The Effect of Broccoli Glucosinolates Hydrolysis Products on Botrytis cinerea: A Potential New Antifungal Agent
Abstract
1. Introduction
2. Results
2.1. Identification of Glucosinolate Hydrolysis Products in Broccoli Inflorescences, Leaves, and Stems Using GC-MS
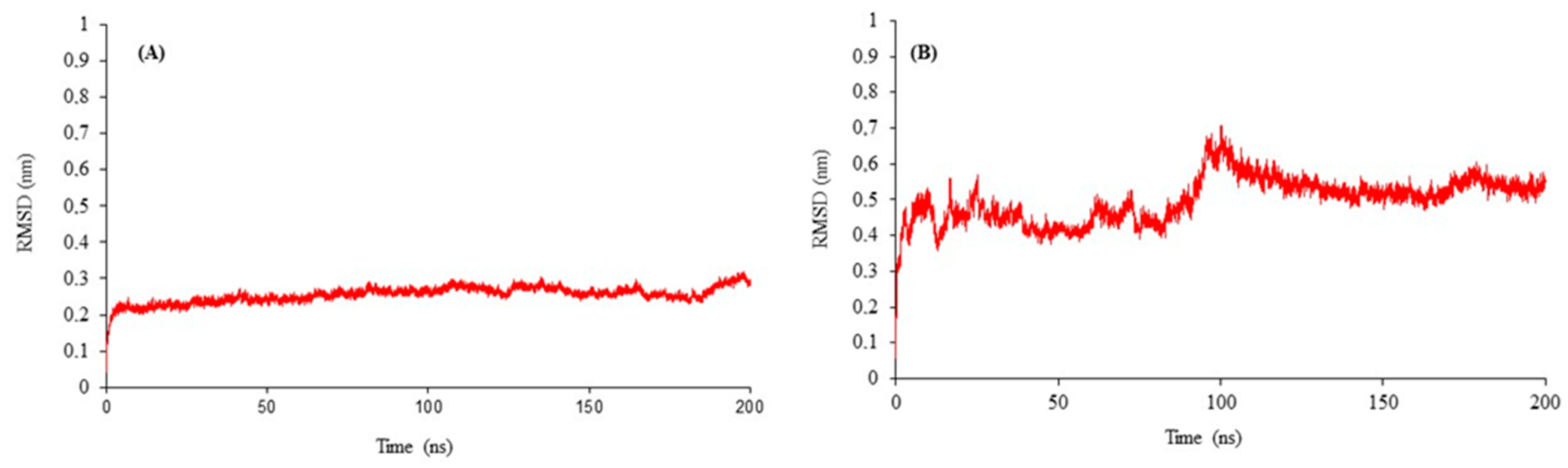
2.2. Structural Modeling and Refinement of B. cinerea CYP51 and GST Enzymes
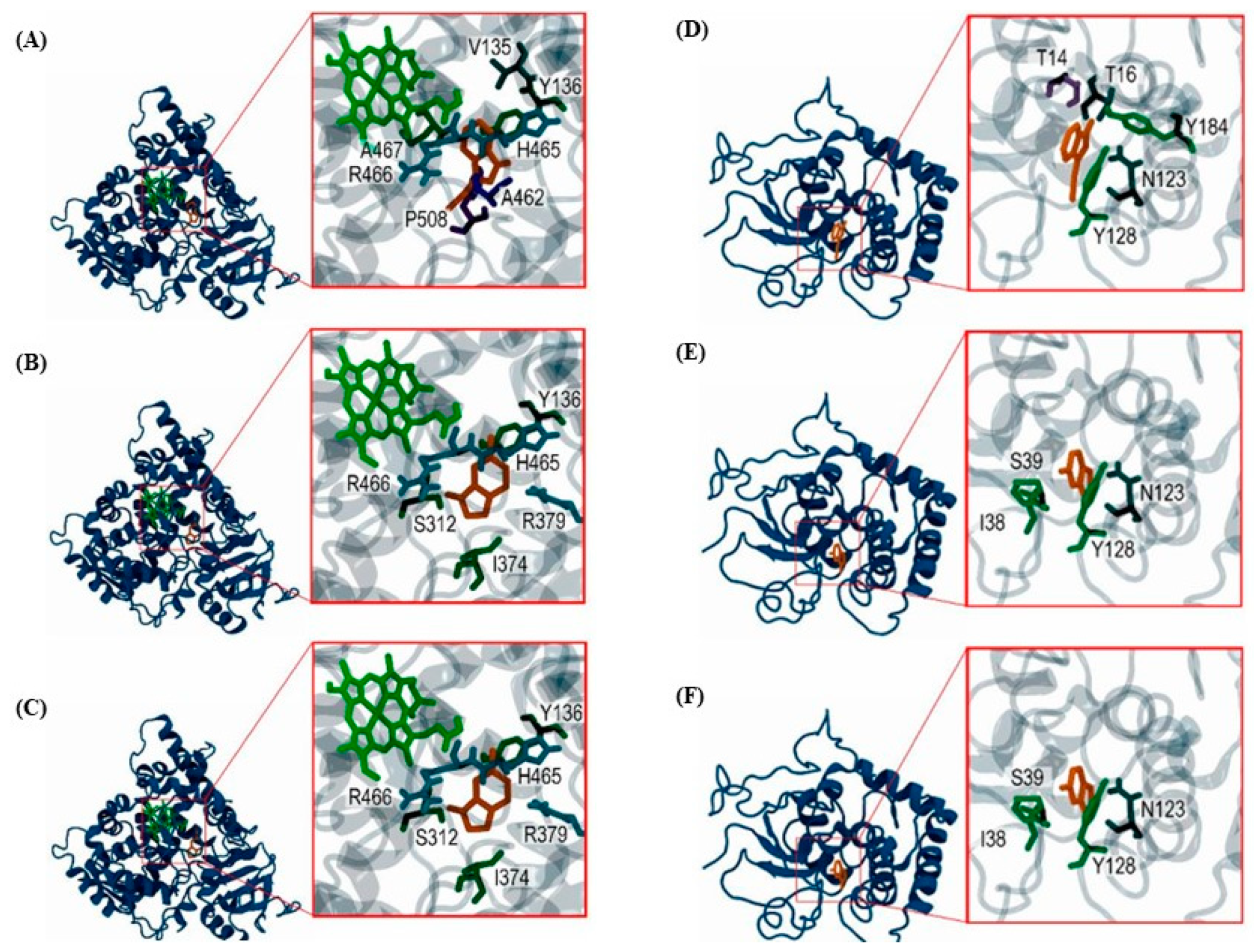
2.3. Molecular Interactions of GHPs with B. cinerea CYP51 and GST Enzymes
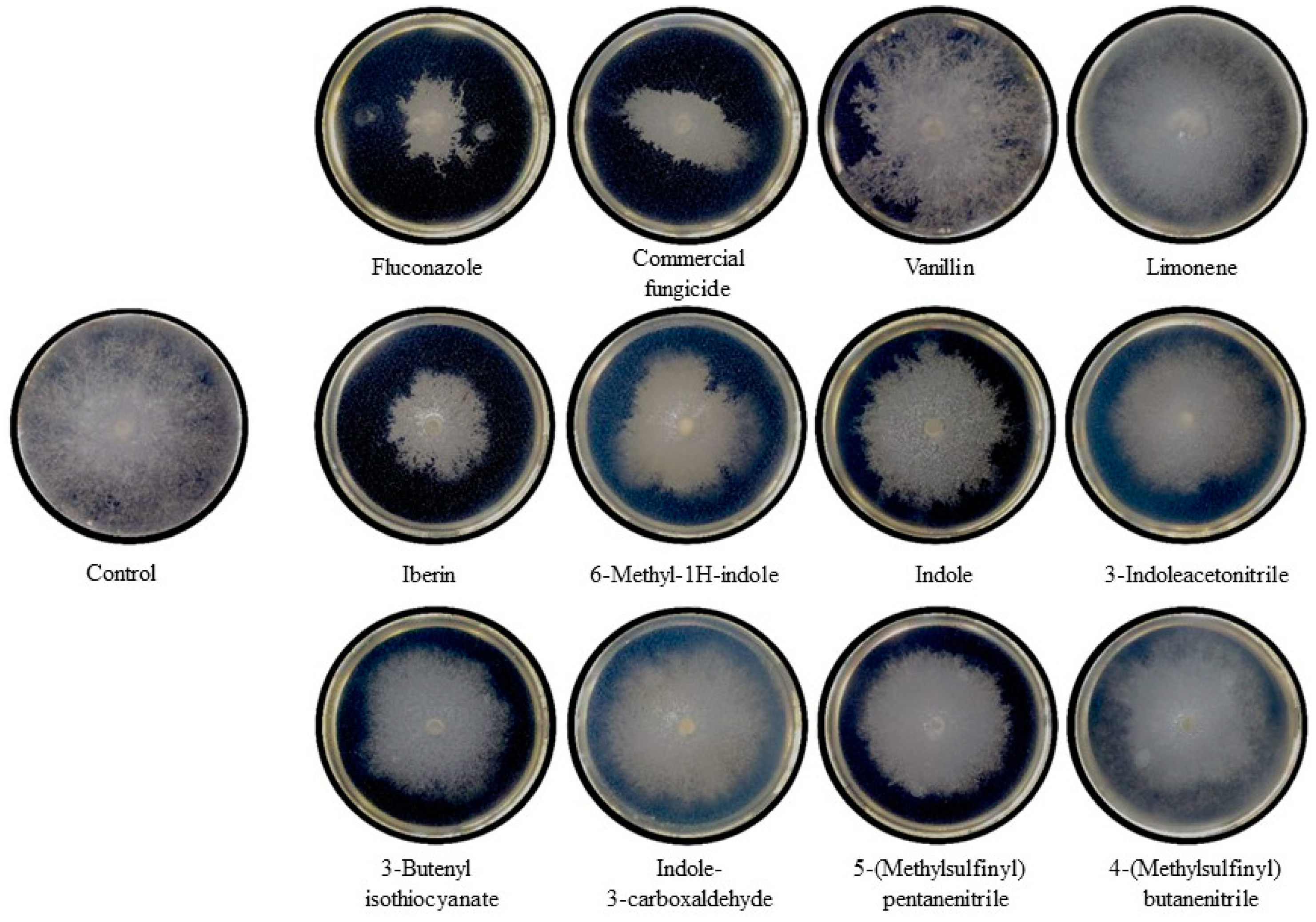
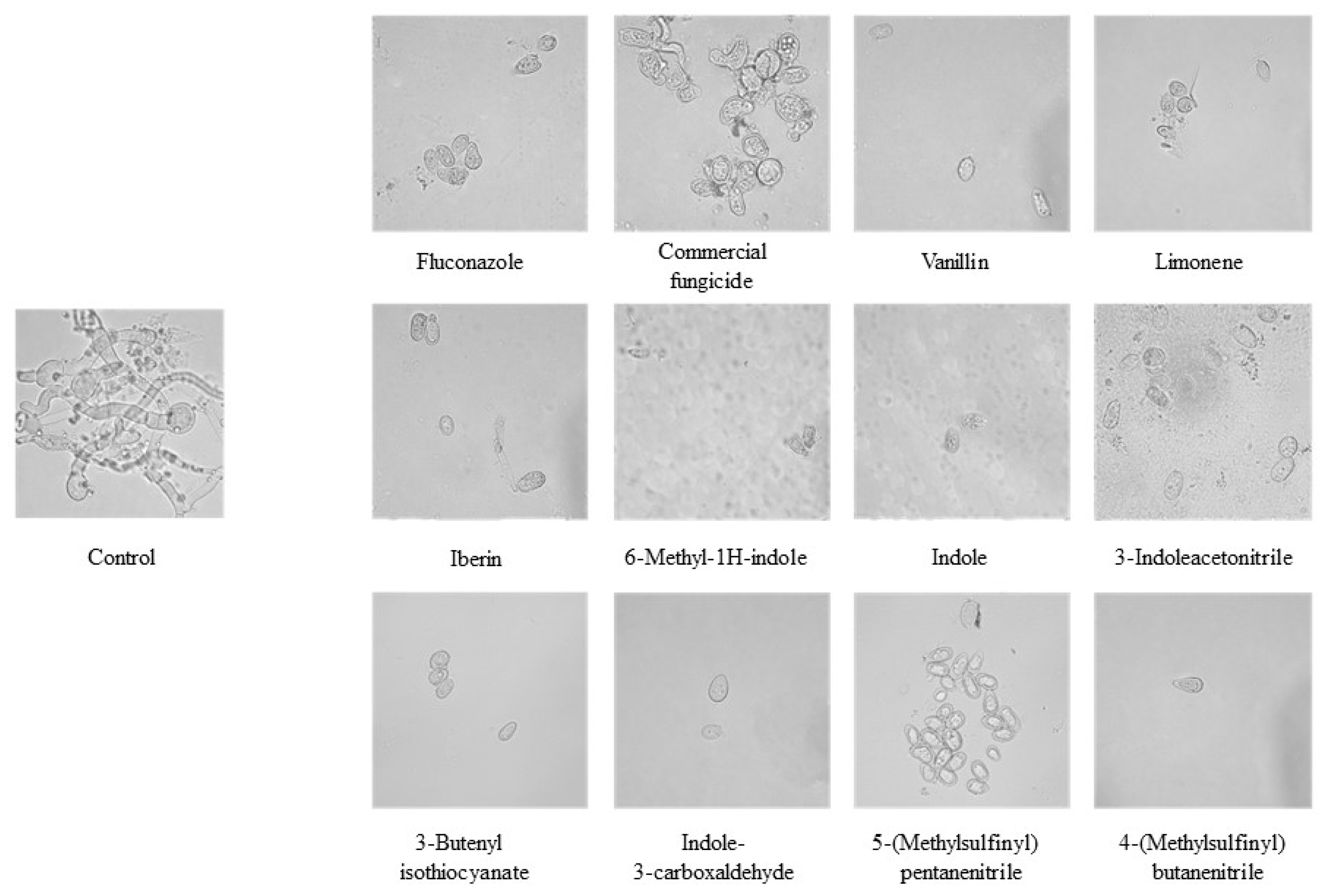
2.4. In Vitro Antifungal Activity of GHPs Against Botrytis cinerea: Mycelial Growth and Conidial Germination Inhibition
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Preparation, Extraction and Analysis of Glucosinolate Hydrolysis Products (GHPs)
4.3. In Silico Analysis of GHP Interactions with B. cinerea Enzymes
4.3.1. Protein Structure Retrieval and Dynamic Modeling of CYP51 and GST
4.3.2. Molecular Docking of GHPs with B. cinerea CYP51 and GST
4.4. In Vitro Analysis of GHPs against B. cinerea
4.4.1. Preparation of GHPs and Cultivation of Botrytis cinerea
4.4.2. Evaluation of the Antifungal Activity of GHPs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jaffar, N.S.; Jawan, R.; Chong, K.P. The potential of lactic acid bacteria in mediating the control of plant diseases and plant growth stimulation in crop production—A mini review. Front. Plant Sci. 2023, 13, 1047945. [Google Scholar] [CrossRef]
- Xu, X.; Chen, Y.; Li, B.; Zhang, Z.; Qin, G.; Chen, T.; Tian, S. Molecular mechanisms underlying multi-level defense responses of horticultural crops to fungal pathogens. Hortic. Res. 2022, 9, uhac066. [Google Scholar] [CrossRef] [PubMed]
- Matrose, N.A.; Obikeze, K.; Belay, Z.A.; Caleb, O.J. Plant extracts and other natural compounds as alternatives for post-harvest management of fruit fungal pathogens: A review. Food Biosci. 2021, 41, 100840. [Google Scholar] [CrossRef]
- Buxdorf, K.; Yaffe, H.; Barda, O.; Levy, M. The Effects of Glucosinolates and Their Breakdown Products on Necrotrophic Fungi. PLoS ONE 2013, 8, e70771. [Google Scholar] [CrossRef]
- Ribes, S.; Fuentes, A.; Talens, P.; Barat, J.M. Prevention of fungal spoilage in food products using natural compounds: A review. Crit. Rev. Food Sci. Nutr. 2018, 58, 2002–2016. [Google Scholar] [CrossRef] [PubMed]
- Ugolini, L.; Martini, C.; Lazzeri, L.; D’Avino, L.; Mari, M. Control of postharvest grey mould (Botrytis cinerea Per.: Fr.) on strawberries by glucosinolate-derived allyl-isothiocyanate treatments. Postharvest Biol. Technol. 2014, 90, 34–39. [Google Scholar] [CrossRef]
- Andriana, Y.; Fajriani, N.A.; Iwansyah, A.C.; Xuan, T.D. Phytochemical Constituents of Indonesian Adlay (Coix lacrima-jobi L.) and Their Potential as Antioxidants and Crop Protection Agents. Agrochemicals 2023, 2, 135–149. [Google Scholar] [CrossRef]
- Sciubba, F.; Chronopoulou, L.; Pizzichini, D.; Lionetti, V.; Fontana, C.; Aromolo, R.; Socciarelli, S.; Gambelli, L.; Bartolacci, B.; Finotti, E.; et al. Olive Mill Wastes: A Source of Bioactive Molecules for Plant Growth and Protection against Pathogens. Biology 2020, 9, 450. [Google Scholar] [CrossRef]
- Del Carmen Martínez-Ballesta, M.; Moreno, D.A.; Carvajal, M. The Physiological Importance of Glucosinolates on Plant Response to Abiotic Stress in Brassica. Int. J. Mol. Sci. 2013, 14, 11607–11625. [Google Scholar] [CrossRef]
- Thomas, M.; Badr, A.; Desjardins, Y.; Gosselin, A.; Angers, P. Characterization of industrial broccoli discards (Brassica oleracea var. italica) for their glucosinolate, polyphenol and flavonoid contents using UPLC MS/MS and spectrophotometric methods. Food Chem. 2018, 245, 1204–1211. [Google Scholar] [CrossRef]
- Lelario, F.; Bianco, G.; Bufo, S.A.; Cataldi, T.R.I. Establishing the occurrence of major and minor glucosinolates in Brassicaceae by LC–ESI-hybrid linear ion-trap and Fourier-transform ion cyclotron resonance mass spectrometry. Phytochemistry 2012, 73, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Sønderby, I.E.; Geu-Flores, F.; Halkier, B.A. Biosynthesis of glucosinolates--gene discovery and beyond. Trends Plant Sci. 2010, 15, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Ladak, Z.; Khairy, M.; Armstrong, E.A.; Yager, J.Y. Chapter 11—Glucosinolates: Paradoxically beneficial in fighting both brain cell death and cancer. In Nutraceuticals in Brain Health and Beyond; Ghosh, D., Ed.; Academic Press: Cambridge, MA, USA, 2021; pp. 155–167. [Google Scholar] [CrossRef]
- Bhat, R. Chapter 12—Glucosinolates. In Nutraceuticals and Health Care; Kour, J., Nayik, G.A., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 233–243. [Google Scholar] [CrossRef]
- Ahuja, I.; de Vos, R.C.H.; Bones, A.M.; Hall, R.D. Plant molecular stress responses face climate change. Trends Plant Sci. 2010, 15, 664–674. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Liang, Z.L.; Zang, Y.X.; Zhu, Z.J.; Yang, J. Diversity of glucosinolates among common Brassicaceae vegetables in China. Hortic. Plant J. 2023, 9, 365–380. [Google Scholar] [CrossRef]
- Latté, K.P.; Appel, K.E.; Lampen, A. Health benefits and possible risks of broccoli—An overview. Food Chem. Toxicol. 2011, 49, 3287–3309. [Google Scholar] [CrossRef]
- Piasecka, A.; Jedrzejczak-Rey, N.; Bednarek, P. Secondary metabolites in plant innate immunity: Conserved function of divergent chemicals. New Phytol. 2015, 206, 948–964. [Google Scholar] [CrossRef]
- Aghajanzadeh, T.A.; Watanabe, M.; Tohge, T.; Hawkesford, M.J.; Fernie, A.R.; Hoefgen, R.; Elzenga, J.T.M.; De Kok, L.J. Necrotrophic fungal infection affects indolic glucosinolate metabolism in Brassica rapa. Acta Physiol. Plant. 2023, 45, 64. [Google Scholar] [CrossRef]
- Madloo, P.; Lema, M.; Cartea, M.E.; Soengas, P. Sclerotinia sclerotiorum Response to Long Exposure to Glucosinolate Hydrolysis Products by Transcriptomic Approach. Microbiol. Spectr. 2021, 9, e0018021. [Google Scholar] [CrossRef] [PubMed]
- Ayaz, M.; Ullah, F.; Sadiq, A.; Ullah, F.; Ovais, M.; Ahmed, J.; Devkota, H.P. Synergistic interactions of phytochemicals with antimicrobial agents: Potential strategy to counteract drug resistance. Chem. Biol. Interact. 2019, 308, 294–303. [Google Scholar] [CrossRef]
- Angelino, D.; Dosz, E.B.; Sun, J.; Hoeflinger, J.L.; Van Tassell, M.L.; Chen, P.; Harnly, J.M.; Miller, M.J.; Jeffery, E.H. Myrosinase-dependent and –independent formation and control of isothiocyanate products of glucosinolate hydrolysis. Front. Plant Sci. 2015, 6, 831. [Google Scholar] [CrossRef]
- Román, J.; González, D.; Inostroza-Ponta, M.; Mahn, A. Molecular Modeling of Epithiospecifier and Nitrile-Specifier Proteins of Broccoli and Their Interaction with Aglycones. Molecules 2020, 25, 772. [Google Scholar] [CrossRef] [PubMed]
- Albertini, C.; Thebaud, G.; Fournier, E.; Leroux, P. Eburicol 14α-demethylase gene (CYP51) polymorphism and speciation in Botrytis cinerea. Mycol. Res. 2002, 106, 1171–1178. [Google Scholar] [CrossRef]
- Zhang, C.; Imran, M.; Liu, M.; Li, Z.; Gao, H.; Duan, H.; Zhou, S.; Liu, X. Two Point Mutations on CYP51 Combined With Induced Expression of the Target Gene Appeared to Mediate Pyrisoxazole Resistance in Botrytis cinerea. Front. Microbiol. 2020, 11, 1396. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Li, T.; Xiao, L.; Zhou, S.; Liu, X. Characterization of tebuconazole resistance in Botrytis cinerea from tomato plants in China. Phytopathol. Res. 2020, 2, 25. [Google Scholar] [CrossRef]
- Vanduchova, A.; Anzenbacher, P.; Anzenbacherova, E. Isothiocyanate from Broccoli, Sulforaphane, and Its Properties. J. Med. Food 2019, 22, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xia, Y.; Liu, H.Y.; Guo, H.; He, X.Q.; Liu, Y.; Wu, D.T.; Mai, Y.H.; Li, H.B.; Zou, L.; et al. Nutritional values, beneficial effects, and food applications of broccoli (Brassica oleracea var. italica Plenck). Trends Food Sci. Technol. 2022, 119, 288–308. [Google Scholar] [CrossRef]
- Li, Z.S.; Zheng, S.N.; Liu, Y.M.; Fang, Z.Y.; Yang, L.M.; Zhuang, M.; Zhang, Y.Y.; Lv, H.H.; Wang, Y.; Xu, D.H. Characterization of glucosinolates in 80 broccoli genotypes and different organs using UHPLC-Triple-TOF-MS method. Food Chem. 2021, 334, 127519. [Google Scholar] [CrossRef] [PubMed]
- Villano, D.; Fernandez-Pan, I.; Arozarena, I.; Ibanez, F.C.; Virseda, P.; Beriain, M.J. Revalorisation of broccoli crop surpluses and field residues: Novel ingredients for food industry uses. Eur. Food Res. Technol. 2023, 249, 3227–3237. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, L.; Ser, S.L.; Cumming, J.R.; Ku, K.M. Comparative Phytonutrient Analysis of Broccoli By-Products: The Potentials for Broccoli By-Product Utilization. Molecules 2018, 23, 900. [Google Scholar] [CrossRef]
- Gudino, I.; Martin, A.; Casquete, R.; Prieto, M.H.; Ayuso, M.C.; Cordoba, M.G. Evaluation of broccoli (Brassica oleracea var. italica) crop by-products as sources of bioactive compounds. Sci. Hortic. 2022, 304, 111284. [Google Scholar] [CrossRef]
- Silva, R.C.; Freitas, H.F.; Campos, J.M.; Kimani, N.M.; Silva, C.; Borges, R.S.; Pita, S.S.R.; Santos, C.B.R. Natural Products-Based Drug Design against SARS-CoV-2 Mpro 3CLpro. Int. J. Mol. Sci. 2021, 22, 11739. [Google Scholar] [CrossRef] [PubMed]
- Don, C.G.; Smieško, M. Microsecond MD simulations of human CYP2D6 wild-type and five allelic variants reveal mechanistic insights on the function. PLoS ONE 2018, 13, e0202534. [Google Scholar] [CrossRef] [PubMed]
- Moriwaki, Y.; Terada, T.; Tsumoto, K.; Shimizu, K. Rapid Heme Transfer Reactions between NEAr Transporter Domains of Staphylococcus aureus: A Theoretical Study Using QM/MM and MD Simulations. PLoS ONE 2015, 10, e0145125. [Google Scholar] [CrossRef] [PubMed]
- Kandeel, M.; Kitade, Y.; Al-Taher, A.; Al-Nazawi, M. The structural basis of unique substrate recognition by Plasmodium thymidylate kinase: Molecular dynamics simulation and inhibitory studies. PLoS ONE 2019, 14, e0212065. [Google Scholar] [CrossRef] [PubMed]
- Basnet, S.; Marahatha, R.; Shrestha, A.; Bhattarai, S.; Katuwal, S.; Sharma, K.R.; Marasini, B.P.; Dahal, S.R.; Basnyat, R.C.; Patching, S.G.; et al. In Vitro and In Silico Studies for the Identification of Potent Metabolites of Some High-Altitude Medicinal Plants from Nepal Inhibiting SARS-CoV-2 Spike Protein. Molecules 2022, 27, 8957. [Google Scholar] [CrossRef] [PubMed]
- Dixon, D.P.; Lapthorn, A.; Edwards, R. Plant glutathione transferases. Genome Biol. 2002, 3, Reviews3004. [Google Scholar] [CrossRef]
- Hargrove, T.Y.; Kim, K.; de Nazaré Correia Soeiro, M.; da Silva, C.F.; Batista, D.D.; Batista, M.M.; Yazlovitskaya, E.M.; Waterman, M.R.; Sulikowski, G.A.; Lepesheva, G.I. CYP51 structures and structure-based development of novel, pathogen-specific inhibitory scaffolds. Int. J. Parasitol. Drugs Drug Resist. 2012, 2, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, Y.; Prosper, P.; Buée, M.; Dumarçay, S.; Favier, F.; Gelhaye, E.; Gérardin, P.; Harvengt, L.; Jacquot, J.P.; Lamant, T.; et al. Characterization of a Phanerochaete chrysosporium glutathione transferase reveals a novel structural and functional class with ligandin properties. J. Biol. Chem. 2012, 287, 39001–39011. [Google Scholar] [CrossRef]
- Ming, Z.-H.; Xu, S.-Z.; Zhou, L.; Ding, M.-W.; Yang, J.-Y.; Yang, S.; Xiao, W.-J. Organocatalytic synthesis and sterol 14α-demethylase binding interactions of enantioriched 3-(1H-1,2,4-triazol-1-yl)butyl benzoates. Bioorganic Med. Chem. Lett. 2009, 19, 3938–3940. [Google Scholar] [CrossRef]
- Meux, E.; Morel, M.; Lamant, T.; Gérardin, P.; Jacquot, J.-P.; Dumarçay, S.; Gelhaye, E. New substrates and activity of Phanerochaete chrysosporium Omega glutathione transferases. Biochimie 2013, 95, 336–346. [Google Scholar] [CrossRef]
- Lepesheva, G.I.; Waterman, M.R. Sterol 14alpha-demethylase cytochrome P450 (CYP51), a P450 in all biological kingdoms. Biochim. Biophys. Acta 2007, 1770, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, L.; Lv, Q.; Yan, L.; Wang, Y.; Jiang, Y. The Fungal CYP51s: Their Functions, Structures, Related Drug Resistance, and Inhibitors. Front. Microbiol. 2019, 10, 691. [Google Scholar] [CrossRef]
- Reglinski, T.; Elmer, P.; Taylor, J.; Wood, P.; Hoyte, S. Inhibition of Botrytis cinerea growth and suppression of botrytis bunch rot in grapes using chitosan. Plant Pathol. 2010, 59, 882–890. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, L.; Chen, Y.; Yun, L.; Liu, S.; Li, Y. Stalk length affects the mineral distribution and floret quality of broccoli (Brassica oleracea L. var. italica) heads during storage. Postharvest Biol. Technol. 2018, 145, 166–171. [Google Scholar] [CrossRef]
- Salas-Millán, J.-Á.; Aznar, A.; Conesa, E.; Conesa-Bueno, A.; Aguayo, E. Functional food obtained from fermentation of broccoli by-products (stalk): Metagenomics profile and glucosinolate and phenolic compounds characterization by LC-ESI-QqQ-MS/MS. LWT 2022, 169, 113915. [Google Scholar] [CrossRef]
- Jeschke, V.; Gershenzon, J.; Vassao, D.G. Insect Detoxification of Glucosinolates and Their Hydrolysis Products. In Glucosinolates; Kopriva, S., Ed.; Academic Press: Cambridge, MA, USA, 2016; Volume 80, pp. 199–245. [Google Scholar]
- Hopkins, R.J.; van Dam, N.M.; van Loon, J.J. Role of glucosinolates in insect-plant relationships and multitrophic interactions. Annu. Rev. Entomol. 2009, 54, 57–83. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef]
- Çevik, U.A.; Celik, I.; Işık, A.; Pillai, R.R.; Tallei, T.E.; Yadav, R.; Özkay, Y.; Kaplancıklı, Z.A. Synthesis, molecular modeling, quantum mechanical calculations and ADME estimation studies of benzimidazole-oxadiazole derivatives as potent antifungal agents. J. Mol. Struct. 2022, 1252, 132095. [Google Scholar] [CrossRef]
- Shi, N.; Zheng, Q.; Zhang, H. Molecular Dynamics Investigations of Binding Mechanism for Triazoles Inhibitors to CYP51. Front. Mol. Biosci. 2020, 7, 586540. [Google Scholar] [CrossRef]
- Gao, Q.; Lu, C.; Wang, X.W.; Zhang, J.W.; Song, Y.; Xue, Y.L. Molecular dynamics simulation and steered molecular dynamics simulation on irisin dimers. J. Mol. Model. 2018, 24, 95. [Google Scholar] [CrossRef]
- Omae, Y.; Saito, H.; Takagi, H.; Nishimura, M.; Iwayama, M.; Kawaguchi, K.; Nagao, H. Molecular Dynamics Study of Glutathione. In Quantum Systems in Chemistry and Physics; Springer: Dordrecht, The Netherlands, 2012; pp. 545–553. [Google Scholar]
- Stella, L.; Di Iorio, E.E.; Nicotra, M.; Ricci, G. Molecular dynamics simulations of human glutathione transferase P1-1: Conformational fluctuations of the apo-structure. Proteins 1999, 37, 10–19. [Google Scholar] [CrossRef]
- de Souza, T.L.F.; Sanches, D.; Gonçalves, R.B.; da RochaPita, S.S.; Pascutti, P.G.; Bianconi, M.L.; de Almeida, F.C.L.; Silva, J.L.; de Oliveira, A.C. Conformational selection, dynamic restriction and the hydrophobic effect coupled to stabilization of the BIR3 domain of the human X-linked inhibitor of apoptosis protein by the tetrapeptide AVPI. Biophys. Chem. 2010, 152, 99–108. [Google Scholar] [CrossRef]
- Da Rocha Pita, S.S.; Batista, P.R.; Albuquerque, M.G.; Pascutti, P.G. Molecular Dynamics Simulations of Peptide Inhibitors Complexed With Trypanosoma cruzi Trypanothione Reductase. Chem. Biol. Drug Des. 2012, 80, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Román, J.; Castillo, A.; Cottet, L.; Mahn, A. Kinetic and structural study of broccoli myrosinase and its interaction with different glucosinolates. Food Chem. 2018, 254, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Yuan, Q.P.; Dong, H.R.; Liu, Y.M. Determination of sulforaphane in broccoli and cabbage by high-performance liquid chromatography. J. Food Compos. Anal. 2006, 19, 473–476. [Google Scholar] [CrossRef]
- Kissen, R.; Bones, A.M. Nitrile-specifier proteins involved in glucosinolate hydrolysis in Arabidopsis thaliana. J. Biol. Chem. 2009, 284, 12057–12070. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Ku, K.M.; Becker, T.M.; Juvik, J.A. Chemopreventive glucosinolate accumulation in various broccoli and collard tissues: Microfluidic-based targeted transcriptomics for by-product valorization. PLoS ONE 2017, 12, e0185112. [Google Scholar] [CrossRef] [PubMed]
- UniProt-Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef]
- Lüthy, R.; Bowie, J.U.; Eisenberg, D. Assessment of protein models with three-dimensional profiles. Nature 1992, 356, 83–85. [Google Scholar] [CrossRef]
- Pontius, J.; Richelle, J.; Wodak, S.J. Deviations from standard atomic volumes as a quality measure for protein crystal structures. J. Mol. Biol. 1996, 264, 121–136. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef]
- Laskowski, R.A. PDBsum new things. Nucleic Acids Res. 2009, 37, D355–D359. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- GROMACS, d.t. GROMACS Documentation. Release 2020.3; Royal Institute of Technology: Stockholm, Sweden, 2020. [Google Scholar] [CrossRef]
- Chang, A.; Jeske, L.; Ulbrich, S.; Hofmann, J.; Koblitz, J.; Schomburg, I.; Neumann-Schaal, M.; Jahn, D.; Schomburg, D. BRENDA, the ELIXIR core data resource in 2021: New developments and updates. Nucleic Acids Res. 2021, 49, D498–D508. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 update. Nucleic Acids Res 2023, 51, D1373–D1380. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Schrödinger, L.; DeLano, W. PyMOL. 2020. Available online: http://www.pymol.org/pymol (accessed on 15 July 2020).
- Ahn, S.Y.; Kim, S.A.; Yun, H.K. Inhibition of Botrytis cinerea and accumulation of stilbene compounds by light-emitting diodes of grapevine leaves and differential expression of defense-related genes. Eur. J. Plant Pathol. 2015, 143, 753–765. [Google Scholar] [CrossRef]
- Jeong, M.-J.; Bae, D.; Bae, H.; Lee, S.I.; Kim, J.A.; Shin, S.C.; Park, S.H.; Park, S.-C. Inhibition of Botrytis cinerea spore germination and mycelia growth by frequency-specific sound. J. Korean Soc. Appl. Biol. Chem. 2013, 56, 377–382. [Google Scholar] [CrossRef]
- Barreto, J.A.R.; Anaguano, A.H. Evaluación del crecimiento y compatibilidad de hongos de la podredumbre blanca Evaluation of Growth and Compatibility of white Rot Fungi. Cienc. Desarro. 2015, 5, 197–205. [Google Scholar]
- Piermann, L.; Fujinawa, M.; Pontes, N.; Galvão, J.; Bettiol, W. Inhibition of mycelial growth, conidial germination, and Botrytis cinerea Pers.: Fr colonization in begonia with biocompatible products. Sci. Agric. 2023, 80, e20210062. [Google Scholar] [CrossRef]

| Peak | GHP Compound Name | Molecular Formula | Molecular Weight (g/mol) | tR. (min) | Peak Area (% of Total) | ||
|---|---|---|---|---|---|---|---|
| Inflorescences | Leaves | Steams | |||||
| 1 | 1-butene, 4-isothiocyanato- | C5H7NS | 113 | 9.29 | 4.03 | 1.19 | 4.92 |
| 2 | Indole | C8H7N | 117 | 13.22 | 9.23 | 8.37 | 5.5 |
| 3 | Iberin Nitrile | C5H9NOS | 131 | 13.98 | 4.39 | n.d. 1 | 3.56 |
| 4 | 5-methylindole | C9H9N | 131 | 14.01 | 4.29 | 5.42 | 4.88 |
| 5 | Sulforaphane nitrile | C6H11NOS | 145 | 15.04 | 33.8 | 9.14 | 46.88 |
| 6 | Iberin | C5H9NOS2 | 163 | 15.74 | 7.08 | 54.39 | 14.85 |
| 7 | 1H-Indole-3-acetonitrile | C10H8N2 | 156 | 16.96 | 22.96 | 17.04 | 12.85 |
| 8 | 1H-Indole-3-carboxaldehyde | C9H7NO | 145 | 17.02 | 14.22 | 4.45 | 6.56 |
| Ligand Type | Ligand Name | Affinity Energy (kcal/mol) | Interacting Residues | ||
|---|---|---|---|---|---|
| CYP51 | GST | CYP51 | GST | ||
| Substrates | Glutathione | - | −6.7 | N16; K19; R56; I57; F73; S75; G120; Q121 | |
| Obtusifoliol | −3.5 | - | E119; E120; D137; P139 | ||
| Inhibitors | Carbendazim | −6.7 | −6.6 | Y136; H311; S312; S313; C467; P508 | N16; K19; I57; Q119; G120; N123; W178; I181 |
| Diniconazole | −4.6 | −6.8 | Y122; V135; Y136; A308; S312; I374; R379; F460; G461; A462; H465; R466; C467; P508 | T12; N16; K19; I38; I57; G120; N123; W178 | |
| Fluconazole | −8.3 | −8.2 | Y122; Y136; S312; I374; P459; F460; A462; H465; R466; C467; P508 | N16; I38; R56; I57; G120; N123; P128 | |
| Limonene | −6.1 | −5.2 | Y122; Y136; S312; R379; A462; H465; P508 | I38; N123 | |
| Tebuconazole | −7.0 | −7.2 | Y122; L125; Y136; F234; I374; S376; F460; G461; A462; R466; L507; P508 | T14; K19; I38; R56; I57; P58; G120; N123; H124; W178 | |
| Triadimefon | −6.4 | −6.3 | V135; Y136; A308; S312; I374; I377; F460; G461; A462; R466; C467; P508 | T12; T14; I38; NT123; PT128 | |
| Vanillin | −5.8 | −5.3 | Y122; S312; R379; A462; P508 | T14; I38; Q121; Y128 | |
| GHPs | 3-Indoleacetonitrile | −6.4 | −6.4 | V135; Y136; A462; H465; R466; C467; P508 | T14; N16; N123; Y128; Y184 |
| 4-(Methylsulfinyl) butane nitrile | −4.3 | −4.2 | S312 | T12; I38; Y128 | |
| 6-Methyl-1H-indole | −5.5 | −5.3 | Y136; S312; I374; R379; HT65; R466 | I38; S39; N123; Y128 | |
| Iberin | −4.1 | −3.8 | Y122; F460 | I57; NT1T23; HT124 | |
| Indole | −5.7 | −5.2 | Y122; Y136; I374; S376; A462; L507; P508 | K19; G120; N123; W178 | |
| Indole-3-carboxaldehyde | −6.2 | −5.7 | Y136; S312; I374; F460; A462; P508 | I38; N123 | |
| 3-Butenyl isothiocyanate | −4.0 | −3.7 | Y122; R379; L507 | I38; G120; N123 | |
| 5-(Methylsulfinyl) pentanenitrile | −4.7 | −4.2 | Y122; S312; I374; F460 | T12; N123; Y128 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Román, J.; Lagos, A.; Mahn, A.; Quintero, J. The Effect of Broccoli Glucosinolates Hydrolysis Products on Botrytis cinerea: A Potential New Antifungal Agent. Int. J. Mol. Sci. 2024, 25, 7945. https://doi.org/10.3390/ijms25147945
Román J, Lagos A, Mahn A, Quintero J. The Effect of Broccoli Glucosinolates Hydrolysis Products on Botrytis cinerea: A Potential New Antifungal Agent. International Journal of Molecular Sciences. 2024; 25(14):7945. https://doi.org/10.3390/ijms25147945
Chicago/Turabian StyleRomán, Juan, Ailine Lagos, Andrea Mahn, and Julián Quintero. 2024. "The Effect of Broccoli Glucosinolates Hydrolysis Products on Botrytis cinerea: A Potential New Antifungal Agent" International Journal of Molecular Sciences 25, no. 14: 7945. https://doi.org/10.3390/ijms25147945
APA StyleRomán, J., Lagos, A., Mahn, A., & Quintero, J. (2024). The Effect of Broccoli Glucosinolates Hydrolysis Products on Botrytis cinerea: A Potential New Antifungal Agent. International Journal of Molecular Sciences, 25(14), 7945. https://doi.org/10.3390/ijms25147945






