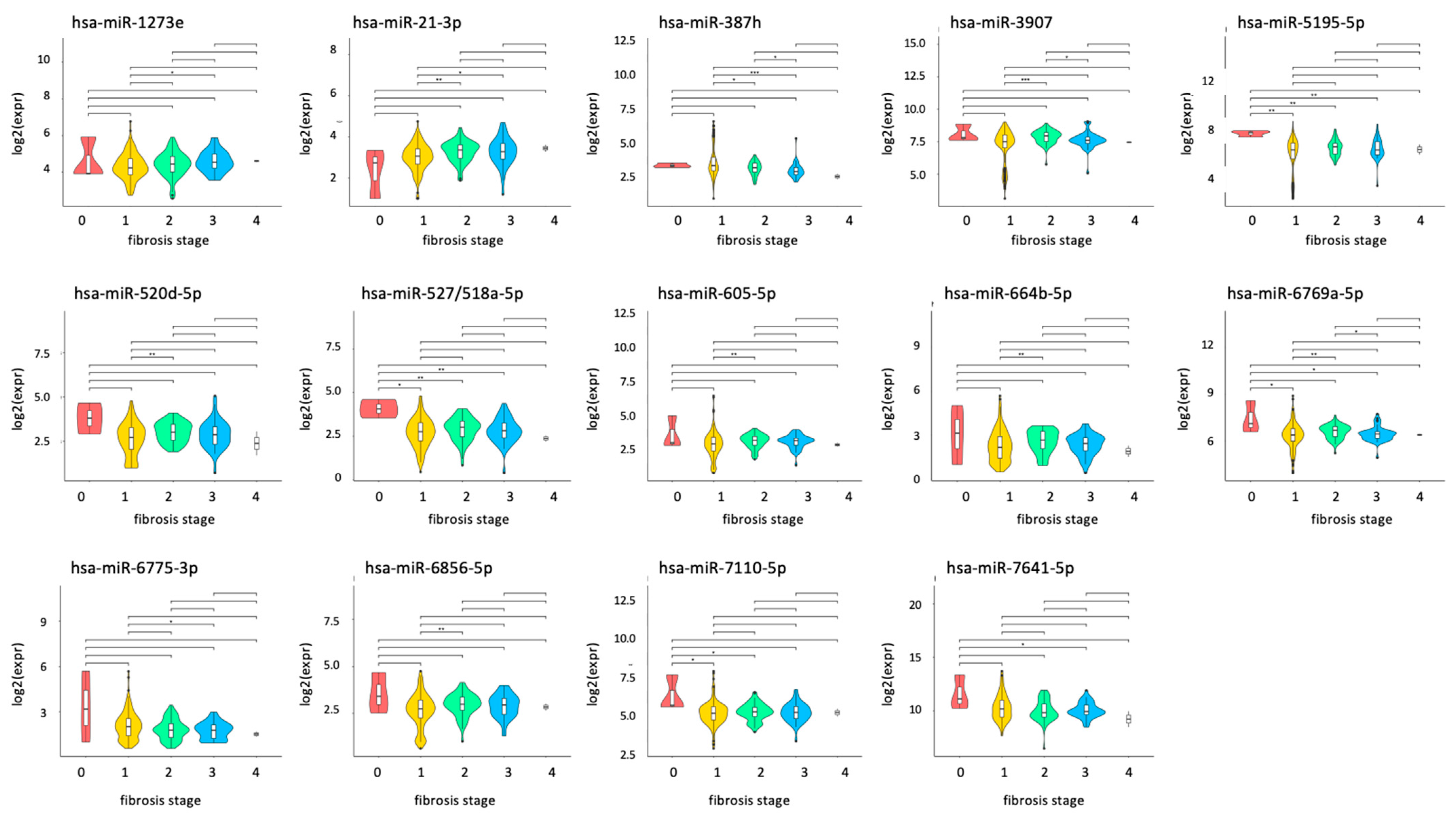
2.4. miRNAs Affected by the Stage of Liver Fibrosis
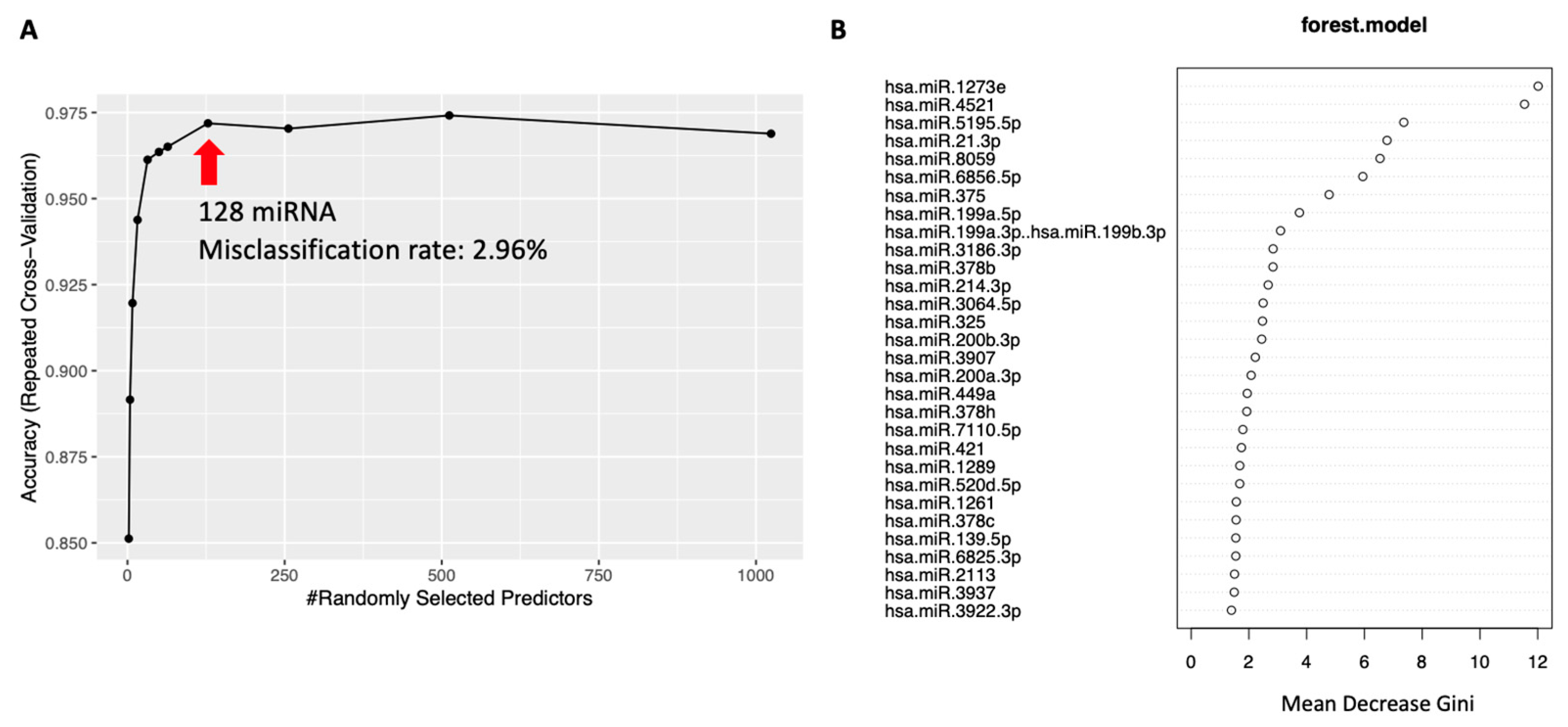
2.5. Disease Classification Using Random Forest
3. Discussion
4. Materials and Methods
4.1. Analysis Strategy
4.2. RNA Preparation and Microarray Analysis
4.3. Statistical Analysis
4.3.1. Clinical Data Comparison among Groups
4.3.2. Analysis of Differences in miRNA between Age Groups for the Same Disease
4.3.3. Analysis of Differences between Diseases in the Same Age Group
4.3.4. Analysis of miRNAs Involved in Liver Fibrosis
4.3.5. Evaluation of Factors That Contributed to Classification when Performing Group Classification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Altekruse, S.F.; McGlynn, K.A.; Reichman, M.E. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J. Clin. Oncol. 2009, 27, 1485–1491. [Google Scholar] [CrossRef] [PubMed]
- Honda, T.; Miyaaki, H.; Ichikawa, T.; Taura, N.; Miuma, S.; Shibata, H.; Isomoto, H.; Takeshima, F.; Nakao, K. Clinical characteristics of hepatocellular carcinoma in elderly patients. Oncol. Lett. 2011, 2, 851–854. [Google Scholar] [CrossRef] [PubMed]
- Balducci, L.; Ershler, W.B. Cancer and ageing: A nexus at several levels. Nat. Rev. Cancer 2005, 5, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Jemal, A.; Siegel, R.; Xu, J.; Ward, E. Cancer statistics, 2010. CA Cancer J. Clin. 2010, 60, 277–300. [Google Scholar] [CrossRef] [PubMed]
- Asahina, Y.; Tsuchiya, K.; Tamaki, N.; Hirayama, I.; Tanaka, T.; Sato, M.; Yasui, Y.; Hosokawa, T.; Ueda, K.; Kuzuya, T.; et al. Effect of aging on risk for hepatocellular carcinoma in chronic hepatitis C virus infection. Hepatology 2010, 52, 518–527. [Google Scholar] [CrossRef]
- Wynne, H.A.; Cope, L.H.; Mutch, E.; Rawlins, M.D.; Woodhouse, K.W.; James, O.F. The effect of age upon liver volume and apparent liver blood flow in healthy man. Hepatology 1989, 9, 297–301. [Google Scholar] [CrossRef] [PubMed]
- Roberts, C.J.; Jackson, L.; Halliwell, M.; Branch, R.A. The relationship between liver volume, antipyrine clearance and indocyanine green clearance before and after phenobarbitone administration in man. Br. J. Clin. Pharmacol. 1976, 3, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Caesar, J.; Shaldon, S.; Chiandussi, L.; Guevara, L.; Sherlock, S. The use of indocyanine green in the measurement of hepatic blood flow and as a test of hepatic function. Clin. Sci. 1961, 21, 43–57. [Google Scholar]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Tutar, Y. miRNA and cancer; computational and experimental approaches. Curr. Pharm. Biotechnol. 2014, 15, 429. [Google Scholar] [CrossRef]
- Ibanez-Ventoso, C.; Yang, M.; Guo, S.; Robins, H.; Padgett, R.W.; Driscoll, M. Modulated microRNA expression during adult lifespan in Caenorhabditis elegans. Aging Cell 2006, 5, 235–246. [Google Scholar] [CrossRef]
- de Lencastre, A.; Pincus, Z.; Zhou, K.; Kato, M.; Lee, S.S.; Slack, F.J. MicroRNAs both promote and antagonize longevity in C. elegans. Curr. Biol. 2010, 20, 2159–2168. [Google Scholar] [CrossRef]
- Kato, M.; Chen, X.; Inukai, S.; Zhao, H.; Slack, F.J. Age-associated changes in expression of small, noncoding RNAs, including microRNAs, in C. elegans. RNA 2011, 17, 1804–1820. [Google Scholar] [CrossRef]
- Ibanez-Ventoso, C.; Vora, M.; Driscoll, M. Sequence relationships among C. elegans, D. melanogaster and human microRNAs highlight the extensive conservation of microRNAs in biology. PLoS ONE 2008, 3, e2818. [Google Scholar] [CrossRef] [PubMed]
- Maes, O.C.; An, J.; Sarojini, H.; Wang, E. Murine microRNAs implicated in liver functions and aging process. Mech. Ageing Dev. 2008, 129, 534–541. [Google Scholar] [CrossRef]
- Li, N.; Muthusamy, S.; Liang, R.; Sarojini, H.; Wang, E. Increased expression of miR-34a and miR-93 in rat liver during aging, and their impact on the expression of Mgst1 and Sirt1. Mech. Ageing Dev. 2011, 132, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Bates, D.J.; Li, N.; Liang, R.; Sarojini, H.; An, J.; Masternak, M.M.; Bartke, A.; Wang, E. MicroRNA regulation in Ames dwarf mouse liver may contribute to delayed aging. Aging Cell 2010, 9, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Young, K.; Eudy, E.; Bell, R.; Loberg, M.A.; Stearns, T.; Sharma, D.; Velten, L.; Haas, S.; Filippi, M.D.; Trowbridge, J.J. Decline in IGF1 in the bone marrow microenvironment initiates hematopoietic stem cell aging. Cell Stem Cell 2021, 28, 1473–1482.e1477. [Google Scholar] [CrossRef]
- Marino, G.; Ugalde, A.P.; Fernandez, A.F.; Osorio, F.G.; Fueyo, A.; Freije, J.M.; Lopez-Otin, C. Insulin-like growth factor 1 treatment extends longevity in a mouse model of human premature aging by restoring somatotroph axis function. Proc. Natl. Acad. Sci. USA 2010, 107, 16268–16273. [Google Scholar] [CrossRef]
- Hou, J.; Lin, L.; Zhou, W.; Wang, Z.; Ding, G.; Dong, Q.; Qin, L.; Wu, X.; Zheng, Y.; Yang, Y.; et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell 2011, 19, 232–243. [Google Scholar] [CrossRef] [PubMed]
- Kutay, H.; Bai, S.; Datta, J.; Motiwala, T.; Pogribny, I.; Frankel, W.; Jacob, S.T.; Ghoshal, K. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J. Cell Biochem. 2006, 99, 671–678. [Google Scholar] [CrossRef] [PubMed]
- Gramantieri, L.; Ferracin, M.; Fornari, F.; Veronese, A.; Sabbioni, S.; Liu, C.G.; Calin, G.A.; Giovannini, C.; Ferrazzi, E.; Grazi, G.L.; et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007, 67, 6092–6099. [Google Scholar] [CrossRef] [PubMed]
- Coulouarn, C.; Factor, V.M.; Andersen, J.B.; Durkin, M.E.; Thorgeirsson, S.S. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene 2009, 28, 3526–3536. [Google Scholar] [CrossRef] [PubMed]
- Murakami, Y.; Yasuda, T.; Saigo, K.; Urashima, T.; Toyoda, H.; Okanoue, T.; Shimotohno, K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene 2006, 25, 2537–2545. [Google Scholar] [CrossRef]
- Murakami, Y.; Tamori, A.; Itami, S.; Tanahashi, T.; Toyoda, H.; Tanaka, M.; Wu, W.; Brojigin, N.; Kaneoka, Y.; Maeda, A.; et al. The expression level of miR-18b in hepatocellular carcinoma is associated with the grade of malignancy and prognosis. BMC Cancer 2013, 13, 99. [Google Scholar] [CrossRef] [PubMed]
- Sato, F.; Hatano, E.; Kitamura, K.; Myomoto, A.; Fujiwara, T.; Takizawa, S.; Tsuchiya, S.; Tsujimoto, G.; Uemoto, S.; Shimizu, K. MicroRNA profile predicts recurrence after resection in patients with hepatocellular carcinoma within the Milan Criteria. PLoS ONE 2011, 6, e16435. [Google Scholar] [CrossRef]
- Yoshida, H.; Shiratori, Y.; Moriyama, M.; Arakawa, Y.; Ide, T.; Sata, M.; Inoue, O.; Yano, M.; Tanaka, M.; Fujiyama, S.; et al. Interferon therapy reduces the risk for hepatocellular carcinoma: National surveillance program of cirrhotic and noncirrhotic patients with chronic hepatitis C in Japan. IHIT Study Group. Inhibition of Hepatocarcinogenesis by Interferon Therapy. Ann. Intern. Med. 1999, 131, 174–181. [Google Scholar] [CrossRef]
- Tateishi, R.; Shiina, S.; Yoshida, H.; Teratani, T.; Obi, S.; Yamashiki, N.; Yoshida, H.; Akamatsu, M.; Kawabe, T.; Omata, M. Prediction of recurrence of hepatocellular carcinoma after curative ablation using three tumor markers. Hepatology 2006, 44, 1518–1527. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Abak, A.; Talebi, S.F.; Shoorei, H.; Branicki, W.; Taheri, M.; Akbari Dilmaghani, N. Role of miRNA and lncRNAs in organ fibrosis and aging. Biomed. Pharmacother. 2021, 143, 112132. [Google Scholar] [CrossRef]
- Li, L.; Ji, Y.; Chen, Y.C.; Zhen, Z.J. MiR-325-3p mediate the CXCL17/CXCR8 axis to regulate angiogenesis in hepatocellular carcinoma. Cytokine 2021, 141, 155436. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Bu, X.; Kan, A.; Luo, L.; Xu, Y.; Chen, H.; Lin, X.; Lai, Z.; Wen, D.; Huang, L.; et al. SP1-induced lncRNA DUBR promotes stemness and oxaliplatin resistance of hepatocellular carcinoma via E2F1-CIP2A feedback. Cancer Lett. 2022, 528, 16–30. [Google Scholar] [CrossRef] [PubMed]
- Nomura, K.; Kitanaka, A.; Iwama, H.; Tani, J.; Nomura, T.; Nakahara, M.; Ohura, K.; Tadokoro, T.; Fujita, K.; Mimura, S.; et al. Association between microRNA-527 and glypican-3 in hepatocellular carcinoma. Oncol. Lett. 2021, 21, 229. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Kaur, R.; Kanthaje, S.; Dhiman, R.K.; Chakraborti, A. Bacterial metabolite butyrate in modulating sorafenib-targeted microRNAs to curtail its resistance in hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2022, 149, 5823–5839. [Google Scholar] [CrossRef] [PubMed]
- Zanetti-Dallenbach, R.; Vuaroqueaux, V.; Wight, E.; Labuhn, M.; Singer, G.; Urban, P.; Eppenberger, U.; Holzgreve, W.; Eppenberger-Castori, S. Comparison of gene expression profiles in core biopsies and corresponding surgical breast cancer samples. Breast Cancer Res. 2006, 8, R51. [Google Scholar] [CrossRef] [PubMed]
- Berghuis, A.M.S.; van Deurzen, C.H.M.; Koffijberg, H.; Terstappen, L.; Sleijfer, S.; IJzerman, I.J. Real-world data on discordance between estrogen, progesterone, and HER2 receptor expression on diagnostic tumor biopsy versus tumor resection material. Breast Cancer Res. Treat. 2019, 175, 451–458. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]

| Group According to Age (Years) | Unit | <50 | 50s | 60s | 70s |
|---|---|---|---|---|---|
| histology (A) | 1.20 ± 0.51 | 1.54 ± 0.64 | 1.59 ± 0.72 | 1.70 ± 0.61 | |
| histology (F) | 1.10 ± 0.58 | 1.45 ± 0.74 | 1.60 ± 0.84 | 1.67 ± 0.68 | |
| HCVRNA | log copies/mL | 6.15 ± 1.03 | 6.07 ± 0.81 | 6.00 ± 0.90 | 5.97 ± 0.78 |
| AST | IU/L | 48.02 ± 46.30 | 57.14 ± 39.59 | 50.75 ± 32.06 | 63.07 ± 44.93 |
| ALT | IU/L | 68.3 ± 73.50 | 71.58 ± 61.95 | 52.91 ± 37.04 | 44.92 ± 75.30 |
| WBC | μL | 6000 ± 1720 | 5440 ± 1680 | 5110 ± 1250 | 4890 ± 1190 |
| PLT | x10^4/μL | 23.48 ± 5.80 | 19.25 ± 6.42 | 16.82 ± 5.19 | 17.35 ± 4.92 |
| T_BIL | mg/dL | 0.69 ± 0.32 | 0.73 ± 0.33 | 0.70 ± 0.26 | 0.63 ± 0.18 |
| ALP | IU/L | 231.41 ± 97.09 | 273.54 ± 110.06 | 301.44 ± 131.39 | 289.00 ± 90.63 |
| GGTP | U/L | 75.68 ± 122.54 | 60.18 ± 60.20 | 50.66 ± 64.45 | 61.07 ± 62.70 |
| Hb | g/dL | 14.08 ± 1.83 | 13.95 ± 1.35 | 13.74 ± 1.33 | 13.82 ± 1.06 |
| ALB | g/dL | 4.41 ± 0.34 | 4.28 ± 0.33 | 4.16 ± 0.35 | 4.25 ± 0.32 |
| AFP | ng/mL | 3.06 ± 7.41 | 4.41 ± 6.06 | 5.75 ± 8.90 | 6.40 ± 7.75 |
| Summary of clinical data by disease and age group (HCC and SNT group) | |||||
| Group according to age (years) | unit | 50s | 60s | 70s | |
| Stage | 1.50 ± 0.71 | 1.82 ± 0.40 | 2.11 ± 0.75 | ||
| AFP | ng/ml | 26.30 ± 20.02 | 129.38 ± 199.08 | 37.77 ± 83.58 | |
| DCP | U/ml | 6940 ± 12,000 | 398.1 ± 990.0 | 3670 ± 13,100 | |
| differentiation (T) | 2.00 ± 0.00 | 1.83 ± 0.58 | 1.64 ± 0.49 | ||
| fibrosis (N) | 3.33 ± 0.58 | 3.38 ± 0.52 | 3.36 ± 0.49 | ||
| Age Group (Years) | Comparison | miRNA Function | |
|---|---|---|---|
| tumor suppressor | oncogene | ||
| 50s | CH vs. SNT | hsa-miR-1180-5p | hsa-miR-296-3p |
| hsa-miR-135a-3p | hsa-miR-671-5p | ||
| hsa-miR-298 | |||
| hsa-miR-4516 | |||
| hsa-miR-4741 | |||
| hsa-miR-486-3p | |||
| hsa-miR-508-5p | |||
| hsa-miR-6071 | |||
| hsa-miR-642a-3p | |||
| hsa-miR-6775-3p | |||
| hsa-miR-8063 | |||
| 50s_60s | CH vs. SNT | hsa-miR-520d-5p | hsa-miR-196b-5p |
| 50s_60s_70s | CH vs. SNT | hsa-miR-325 | hsa-miR-3064-5p |
| hsa-miR-4521 | |||
| 60s | CH vs. SNT | hsa-miR-1248 | hsa-miR-3907 |
| hsa-miR-378a-5p | hsa-miR-421 | ||
| hsa-miR-495-3p | hsa-miR-7641 | ||
| hsa-miR-5096 | |||
| hsa-miR-605-5p | |||
| hsa-miR-619-5p | |||
| hsa-miR-664b-5p | |||
| 60s_70s | CH vs. SNT | hsa-miR-527, hsa-miR-518a-5p | hsa-miR-21-3p |
| 70s | SNT vs. HCC | hsa-miR-217 | hsa-miR-1269b |
| hsa-miR-449a | hsa-miR-216b-5p | ||
| hsa-miR-130a-3p | hsa-miR-10a-5p | ||
| hsa-miR-139-5p | hsa-miR-141-3p | ||
| hsa-miR-142-5p | hsa-miR-146a-5p | ||
| hsa-miR-144-3p | hsa-miR-376c-3p | ||
| hsa-miR-145-5p | |||
| hsa-miR-150-5p | |||
| hsa-miR-199a-3p, hsa-miR-199b-3p | |||
| hsa-miR-199a-5p | |||
| hsa-miR-200a-3p | |||
| hsa-miR-200b-3p | |||
| hsa-miR-200c-3p | |||
| hsa-miR-214-3p | |||
| hsa-miR-214-5p | |||
| hsa-miR-325 | |||
| hsa-miR-375 | |||
| hsa-miR-377-3p | |||
| hsa-miR-424-5p | |||
| hsa-miR-429 | |||
| hsa-miR-450a-5p | |||
| hsa-miR-451a |
| Age (Years) | CH | HCC | SNT | Subtotal |
|---|---|---|---|---|
| 50> | 93 | 0 | 0 | 93 |
| 51–60 | 115 | 3 | 3 | 118 |
| 61–70 | 134 | 12 | 12 | 146 |
| 71–80 | 18 | 28 | 28 | 46 |
| Total | 360 | 43 | 43 | 403 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Umezu, T.; Mori, T.; Toyoda, H.; Kanekura, K.; Tamori, A.; Ochiya, T.; Kuroda, M.; Akutsu, T.; Murakami, Y. Analysis of Carcinogenic Involvement of MicroRNA Pattern in Peripheral Non-Cancerous Tissues and Chronic Viral Liver Injury. Int. J. Mol. Sci. 2024, 25, 7858. https://doi.org/10.3390/ijms25147858
Umezu T, Mori T, Toyoda H, Kanekura K, Tamori A, Ochiya T, Kuroda M, Akutsu T, Murakami Y. Analysis of Carcinogenic Involvement of MicroRNA Pattern in Peripheral Non-Cancerous Tissues and Chronic Viral Liver Injury. International Journal of Molecular Sciences. 2024; 25(14):7858. https://doi.org/10.3390/ijms25147858
Chicago/Turabian StyleUmezu, Tomohiro, Tomoya Mori, Hidenori Toyoda, Kohsuke Kanekura, Akihiro Tamori, Takahiro Ochiya, Masahiko Kuroda, Tatsuya Akutsu, and Yoshiki Murakami. 2024. "Analysis of Carcinogenic Involvement of MicroRNA Pattern in Peripheral Non-Cancerous Tissues and Chronic Viral Liver Injury" International Journal of Molecular Sciences 25, no. 14: 7858. https://doi.org/10.3390/ijms25147858
APA StyleUmezu, T., Mori, T., Toyoda, H., Kanekura, K., Tamori, A., Ochiya, T., Kuroda, M., Akutsu, T., & Murakami, Y. (2024). Analysis of Carcinogenic Involvement of MicroRNA Pattern in Peripheral Non-Cancerous Tissues and Chronic Viral Liver Injury. International Journal of Molecular Sciences, 25(14), 7858. https://doi.org/10.3390/ijms25147858







