Genome-Wide Identification and Interaction Analysis of Turbot Heat Shock Protein 40 and 70 Families Suggest the Mechanism of Chaperone Proteins Involved in Immune Response after Bacterial Infection
Abstract
:1. Introduction
2. Results
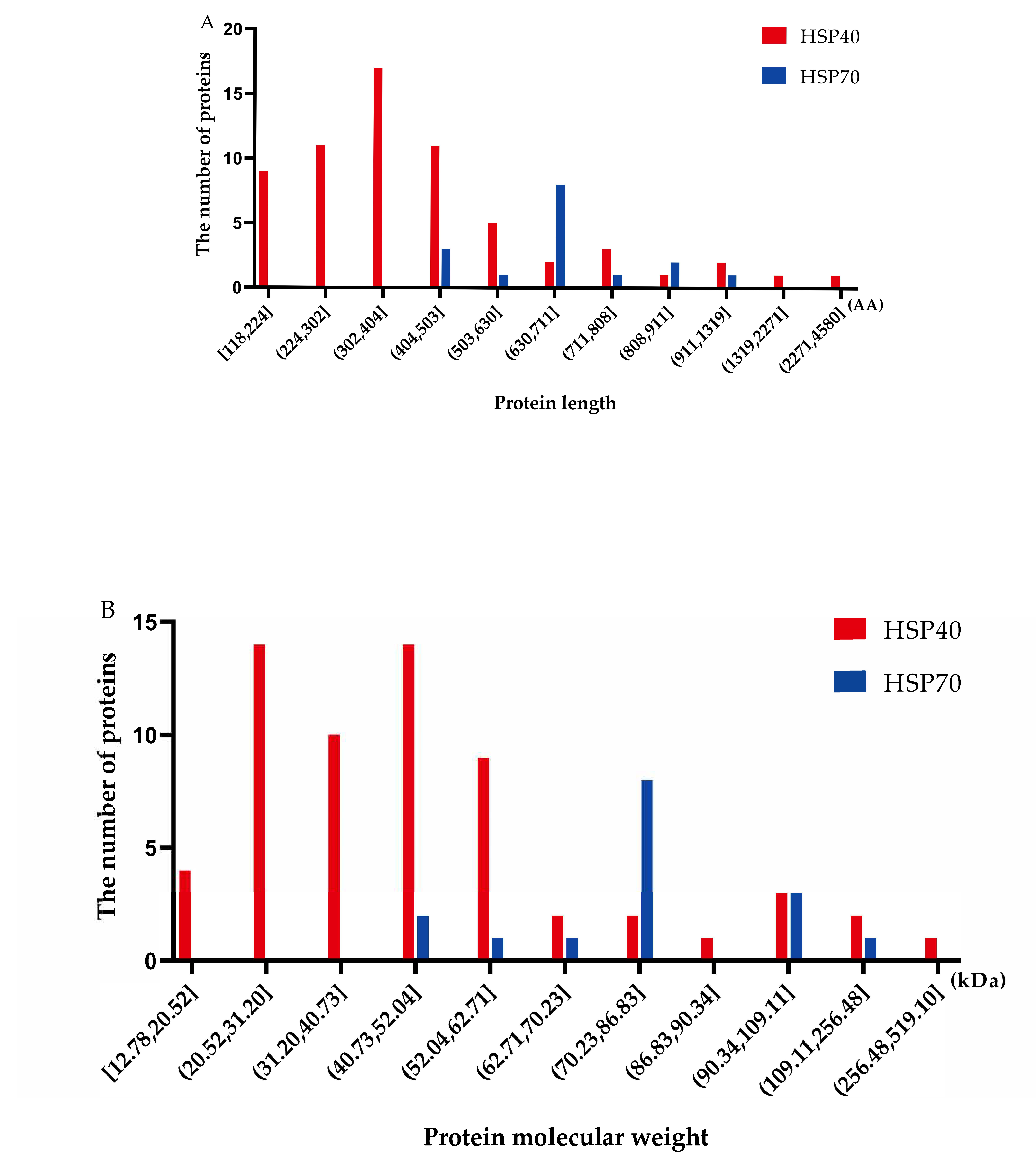
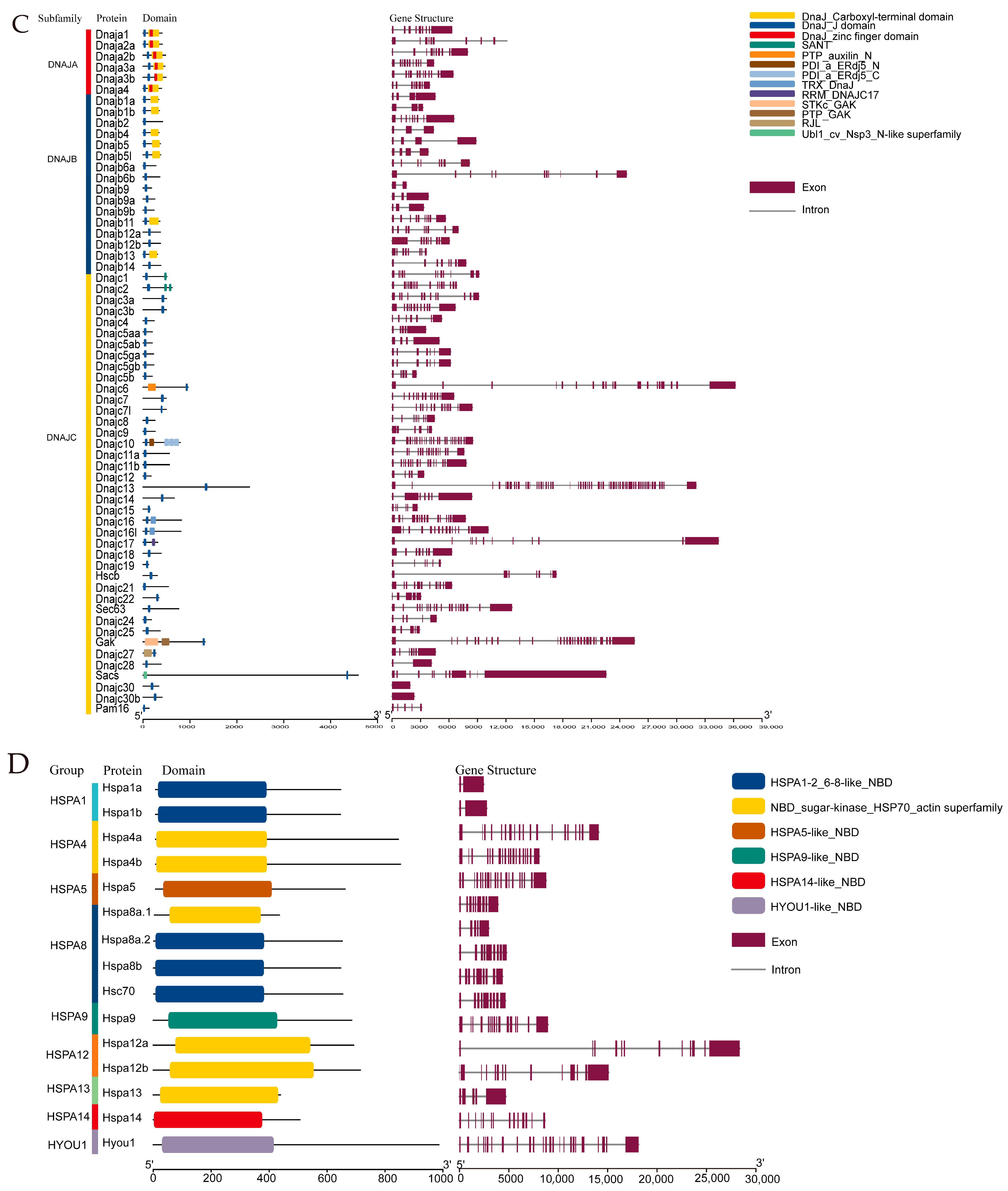
2.1. Identification of Hsp40 and Hsp70 Genes in the Turbot Genome
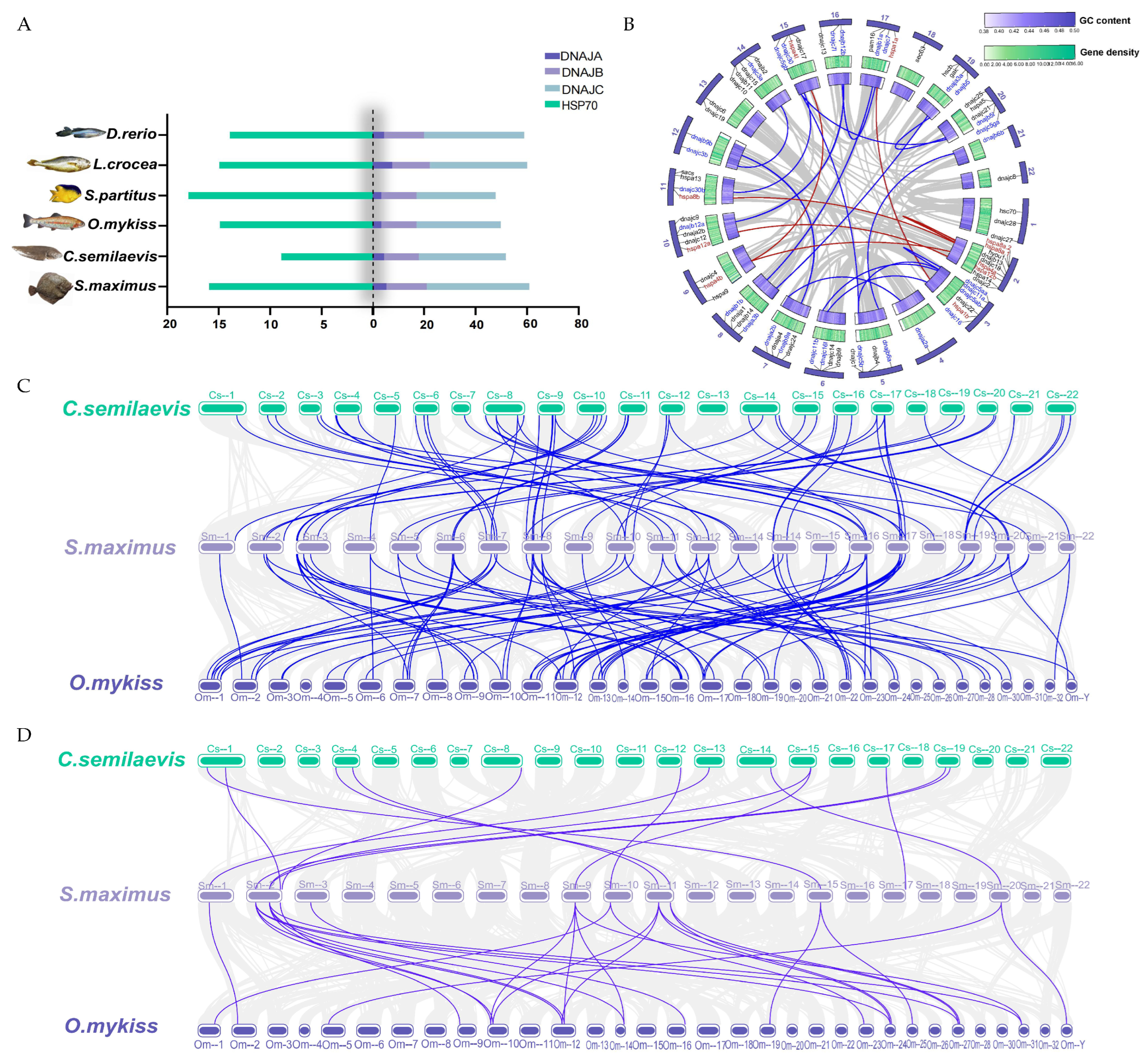
2.2. Chromosome Distribution Analysis
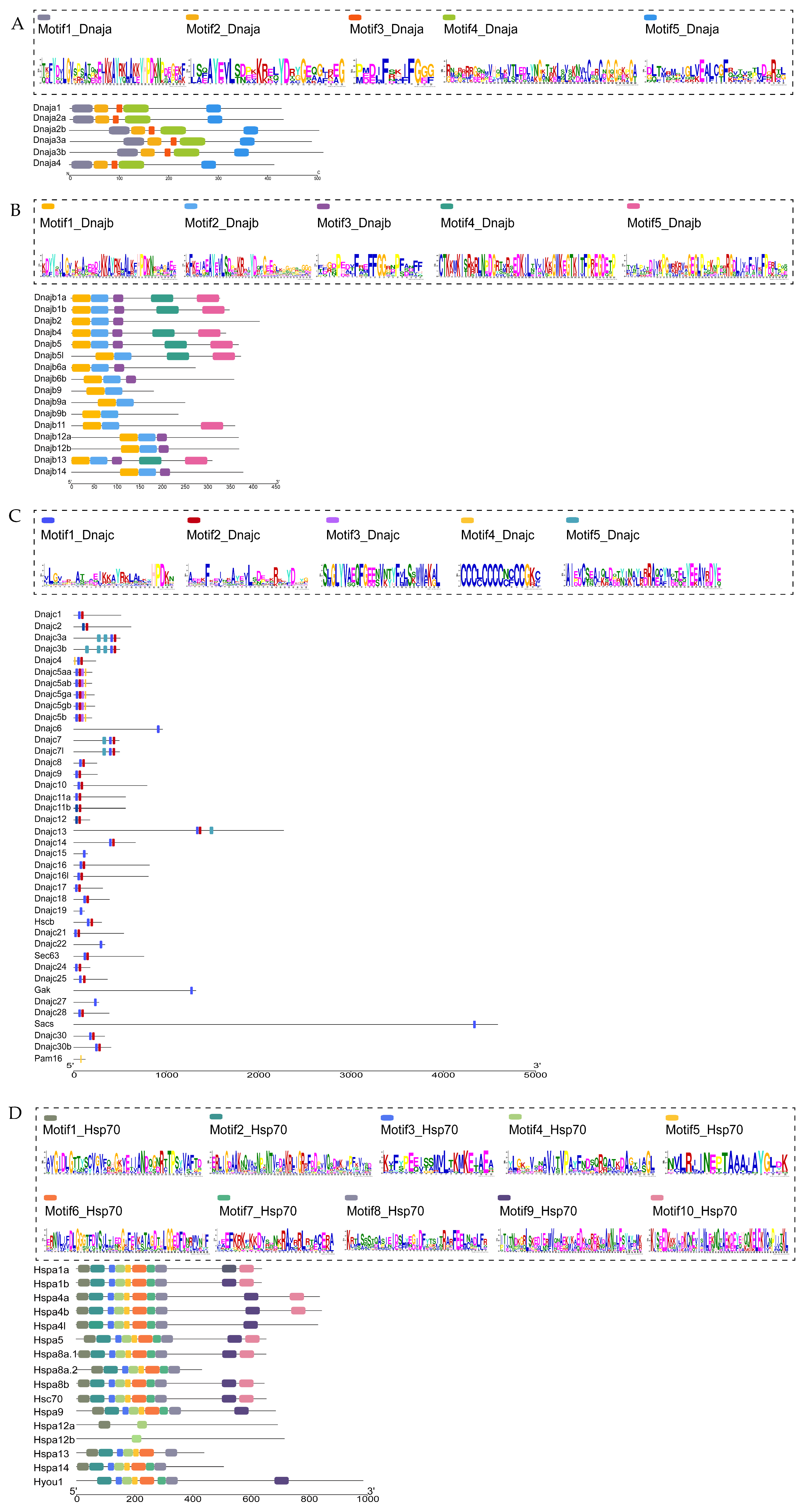
2.3. Motif Analysis of the Identified Hsp40 and Hsp70 Proteins
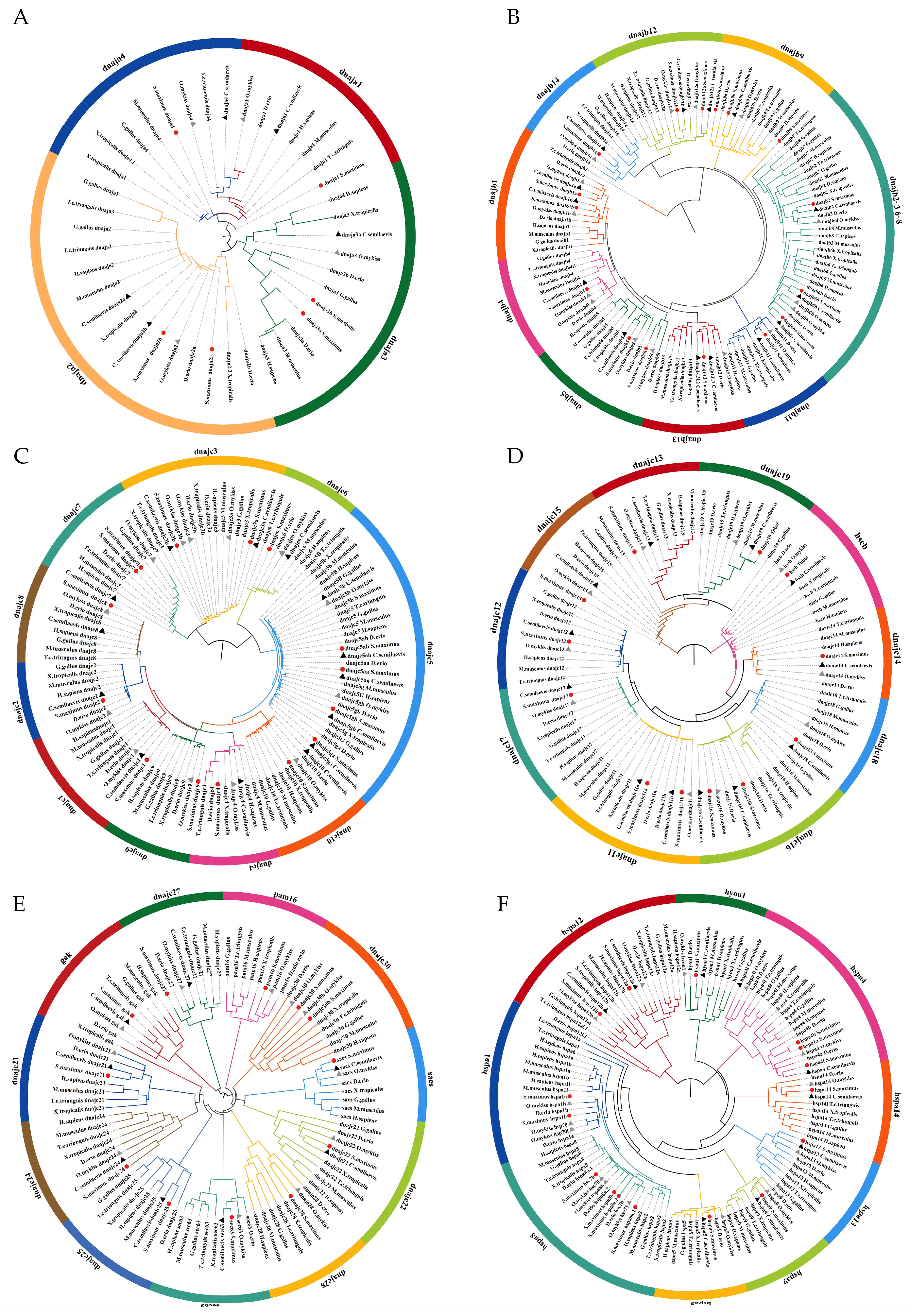
2.4. Phylogenetic Analysis
2.5. Synteny and Gene Duplication Analysis
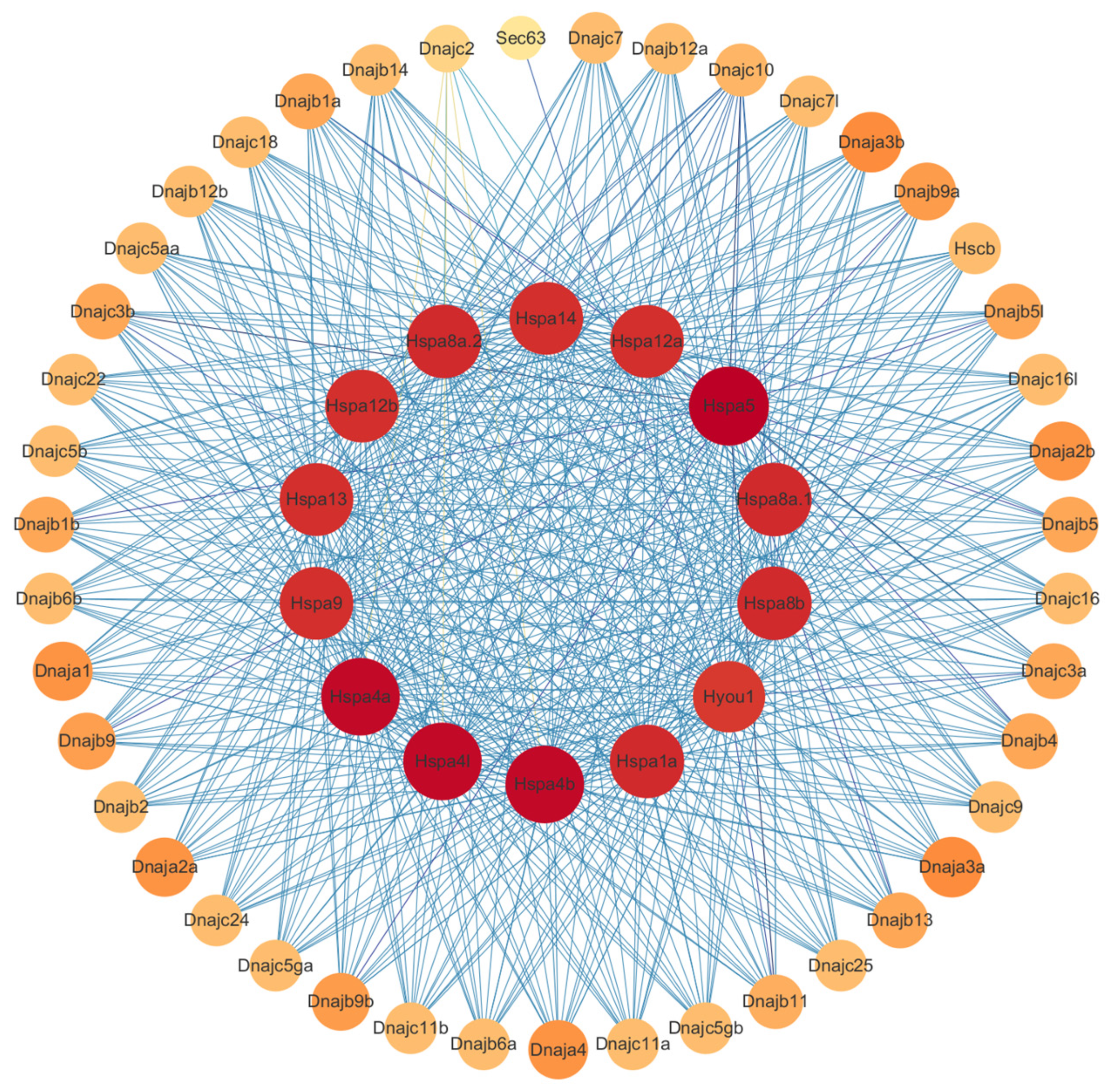
2.6. Protein-Protein Interaction Analysis
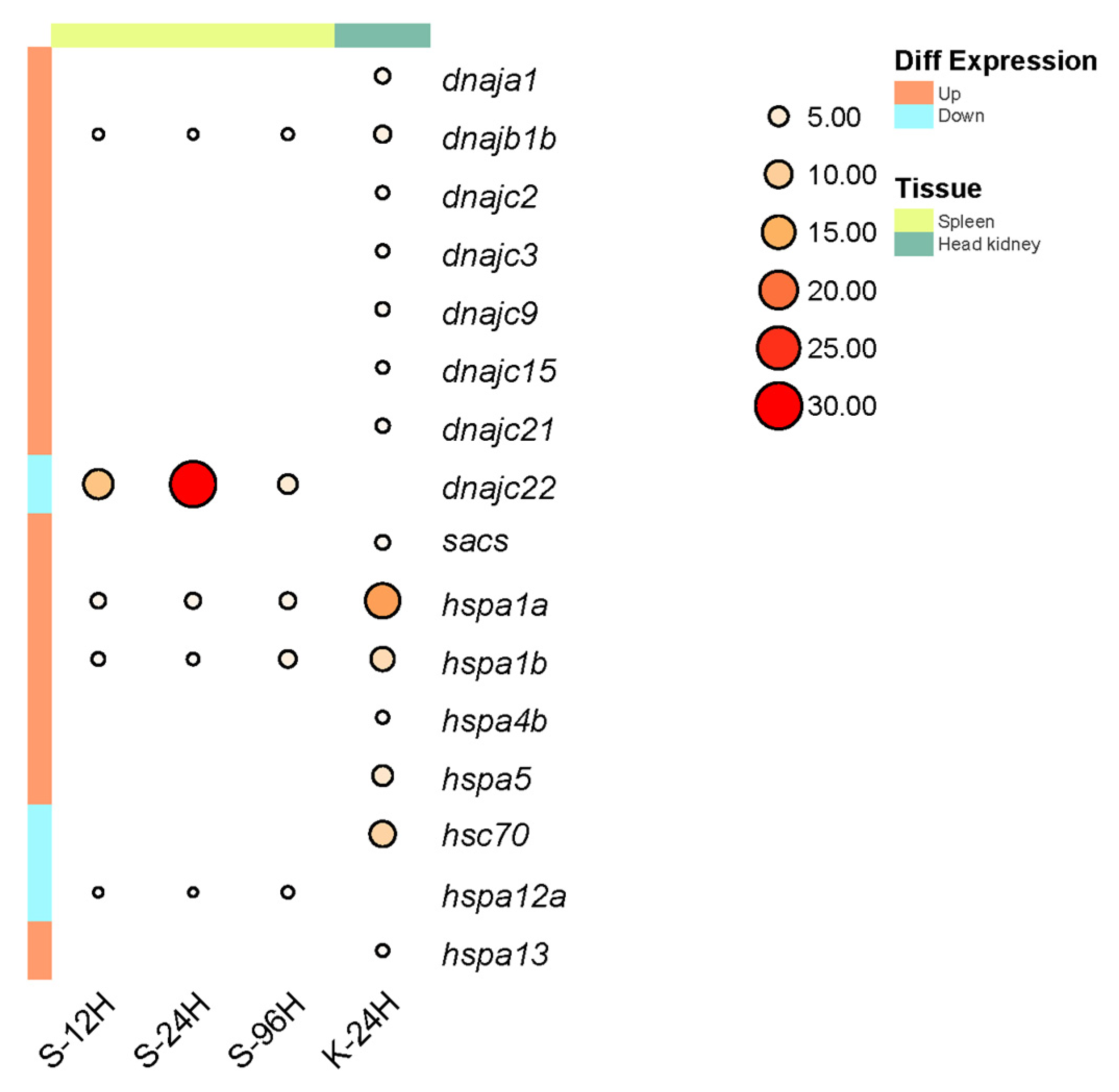
2.7. Expression Analysis
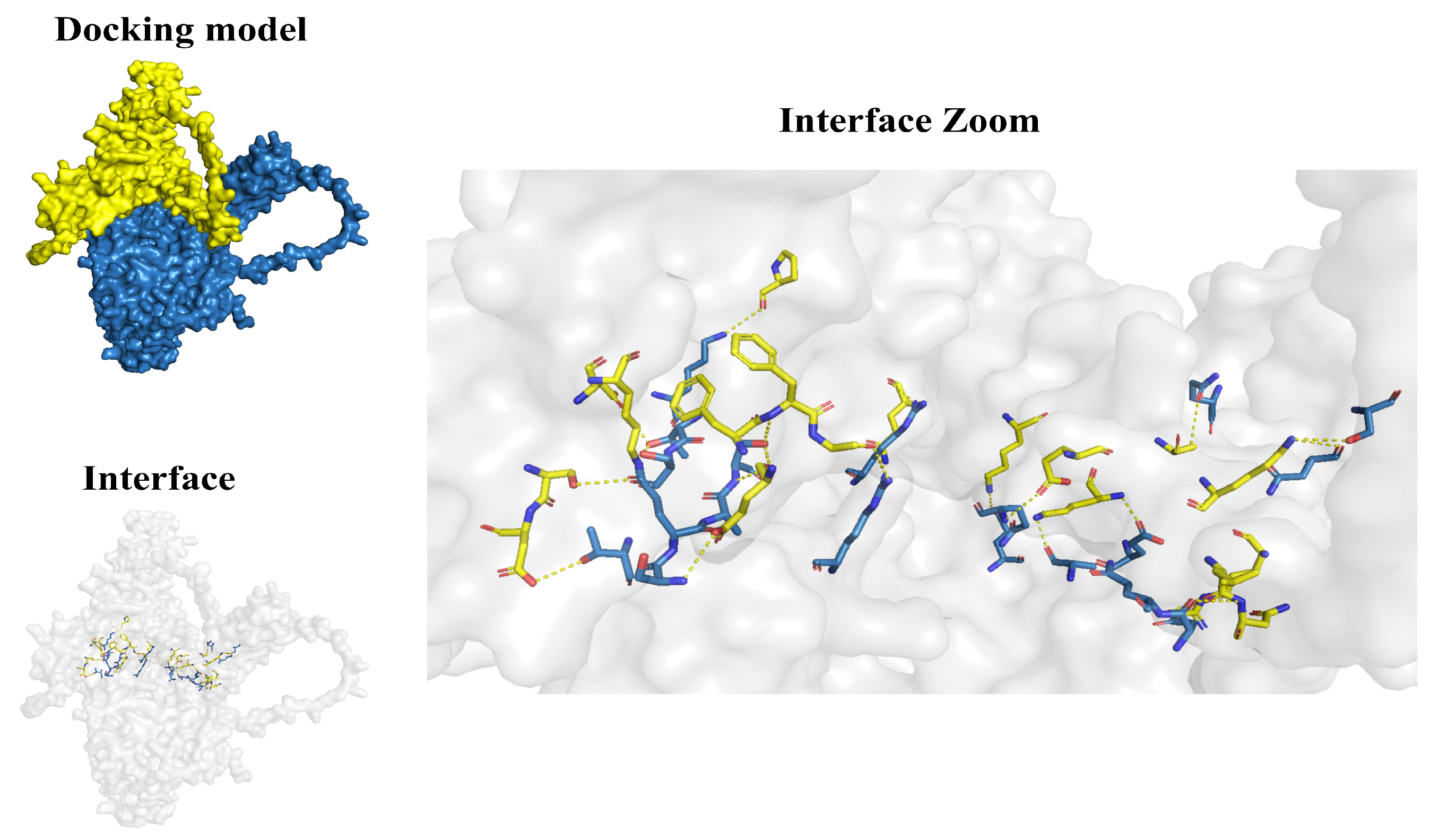
2.8. Molecular Docking Analysis
3. Discussion
4. Materials and Methods
4.1. Identification and Characterization of Hsp40 and Hsp70 Genes in Turbot
4.2. Chromosomal Distribution Analysis of Hsp40 and Hsp70 Genes
4.3. Motif Analysis of Hsp40 and Hsp70
4.4. Phylogenetic Analysis of Hsp40 and Hsp70 Proteins
4.5. Synteny Analysis of Hsp40 and Hsp70 Genes
4.6. Interaction Analysis of Hsp40 and Hsp70
4.7. Expression Analysis of Hsp40 and Hsp70
4.8. Molecular Docking of Dnajb1b-Hspa1a
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pereiro, P.; Figueras, A.; Novoa, B. Turbot (Scophthalmus maximus) vs. VHSV (Viral Hemorrhagic Septicemia Virus): A Review. Front. Physiol. 2016, 7, 192. [Google Scholar] [CrossRef] [PubMed]
- Bjørndal, T.; Øiestad, V. The Development of a New Farmed Species: Production Technology and Markets for Turbot; Working Paper; Institute for Research in Economics and Business Administration: Bergen, Norway, 2010. [Google Scholar]
- Dallaire-Dufresne, S.; Tanaka, K.H.; Trudel, M.V.; Lafaille, A.; Charette, S.J. Virulence, genomic features, and plasticity of Aeromonas salmonicida subsp. salmonicida, the causative agent of fish furunculosis. Vet. Microbiol. 2014, 169, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef]
- Li, J.; Qian, X.; Sha, B. Heat shock protein 40: Structural studies and their functional implications. Protein Pept. Lett. 2009, 16, 606–612. [Google Scholar] [CrossRef]
- Xu, H. Cochaperones enable Hsp70 to use ATP energy to stabilize native proteins out of the folding equilibrium. Sci. Rep. 2018, 8, 13213. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kumada, K.; Fuse, N.; Tamura, T.; Okamori, C.; Kurata, S. HSP70/DNAJA3 chaperone/cochaperone regulates NF-κB activity in immune responses. Biochem. Biophys. Res. Commun. 2019, 513, 947–951. [Google Scholar] [CrossRef] [PubMed]
- Ghazaei, C. Role and mechanism of the Hsp70 molecular chaperone machines in bacterial pathogens. J. Med. Microbiol. 2017, 66, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Iyer, K.; Chand, K.; Mitra, A.; Trivedi, J.; Mitra, D. Diversity in heat shock protein families: Functional implications in virus infection with a comprehensive insight of their role in the HIV-1 life cycle. Cell Stress Chaperones 2021, 26, 743–768. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, J.Z.; Whitham, S.A. Overexpression of a soybean nuclear localized type-III DnaJ domain-containing HSP40 reveals its roles in cell death and disease resistance. Plant J. 2013, 74, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, Y.; Liu, Y.; Zhang, Y.; Xiao, S.; Yu, Z. Co-expression of heat shock protein (HSP) 40 and HSP70 in Pinctada martensii response to thermal, low salinity and bacterial challenges. Fish Shellfish Immunol. 2016, 48, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Liang, C.; Zhou, L. Structural and functional analysis of the Hsp70/Hsp40 chaperone system. Protein Sci. 2020, 29, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Kampinga, H.H.; Craig, E.A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010, 11, 579–592. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.Y.; Lee, S.; Cyr, D.M. Mechanisms for regulation of Hsp70 function by Hsp40. Cell Stress Chaperones 2003, 8, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Alderson, T.R.; Kim, J.H.; Markley, J.L. Dynamical structures of Hsp70 and Hsp70-Hsp40 complexes. Structure 2016, 24, 1014–1030. [Google Scholar] [CrossRef] [PubMed]
- Karzai, A.W.; McMacken, R. A bipartite signaling mechanism involved in DnaJ-mediated activation of the Escherichia coli DnaK protein. J. Biol. Chem. 1996, 271, 11236–11246. [Google Scholar] [CrossRef] [PubMed]
- Kelley, W.L. The J-domain family and the recruitment of chaperone power. Trends Biochem. Sci. 1998, 23, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Hong, L.; Wang, Y.; Yu, J.; Yang, J.; Yang, J.; Zhang, H.; Perrett, S. Kinetics of the conformational cycle of Hsp70 reveals the importance of the dynamic and heterogeneous nature of Hsp70 for its function. Proc. Natl. Acad. Sci. USA 2020, 117, 7814–7823. [Google Scholar] [CrossRef] [PubMed]
- Faust, O.; Abayev-Avraham, M.; Wentink, A.S.; Maurer, M.; Nillegoda, N.B.; London, N.; Bukau, B.; Rosenzweig, R. HSP40 proteins use class-specific regulation to drive HSP70 functional diversity. Nature 2020, 587, 489–494. [Google Scholar] [CrossRef]
- Freeman, B.C.; Myers, M.P.; Schumacher, R.; Morimoto, R.I. Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. EMBO J. 1995, 14, 2281–2292. [Google Scholar] [CrossRef]
- Allan, R.K.; Ratajczak, T. Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress Chaperones 2011, 16, 353–367. [Google Scholar] [CrossRef]
- Kuppner, M.C.; Gastpar, R.; Gelwer, S.; Nössner, E.; Ochmann, O.; Scharner, A.; Issels, R.D. The role of heat shock protein (hsp70) in dendritic cell maturation: Hsp70 induces the maturation of immature dendritic cells but reduces DC differentiation from monocyte precursors. Eur. J. Immunol. 2001, 31, 1602–1609. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Cao, X. Stress for maintaining memory: HSP70 as a mobile messenger for innate and adaptive immunity. Eur. J. Immunol. 2010, 40, 1541–1544. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, R.; Nillegoda, N.B.; Mayer, M.P.; Bukau, B. The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 2019, 20, 665–680. [Google Scholar] [CrossRef] [PubMed]
- Borges, T.J.; Wieten, L.; Van Herwijnen, M.J.; Broere, F.; Van Der Zee, R.; Bonorino, C.; Van Eden, W. The anti-inflammatory mechanisms of Hsp70. Front. Immunol. 2012, 3, 95. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.-C.; Zhou, C.-J.; Zhou, Z.-R.; Wu, M.; Cao, C.-Y.; Hu, H.-Y. The C-terminal Helices of Heat Shock Protein 70 Are Essential for J-domain Binding and ATPase Activation*. J. Biol. Chem. 2012, 287, 6044–6052. [Google Scholar] [CrossRef]
- Ko, S.-K.; Kim, J.; Na, D.C.; Park, S.; Park, S.-H.; Hyun, J.Y.; Baek, K.-H.; Kim, N.D.; Kim, N.-K.; Park, Y.N.; et al. A Small Molecule Inhibitor of ATPase Activity of HSP70 Induces Apoptosis and Has Antitumor Activities. Chem. Biol. 2015, 22, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Faust, O.; Rosenzweig, R. Structural and Biochemical Properties of Hsp40/Hsp70 Chaperone System. Adv. Exp. Med. Biol. 2020, 1243, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wang, Q.; Zhao, H.; Chen, C. Transcriptome profiles revealed high- and low-salinity water altered gill homeostasis in half-smooth tongue sole (Cynoglossus semilaevis). Comp. Biochem. Physiol. Part D Genom. Proteom. 2022, 42, 100989. [Google Scholar] [CrossRef]
- Huang, J.; Li, Y.; Liu, Z.; Kang, Y.; Wang, J. Transcriptomic responses to heat stress in rainbow trout Oncorhynchus mykiss head kidney. Fish Shellfish Immunol. 2018, 82, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Li, C.; Xie, Y.; Liu, S.; Zhang, J.; Yao, J.; Jiang, C.; Li, Y.; Liu, Z. Genome-wide identification of Hsp70 genes in channel catfish and their regulated expression after bacterial infection. Fish Shellfish Immunol. 2016, 49, 154–162. [Google Scholar] [CrossRef]
- Du, Y.; Tang, X.; Sheng, X.; Xing, J.; Zhan, W. The influence of concentration of inactivated Edwardsiella tarda bacterin and immersion time on antigen uptake and expression of immune-related genes in Japanese flounder (Paralichthys olivaceus). Microb. Pathog. 2017, 103, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.-W.; Zheng, W.; Meng, Z.; Xu, W.; Liu, Y.; Chen, S. Identification of stress-related genes by co-expression network analysis based on the improved turbot genome. Sci. Data 2022, 9, 374. [Google Scholar] [CrossRef] [PubMed]
- Xue, T.; Liu, Y.; Cao, M.; Li, J.; Tian, M.; Zhang, L.; Wang, B.; Liu, X.; Li, C. Transcriptome analysis reveals deep insights into the early immune response of turbot (Scophthalmus maximus) induced by inactivated Aeromonas salmonicida vaccine. Fish Shellfish Immunol. 2021, 119, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Romero, A.; Rey-Campos, M.; Pereiro, P.; Librán-Pérez, M.; Figueras, A.; Novoa, B. Transcriptomic analysis of turbot (Scophthalmus maximus) treated with zymosan a reveals that lncRNAs and inflammation-related genes mediate the protection conferred against Aeromonas salmonicida. Fish Shellfish Immunol. 2024, 147, 109456. [Google Scholar] [CrossRef] [PubMed]
- Roufayel, R.; Kadry, S. Molecular Chaperone HSP70 and Key Regulators of Apoptosis—A Review. Curr. Mol. Med. 2019, 19, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Nunes, K.P.; de Oliveira, A.A. HSP70: From Signaling Mechanisms to Therapeutics. Biomolecules 2023, 13, 1141. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cyr, D.M.; Ramos, C.H. Specification of Hsp70 Function by Hsp40 Co-chaperones. Subcell. Biochem. 2023, 101, 127–139. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wang, Z. The roles of HSP40/DNAJ protein family in neurodegenerative diseases. Zhejiang Da Xue Xue Bao Yi Xue Ban 2022, 51, 640–646. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chang, Y.L.; Yang, C.C.; Huang, Y.Y.; Chen, Y.A.; Yang, C.W.; Liao, C.Y.; Li, H.; Wu, C.S.; Lin, C.H.; Teng, S.C. The HSP40 family chaperone isoform DNAJB6b prevents neuronal cells from tau aggregation. BMC Biol. 2023, 21, 293. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cyr, D.M.; Ramos, C.H. Specification of Hsp70 function by Type I and Type II Hsp40. Subcell. Biochem. 2015, 78, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Malinverni, D.; Jost Lopez, A.; De Los Rios, P.; Hummer, G.; Barducci, A. Modeling Hsp70/Hsp40 interaction by multi-scale molecular simulations and coevolutionary sequence analysis. eLife 2017, 6, e23471. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Velasco-Carneros, L.; Cuéllar, J.; Dublang, L.; Santiago, C.; Maréchal, J.D.; Martín-Benito, J.; Maestro, M.; Fernández-Higuero, J.Á.; Orozco, N.; Moro, F.; et al. The self-association equilibrium of DNAJA2 regulates its interaction with unfolded substrate proteins and with Hsc70. Nat. Commun. 2023, 14, 5436, Erratum in Nat. Commun. 2024, 15, 99. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bascos, N.A.D.; Mayer, M.P.; Bukau, B.; Landry, S.J. The Hsp40 J-domain modulates Hsp70 conformation and ATPase activity with a semi-elliptical spring. Protein Sci. 2017, 26, 1838–1851. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ciesielski, S.J.; Young, C.; Ciesielska, E.J.; Ciesielski, G.L. The Hsp70 and JDP proteins: Structure-function perspective on molecular chaperone activity. Enzymes 2023, 54, 221–245. [Google Scholar] [CrossRef] [PubMed]
- Wickramaratne, A.C.; Liao, J.Y.; Doyle, S.M.; Hoskins, J.R.; Puller, G.; Scott, M.L.; Alao, J.P.; Obaseki, I.; Dinan, J.C.; Maity, T.K.; et al. J-domain Proteins form Binary Complexes with Hsp90 and Ternary Complexes with Hsp90 and Hsp70. J. Mol. Biol. 2023, 435, 168184. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Malinverni, D.; Zamuner, S.; Rebeaud, M.E.; Barducci, A.; Nillegoda, N.B.; De Los Rios, P. Data-driven large-scale genomic analysis reveals an intricate phylogenetic and functional landscape in J-domain proteins. Proc. Natl. Acad. Sci. USA 2023, 8, e2218217120. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Horne, B.E.; Li, T.; Genevaux, P.; Georgopoulos, C.; Landry, S.J. The Hsp40 J-domain stimulates Hsp70 when tethered by the client to the ATPase domain. J. Biol. Chem. 2010, 285, 21679–21688. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hou, G.; Zhang, Q.; Li, C.; Ding, G.; Hu, L.; Chen, X.; Lv, Z.; Fan, Y.; Zou, J.; Xiao, T.; et al. An Aquareovirus Exploits Membrane-Anchored HSP70 To Promote Viral Entry. Microbiol. Spectr. 2023, 11, e0405522. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pham, T.H.; Cheng, T.C.; Wang, P.C.; Chen, S.C. Protective efficacy of four heat-shock proteins as recombinant vaccines against photobacteriosis in Asian seabass (Lates calcarifer). Fish Shellfish Immunol. 2021, 111, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Fan, K.; Liu, Y.; Liu, Y.; Liu, P.F. Administration of a recombinant ALDH7A1 (rA7) indicates potential regulation of the metabolite and immunology pathways in Atlantic salmon infected with Aeromonas salmonicida. J. Fish Dis. 2021, 44, 961–977. [Google Scholar] [CrossRef] [PubMed]
- Pham, P.H.; Sokeechand, B.S.H.; Hamilton, M.E.; Misk, E.; Jones, G.; Lee, L.E.J.; Lumsden, J.S.; Bols, N.C. VER-155008 induced Hsp70 proteins expression in fish cell cultures while impeding replication of two RNA viruses. Antivir. Res. 2019, 162, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Rao, Y.; Wan, Q.; Su, H.; Xiao, X.; Liao, Z.; Ji, J.; Yang, C.; Lin, L.; Su, J. ROS-induced HSP70 promotes cytoplasmic translocation of high-mobility group box 1b and stimulates antiviral autophagy in grass carp kidney cells. J. Biol. Chem. 2018, 293, 17387–17401. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shan, L.P.; Chen, X.H.; Ling, F.; Zhu, B.; Wang, G.X. Targeting Heat Shock Protein 70 as an antiviral strategy against grass carp reovirus infection. Virus Res. 2018, 247, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Vahdatiraad, L.; Heidari, B.; Zarei, S.; Sohrabi, T.; Ghafouri, H. Protective effects of HSP Inducer on diazinon-exposed stellate sturgeon (Acipenser stellatus) fry: Insights on HSP70 gene expression, immune response, and enzyme indices. PLoS ONE 2023, 18, e0294188. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nitzsche, B.; Höpfner, M.; Biersack, B. Synthetic Small Molecule Modulators of Hsp70 and Hsp40 Chaperones as Promising Anticancer Agents. Int. J. Mol. Sci. 2023, 24, 4083. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Theivanthiran, B.; Yarla, N.; Haykal, T.; Nguyen, Y.V.; Cao, L.; Ferreira, M.; Holtzhausen, A.; Al-Rohil, R.; Salama, A.K.S.; Beasley, G.M.; et al. Tumor-intrinsic NLRP3-HSP70-TLR4 axis drives premetastatic niche development and hyperprogression during anti-PD-1 immunotherapy. Sci. Transl. Med. 2022, 14, eabq7019. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhao, K.; Zhou, G.; Liu, Y.; Zhang, J.; Chen, Y.; Liu, L.; Zhang, G. HSP70 Family in Cancer: Signaling Mechanisms and Therapeutic Advances. Biomolecules 2023, 13, 601. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Elmallah, M.I.Y.; Cordonnier, M.; Vautrot, V.; Chanteloup, G.; Garrido, C.; Gobbo, J. Membrane-anchored heat-shock protein 70 (Hsp70) in cancer. Cancer Lett. 2020, 469, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Perez, V.A.; Sanders, D.W.; Mendoza-Oliva, A.; Stopschinski, B.E.; Mullapudi, V.; White, C.L.; Joachimiak, L.A.; Diamond, M.I. DnaJC7 specifically regulates tau seeding. eLife 2023, 12, e86936. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dong, Y.; Li, T.; Ma, Z.; Zhou, C.; Wang, X.; Li, J. HSPA1A, HSPA2, and HSPA8 Are Potential Molecular Biomarkers for Prognosis among HSP70 Family in Alzheimer’s Disease. Dis. Markers 2022, 2022, 9480398. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chanteloup, G.; Cordonnier, M.; Moreno-Ramos, T.; Pytel, V.; Matías-Guiu, J.; Gobbo, J.; Cabrera-Martín, M.N.; Gómez-Pinedo, U.; Garrido, C.; Matías-Guiu, J.A. Exosomal HSP70 for Monitoring of Frontotemporal Dementia and Alzheimer’s Disease: Clinical and FDG-PET Correlation. J. Alzheimer’s Dis. 2019, 71, 1263–1269. [Google Scholar] [CrossRef] [PubMed]
- Niu, L.; Lou, F.; Sun, Y.; Sun, L.; Cai, X.; Liu, Z.; Zhou, H.; Wang, H.; Wang, Z.; Bai, J.; et al. A micropeptide encoded by lncRNA MIR155HG suppresses autoimmune inflammation via modulating antigen presentation. Sci. Adv. 2020, 6, eaaz2059. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- van Eden, W. Immune tolerance therapies for autoimmune diseases based on heat shock protein T-cell epitopes. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20160531. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rodriguez-Iturbe, B.; Lanaspa, M.A.; Johnson, R.J. The role of autoimmune reactivity induced by heat shock protein 70 in the pathogenesis of essential hypertension. Br. J. Pharmacol. 2019, 176, 1829–1838. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tukaj, S.; Mantej, J.; Sitko, K.; Zillikens, D.; Ludwig, R.J.; Bieber, K.; Kasperkiewicz, M. Pathological Relevance of Anti-Hsp70 IgG Autoantibodies in Epidermolysis Bullosa Acquisita. Front. Immunol. 2022, 13, 877958. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, L.; Fu, Y.; Yu, B.; Jiang, X.; Liu, H.; Liu, J.; Zha, B.; Chu, Y. HSP70, a Novel Regulatory Molecule in B Cell-Mediated Suppression of Autoimmune Diseases. J. Mol. Biol. 2021, 433, 166634. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System. 2002. Available online: http://www.pymol.org (accessed on 20 October 2023).


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geng, Y.; Gai, Y.; Zhang, Y.; Zhao, S.; Jiang, A.; Li, X.; Deng, K.; Zhang, F.; Tan, L.; Song, L. Genome-Wide Identification and Interaction Analysis of Turbot Heat Shock Protein 40 and 70 Families Suggest the Mechanism of Chaperone Proteins Involved in Immune Response after Bacterial Infection. Int. J. Mol. Sci. 2024, 25, 7963. https://doi.org/10.3390/ijms25147963
Geng Y, Gai Y, Zhang Y, Zhao S, Jiang A, Li X, Deng K, Zhang F, Tan L, Song L. Genome-Wide Identification and Interaction Analysis of Turbot Heat Shock Protein 40 and 70 Families Suggest the Mechanism of Chaperone Proteins Involved in Immune Response after Bacterial Infection. International Journal of Molecular Sciences. 2024; 25(14):7963. https://doi.org/10.3390/ijms25147963
Chicago/Turabian StyleGeng, Yuanwei, Yuxuan Gai, Yanping Zhang, Shengwei Zhao, Anlan Jiang, Xueqing Li, Kaiqing Deng, Fuxuan Zhang, Lingling Tan, and Lin Song. 2024. "Genome-Wide Identification and Interaction Analysis of Turbot Heat Shock Protein 40 and 70 Families Suggest the Mechanism of Chaperone Proteins Involved in Immune Response after Bacterial Infection" International Journal of Molecular Sciences 25, no. 14: 7963. https://doi.org/10.3390/ijms25147963
APA StyleGeng, Y., Gai, Y., Zhang, Y., Zhao, S., Jiang, A., Li, X., Deng, K., Zhang, F., Tan, L., & Song, L. (2024). Genome-Wide Identification and Interaction Analysis of Turbot Heat Shock Protein 40 and 70 Families Suggest the Mechanism of Chaperone Proteins Involved in Immune Response after Bacterial Infection. International Journal of Molecular Sciences, 25(14), 7963. https://doi.org/10.3390/ijms25147963




