Genome-Wide Identification and Characterization of U-Box Gene Family Members and Analysis of Their Expression Patterns in Phaseolus vulgaris L. under Cold Stress
Abstract
1. Introduction
2. Results
2.1. Identification and Characterization of the U-Box Gene Family in Phaseolus vulgaris
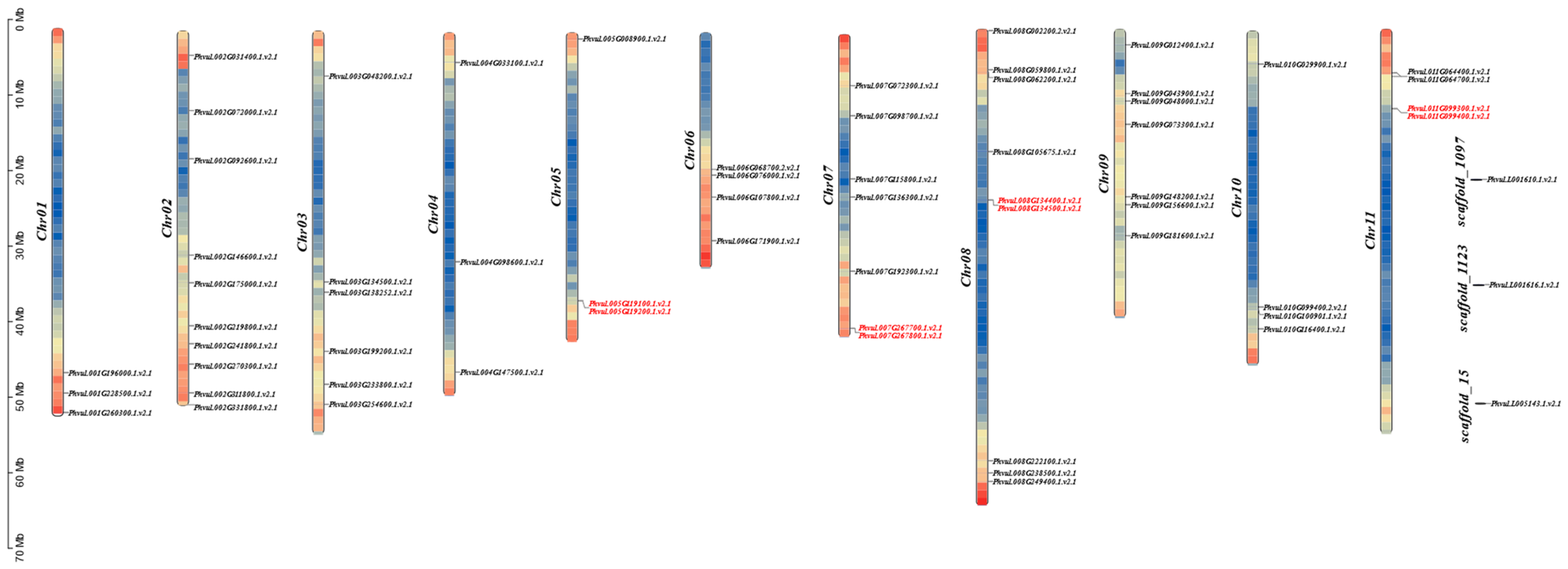
2.2. Chromosomal Localization of the U-Box Gene Family in Phaseolus vulgaris
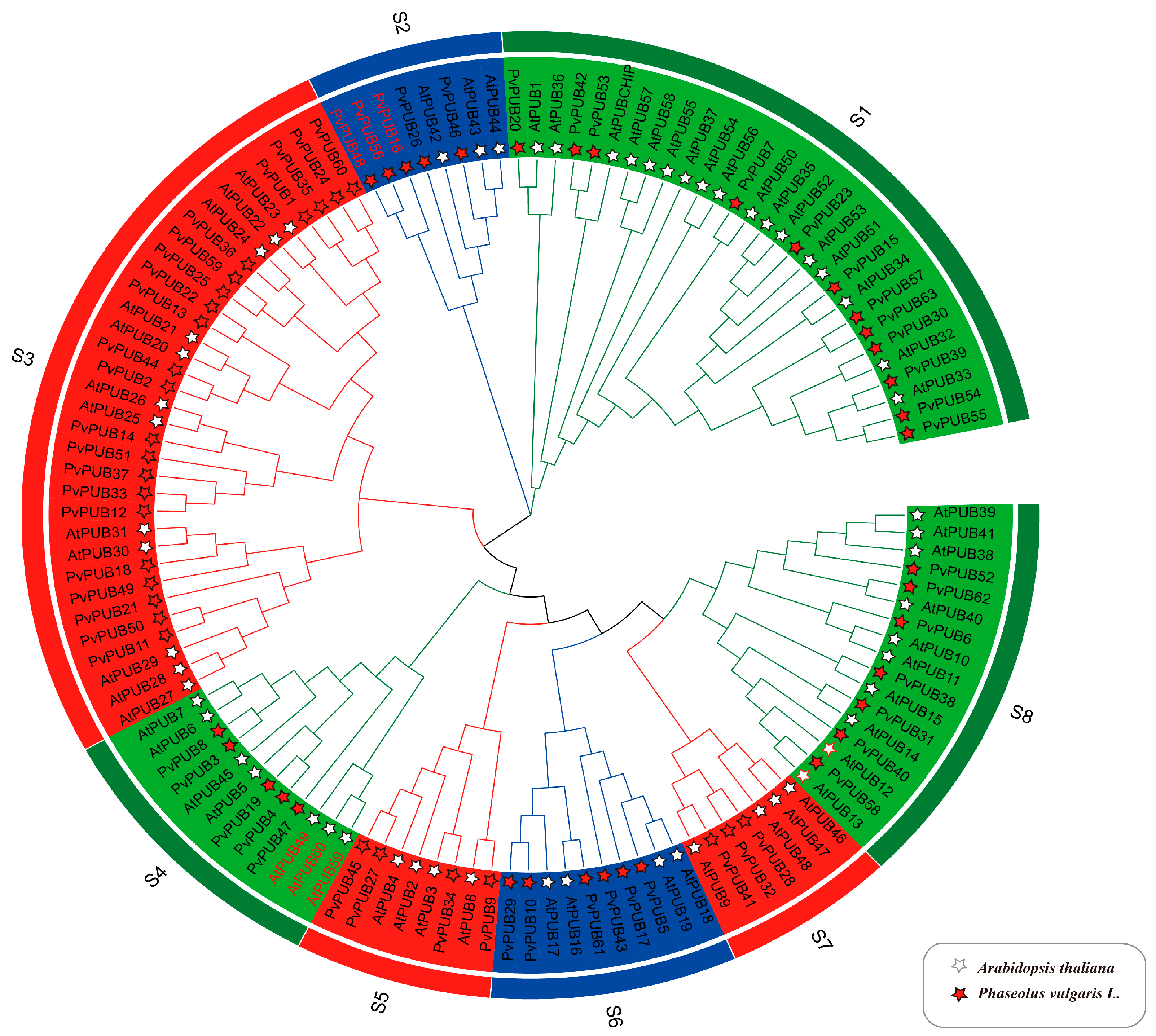
2.3. Phylogenetic Analysis and Classification of the U-Box Gene Family in Phaseolus vulgaris
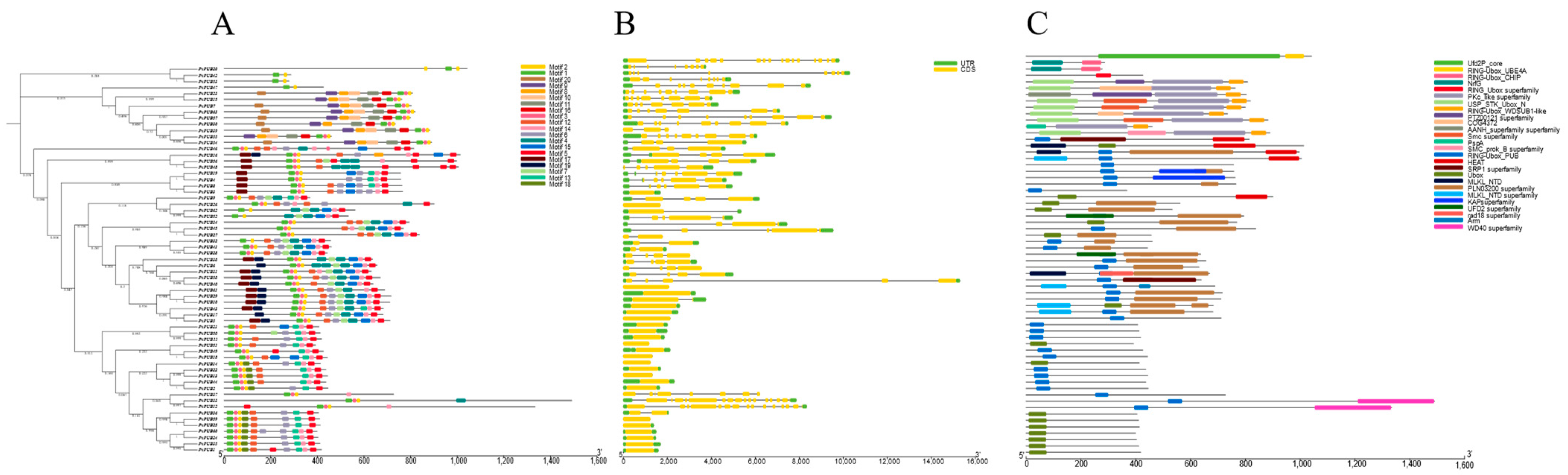
2.4. Analysis of Conserved Motifs, Domains, and Structure of U-Box Genes in Phaseolus vulgaris
2.5. Subcellular Localization and Cis-Acting Element Analysis of U-Box Genes in Phaseolus vulgaris
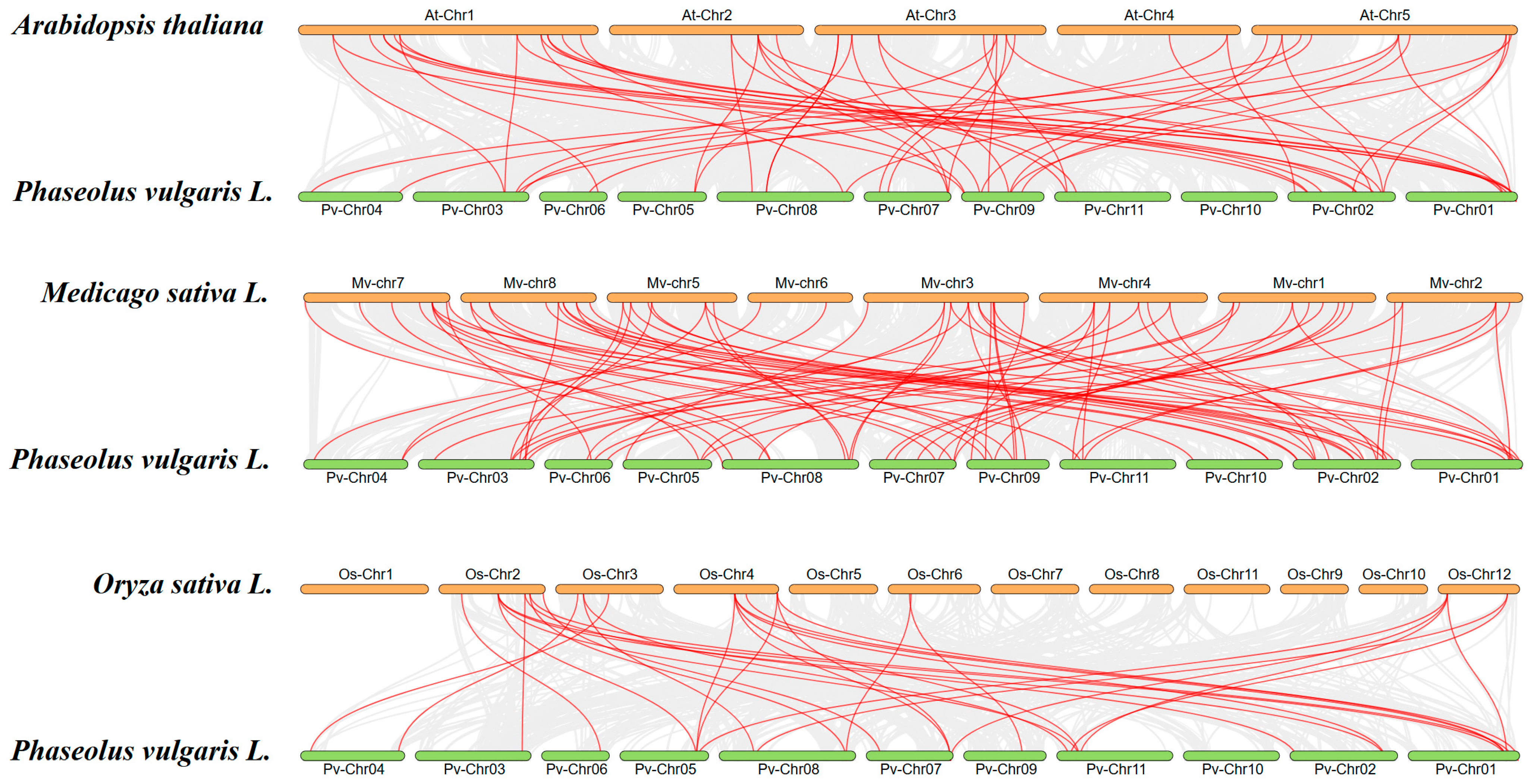
2.6. Tandem Duplication and Collinearity Analysis of U-Box Genes in Phaseolus vulgaris
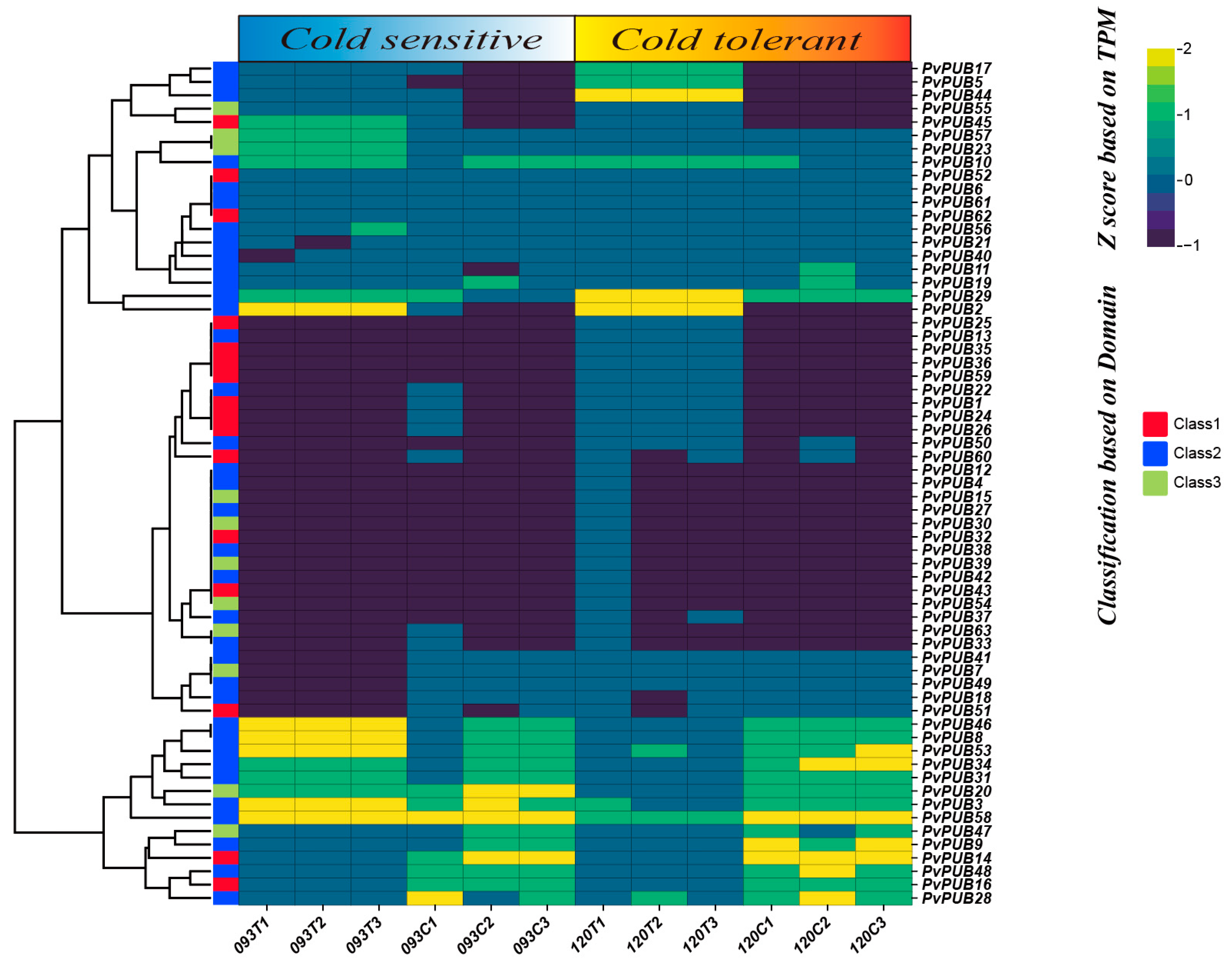
2.7. Expression Patterns of PvPUB Genes under Low-Temperature Stress
3. Discussion
4. Materials and Methods
4.1. Identification of the U-Box Gene Family in Phaseolus vulgaris
4.2. Phylogenetic Analysis and Classification of the U-Box Gene Family in Phaseolus vulgaris
4.3. Analysis of Conserved Motif, Domain, and Gene Structure of the U-Box Gene in Phaseolus vulgaris
4.4. Subcellular Localization and Cis-Acting Element Analysis of the U-Box Gene in Phaseolus vulgaris
4.5. Chromosome Localization and Collinearity Analysis of the U-Box Gene in Phaseolus vulgaris
4.6. RNA-Seq Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kajla, M.; Yadav, V.K.; Khokhar, J.; Singh, S.; Chhokar, R.S.; Meena, R.P.; Sharma, R.K. Increase in wheat production through management of abiotic stresses: A review. J. Appl. Nat. Sci. 2015, 7, 1070–1080. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, K.; Alam, M.M.; Roychowdhury, R.; Fujita, M. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 2013, 14, 9643–9684. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 2014, 5, 170. [Google Scholar] [CrossRef] [PubMed]
- Staiger, D.; Brown, J.W. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 2013, 25, 3640–3656. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; To, T.K.; Nishioka, T.; Seki, M. Chromatin regulation functions in plant abiotic stress responses. Plant Cell Environ. 2010, 33, 604–611. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Mazzucotelli, E.; Mastrangelo, A.M.; Crosatti, C.; Guerra, D.; Stanca, A.M.; Cattivelli, L. Abiotic stress response in plants: When post-transcriptional and post-translational regulations control transcription. Plant Sci. 2008, 174, 420–431. [Google Scholar] [CrossRef]
- Hirayama, T.; Shinozaki, K. Research on plant abiotic stress responses in the post-genome era: Past, present and future. Plant J. 2010, 61, 1041–1052. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.-R.; Park, C.H.; Venu, R.; Gough, J.; Wang, G.-L. Classification, expression pattern, and E3 ligase activity assay of rice U-box-containing proteins. Mol. Plant 2008, 1, 800–815. [Google Scholar] [CrossRef]
- Mazzucotelli, E.; Belloni, S.; Marone, D.; De Leonardis, A.M.; Guerra, D.; Di Fonzo, N.; Cattivelli, L.; Mastrangelo, A.M. The E3 ubiquitin ligase gene family in plants: Regulation by degradation. Curr. Genom. 2006, 7, 509–522. [Google Scholar] [CrossRef]
- Aravind, L.; Koonin, E.V. The U box is a modified RING finger—A common domain in ubiquitination. Curr. Biol. 2000, 10, R132–R134. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.; Joshi, D.; Yadav, P.K.; Gupta, A.K.; Bhatt, T.K. Role of ubiquitin-mediated degradation system in plant biology. Front. Plant Sci. 2016, 7, 806. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, S.; Nakayama, K.I. Nakayama. U-box proteins as a new family of ubiquitin ligases. Biochem. Biophys. Res. Commun. 2003, 302, 635–645. [Google Scholar] [CrossRef]
- Koegl, M.; Hoppe, T.; Schlenker, S.; Ulrich, H.D.; Mayer, T.U.; Jentsch, S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 1999, 96, 635–644. [Google Scholar] [CrossRef]
- Ohi, M.D.; Vander Kooi, C.W.; Rosenberg, J.A.; Chazin, W.J.; Gould, K.L. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat. Struct. Biol. 2003, 10, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.L.; Hauksdóttir, H.; Troy, A.; Herschleb, J.; Kraft, E.; Callis, J. The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. Plant J. 2003, 35, 729–742. [Google Scholar]
- Stone, S.L.; Hauksdóttir, H.; Troy, A.; Herschleb, J.; Kraft, E.; Callis, J. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 2005, 137, 13–30. [Google Scholar] [CrossRef]
- Smalle, J.; Vierstra, R.D. The ubiquitin 26s proteasome proteolytic pathway. Annu. Rev. Plant Biol. 2004, 55, 555–590. [Google Scholar] [CrossRef]
- Hatzfeld, M. The armadillo family at structural proteins, in International Review of Cytology. In A Survey of Cell Biology; Jeon, K.W., Ed.; Elsevier: Amsterdam, The Netherlands, 1999; Volume 186, pp. 179–224. [Google Scholar]
- Wang, C.; Song, B.; Dai, Y.; Zhang, S.; Huang, X. Genome-wide identification and functional analysis of U-box E3 ubiquitin ligases gene family related to drought stress response in Chinese white pear (Pyrus bretschneideri). BMC Plant Biol. 2021, 21, 235. [Google Scholar] [CrossRef]
- Wang, D.-R.; Zhang, X.-W.; Xu, R.-R.; Wang, G.-L.; You, C.-X.; An, J.-P. Apple U-box-type E3 ubiquitin ligase MdPUB23 reduces cold-stress tolerance by degrading the cold-stress regulatory protein MdICE1. Hortic. Res. 2022, 9, uhac171. [Google Scholar] [CrossRef]
- Yu, T.; Zhou, H.; Liu, Z.; Zhai, H.; Liu, Q. The sweet potato transcription factor IbbHLH33 enhances chilling tolerance in transgenic tobacco. Czech J. Genet. Plant Breed. 2022, 58, 210–222. [Google Scholar] [CrossRef]
- Muhammad, N.; Uddin, N.; Khan, M.K.U.; Ali, N.; Ali, K.; Jones, D.A. Diverse role of basic helix-loop-helix (bhlh) transcription factor superfamily genes in the fleshy fruit-bearing plant species. Czech J. Genet. Plant Breed. 2023, 59, 1–13. [Google Scholar] [CrossRef]
- Kaur, N.; Gupta, A.K. Signal transduction pathways under abiotic stresses in plants. Curr. Sci. 2005, 88, 1771–1780. [Google Scholar]
- Jiao, L.; Fu, S.; Zhang, Y.; Lu, J. U-box e3 ubiquitin ligases regulate stress tolerance and growth of plants. Chin. Bull. Bot. 2016, 51, 724–735. [Google Scholar]
- Wiborg, J.; O’Shea, C.; Skriver, K. Biochemical function of typical and variant arabidapsis thaliana u-box e3 ubiquitin-protein ligases. Biochem. J. 2008, 413, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Liu, C.; Li, M.; Li, Y.; Yan, Z.; Feng, G.; Liu, D. Integrated transcriptomics and metabolomics analysis reveals key regulatory network that response to cold stress in common bean (Phaseolus vulgaris L.). BMC Plant Biol. 2023, 23, 85. [Google Scholar] [CrossRef]
- Penfield, S. Temperature perception and signal transduction in plants. New Phytol. 2008, 179, 615–628. [Google Scholar] [CrossRef]
- Liu, J.; Shi, Y.; Yang, S. Insights into the regulation of c-repeat binding factors in plant cold signaling. J. Integr. Plant Biol. 2018, 60, 780–795. [Google Scholar] [CrossRef]
- Raab, S.D.G.; Zarepour, M.; Hartung, W.; Koshiba, T.; Bittner, F.; Hoth, S. The ubiquitin ligase atpub22 regulates cell death in arabidopsis under drought stress. Plant Cell 2009, 21, 1195–1211. [Google Scholar]
- Seo, P.J.; Lee, S.B.; Suh, M.C.; Park, M.-J.; Go, Y.S.; Park, C.-M. The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 2011, 23, 1138–1152. [Google Scholar] [CrossRef]
- Yee, D. The Expanding diversity of Plant u-box E3 Ubiquitin Ligases in Arabidopsis: Identifying atpub18 and atpub19 Function during Abiotic Stress Responses. Ph.D. Thesis, University of Toronto, Toronto, ON, Canada, 2010. [Google Scholar]
- Jiao, L.; Zhang, Y.; Wu, J.; Zhang, H.; Lu, J. A novel u-box protein gene from “zuoshanyi” grapevine (Vitis amurensis rupr. cv.) involved in cold responsive gene expression in Arabidopsis thanliana. Plant Mol. Biol. Rep. 2015, 33, 557–568. [Google Scholar] [CrossRef]
- Pickart, C.M.; Eddins, C.M. Ubiquitin: Structures, functions, mechanisms. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2004, 1695, 55–72. [Google Scholar] [CrossRef]
- Sadowski, M.; Sarcevic, B. Mechanisms of mono- and poly-ubiquitination: Ubiquitination specificity depends on compatibility between the e2 catalytic core and amino acid residues proximal to the lysine. Cell 2010, 5, 19. [Google Scholar] [CrossRef] [PubMed]
- Damgaard, R.B. The ubiquitin system: From cell signalling to disease biology and new therapeutic opportunities. Cell Death Differ. 2021, 28, 423–426. [Google Scholar] [CrossRef] [PubMed]
- Vierstra, R.D. The expanding universe of ubiquitin and ubiquitin-like modifiers. Plant Physiol. 2012, 160, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.-Q.; Xue, H.-W. The ubiquitin-proteasome system in plant responses to environments. Plant Cell Environ. 2019, 42, 2931–2944. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, C.; Li, Y.; Zheng, N.; Chen, H.; Zhao, Q.; Gao, T.; Guo, H.; Xie, Q. SDIR1 is a Ring finger e3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 2007, 19, 1912–1929. [Google Scholar] [CrossRef]
- Qi, S.; Lin, Q.; Zhu, H.; Gao, F.; Zhang, W.; Hua, X. The RING Finger E3 Ligase SpRing is a Positive Regulator of Salt Stress Signaling in Salt-Tolerant Wild Tomato Species. Plant Cell Physiol. 2016, 57, 528–539. [Google Scholar] [CrossRef]
- Qin, F.; Sakuma, Y.; Tran, L.-S.P.; Maruyama, K.; Kidokoro, S.; Fujita, Y.; Fujita, M.; Umezawa, T.; Sawano, Y.; Miyazono, K.-I.; et al. Arabidopsis DREB2A-interacting proteins function as ring e3 ligases and negatively regulate plant drought stress–responsive gene expression. Plant Cell 2008, 20, 1693–1707. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, J.; Shi, K.; Xia, X.J.; Zhou, Y.H.; Yu, J.Q. Hydrogen peroxide is involved in the cold acclimation-induced chilling tolerance of tomato plants. Plant Physiol. Biochem. 2012, 60, 141–149. [Google Scholar] [CrossRef]
- Dou, H.; Buetow, L.; Hock, A.; Sibbet, G.J.; Vousden, K.H.; Huang, D.T. Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl. Nat. Struct. Mol. Biol. 2012, 19, 184–192. [Google Scholar] [CrossRef]
- Kang, R.S.; Daniels, C.M.; Francis, S.A.; Shih, S.C.; Salerno, W.J.; Hicke, L.; Radhakrishnan, I. Solution structure of a cue-ubiquitin complex reveals a conserved mode of ubiquitin binding. Cell 2003, 113, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]



| Genomic ID | Gene Name | Class | Isoelectric Point | Molecular Weight | Cellular Localization | Instability Index | Aliphatic Index | GRAVY | Protein Length (aa) | Gene Size | Chromosomal Position |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Phvul.001G196000.1.v2.1 | PvPUB1 | Class 1 | 8.7 | 46,436.14 | Cytoplasm | 35.56 | 102.15 | −0.038 | 418 | 1604 | 1 |
| Phvul.001G228500.1.v2.1 | PvPUB2 | Class 2 | 8.26 | 49,453.27 | Mitochondrion | 34.74 | 99.6 | −0.099 | 446 | 1660 | 1 |
| Phvul.001G260300.1.v2.1 | PvPUB3 | Class 2 | 5.7 | 84,556.93 | Nucleus | 47.29 | 93.98 | −0.171 | 766 | 4958 | 1 |
| Phvul.002G031400.1.v2.1 | PvPUB4 | Class 2 | 6.32 | 85,204.42 | Nucleus | 48.49 | 95.53 | −0.294 | 758 | 5400 | 2 |
| Phvul.002G072000.1.v2.1 | PvPUB5 | Class 2 | 8.77 | 79,577.39 | Chloroplast | 46.14 | 100.62 | −0.056 | 712 | 2138 | 2 |
| Phvul.002G092600.1.v2.1 | PvPUB6 | Class 2 | 6.85 | 71,532.82 | Nucleus | 45.97 | 102.36 | −0.124 | 656 | 3345 | 2 |
| Phvul.002G146600.1.v2.1 | PvPUB7 | Class 3 | 6.92 | 92,027.45 | Chloroplast | 48.94 | 85.11 | −0.506 | 803 | 4315 | 2 |
| Phvul.002G175000.1.v2.1 | PvPUB8 | Class 2 | 5.22 | 84,772.85 | Nucleus | 52.2 | 89.7 | −0.199 | 763 | 4688 | 2 |
| Phvul.002G219800.1.v2.1 | PvPUB9 | Class 2 | 8.48 | 40,001.25 | Plasma membrane | 49.11 | 110.82 | 0.083 | 368 | 1693 | 2 |
| Phvul.002G241800.1.v2.1 | PvPUB10 | Class 2 | 7.17 | 78,518.64 | Chloroplast | 47.7 | 97.99 | 0.036 | 711 | 3759 | 2 |
| Phvul.002G270300.1.v2.1 | PvPUB11 | Class 2 | 7.99 | 46,301.58 | Nucleus | 49.56 | 106.03 | 0.017 | 418 | 1880 | 2 |
| Phvul.002G311800.1.v2.1 | PvPUB12 | Class 2 | 5.49 | 148,371.85 | Chloroplast | 50.3 | 96.57 | −0.14 | 1334 | 8342 | 2 |
| Phvul.002G331800.1.v2.1 | PvPUB13 | Class 2 | 8.26 | 49,892.08 | Chloroplast | 41.96 | 106.24 | −0.035 | 444 | 1334 | 2 |
| Phvul.003G048200.1.v2.1 | PvPUB14 | Class 1 | 7.15 | 45,242.43 | Cytoplasm | 51.12 | 109.83 | 0.121 | 413 | 1241 | 3 |
| Phvul.003G134500.1.v2.1 | PvPUB15 | Class 3 | 5.71 | 85,778.69 | Chloroplast | 40.3 | 91.73 | −0.294 | 763 | 5302 | 3 |
| Phvul.003G138252.1.v2.1 | PvPUB16 | Class 1 | 5.49 | 112,675.95 | Chloroplast | 41.07 | 110.43 | −0.095 | 1013 | 4644 | 3 |
| Phvul.003G199200.1.v2.1 | PvPUB17 | Class 2 | 8.48 | 75,024.85 | Plasma membrane | 39.69 | 104.81 | 0.019 | 682 | 2491 | 3 |
| Phvul.003G233800.1.v2.1 | PvPUB18 | Class 2 | 6.12 | 48,522.88 | Cytoplasm | 41.53 | 117.65 | 0.249 | 443 | 1331 | 3 |
| Phvul.003G254600.1.v2.1 | PvPUB19 | Class 2 | 6.27 | 84,773.76 | Nucleus | 42.56 | 95.32 | −0.202 | 757 | 4089 | 3 |
| Phvul.004G033100.1.v2.1 | PvPUB20 | Class 3 | 5.48 | 117,694.74 | Nucleus | 43.27 | 92.7 | −0.223 | 1042 | 9823 | 4 |
| Phvul.004G098600.1.v2.1 | PvPUB21 | Class 2 | 8.77 | 44,220.42 | Chloroplast | 44.05 | 103.27 | 0.107 | 407 | 2018 | 4 |
| Phvul.004G147500.1.v2.1 | PvPUB22 | Class 2 | 6.64 | 49,159.99 | Chloroplast | 46.64 | 102.75 | −0.116 | 437 | 1703 | 4 |
| Phvul.005G008900.1.v2.1 | PvPUB23 | Class 3 | 6.72 | 89,958.55 | Chloroplast | 44.36 | 88.97 | −0.316 | 808 | 8517 | 5 |
| Phvul.005G119100.1.v2.1 | PvPUB24 | Class 1 | 8.96 | 45,111.03 | Cytoplasm | 47.53 | 113.23 | 0.137 | 403 | 1484 | 5 |
| Phvul.005G119200.1.v2.1 | PvPUB25 | Class 1 | 8.79 | 46,597.19 | Cytoplasm | 48.56 | 102.69 | −0.073 | 413 | 1389 | 5 |
| Phvul.006G068700.2.v2.1 | PvPUB26 | Class 1 | 6.19 | 99,632.12 | Nucleus | 40.87 | 101.29 | −0.129 | 901 | 6187 | 6 |
| Phvul.006G076000.1.v2.1 | PvPUB27 | Class 2 | 5.45 | 90,667.38 | Chloroplast | 51.18 | 96.3 | −0.248 | 838 | 9544 | 6 |
| Phvul.006G107800.1.v2.1 | PvPUB28 | Class 2 | 7.19 | 49,097.58 | Nucleus | 39.85 | 105.51 | −0.207 | 443 | 1968 | 6 |
| Phvul.006G171900.1.v2.1 | PvPUB29 | Class 2 | 6.85 | 78,355.28 | Chloroplast | 45.26 | 98.98 | 0.014 | 716 | 3290 | 6 |
| Phvul.007G072300.1.v2.1 | PvPUB30 | Class 3 | 5.8 | 83,383.05 | Nucleus | 38.33 | 92.87 | −0.298 | 735 | 9461 | 7 |
| Phvul.007G098700.1.v2.1 | PvPUB31 | Class 2 | 6.17 | 68,934.26 | Cytoplasm | 33.92 | 102.23 | −0.205 | 631 | 3559 | 7 |
| Phvul.007G115800.1.v2.1 | PvPUB32 | Class 1 | 6.23 | 50,705.33 | Chloroplast | 35.65 | 106.3 | −0.107 | 457 | 1786 | 7 |
| Phvul.007G136300.1.v2.1 | PvPUB33 | Class 2 | 5.43 | 167,076.4 | Nucleus | 41.38 | 90.79 | −0.282 | 1491 | 7869 | 7 |
| Phvul.007G192300.1.v2.1 | PvPUB34 | Class 2 | 7.25 | 87,205.42 | Chloroplast | 49.48 | 94.19 | −0.292 | 794 | 4975 | 7 |
| Phvul.007G267700.1.v2.1 | PvPUB35 | Class 1 | 8.64 | 45,041.84 | Cytoplasm | 39.69 | 113.19 | 0.118 | 411 | 1698 | 7 |
| Phvul.007G267800.1.v2.1 | PvPUB36 | Class 1 | 8.73 | 45,836.99 | Cytoplasm | 34.52 | 107.83 | 0.033 | 405 | 2056 | 7 |
| Phvul.008G002200.2.v2.1 | PvPUB37 | Class 2 | 5.64 | 81,145.87 | Nucleus | 42.42 | 94.94 | −0.199 | 727 | 6199 | 8 |
| Phvul.008G059800.1.v2.1 | PvPUB38 | Class 2 | 5.47 | 71,753.7 | Golgi apparatus | 51.11 | 100.74 | −0.256 | 637 | 3036 | 8 |
| Phvul.008G062200.1.v2.1 | PvPUB39 | Class 3 | 5.82 | 98,854 | Cytoplasm | 52.14 | 87.38 | −0.419 | 883 | 7490 | 8 |
| Phvul.008G105675.1.v2.1 | PvPUB40 | Class 2 | 5.69 | 70,209.67 | Endoplasmic Reticulum | 36.6 | 103.35 | −0.05 | 639 | 15,298 | 8 |
| Phvul.008G134400.1.v2.1 | PvPUB41 | Class 2 | 8.55 | 51,408.65 | Cytoplasm | 43.35 | 102.39 | −0.234 | 460 | 3437 | 8 |
| Phvul.008G134500.1.v2.1 | PvPUB42 | Class 2 | 5.21 | 33,113.88 | Cytoplasm | 39.73 | 89.37 | −0.385 | 287 | 3755 | 8 |
| Phvul.008G222100.1.v2.1 | PvPUB43 | Class 1 | 7.15 | 74,729.23 | Nucleus | 43.05 | 105.52 | −0.059 | 683 | 2575 | 8 |
| Phvul.008G238500.1.v2.1 | PvPUB44 | Class 2 | 8.59 | 48,350.49 | Plasma membrane | 38.02 | 100.76 | 0.022 | 437 | 2326 | 8 |
| Phvul.008G249400.1.v2.1 | PvPUB45 | Class 1 | 5.74 | 83,718.27 | Peroxisome | 41.15 | 101.09 | −0.186 | 769 | 7450 | 8 |
| Phvul.009G012400.1.v2.1 | PvPUB46 | Class 2 | 5.15 | 89,127.63 | Plasma membrane | 41.36 | 110.1 | 0.043 | 814 | 5590 | 9 |
| Phvul.009G043900.1.v2.1 | PvPUB47 | Class 3 | 4.72 | 46,563.18 | Nucleus | 56.78 | 60.4 | −0.709 | 426 | 4903 | 9 |
| Phvul.009G048000.1.v2.1 | PvPUB48 | Class 2 | 5.91 | 112,968.0 | Golgi apparatus | 47.48 | 111.79 | −0.068 | 1005 | 6044 | 9 |
| Phvul.009G073300.1.v2.1 | PvPUB49 | Class 2 | 6.94 | 46,648.8 | Chloroplast | 48.4 | 117.58 | 0.21 | 426 | 2138 | 9 |
| Phvul.009G148200.1.v2.1 | PvPUB50 | Class 2 | 8.87 | 45,016.58 | Chloroplast | 42.47 | 111.02 | 0.15 | 412 | 2002 | 9 |
| Phvul.009G156600.1.v2.1 | PvPUB51 | Class 1 | 7.53 | 43,758.33 | Cytoplasm | 41.98 | 110.94 | 0.152 | 392 | 1178 | 9 |
| Phvul.009G181600.1.v2.1 | PvPUB52 | Class 1 | 8.01 | 58,455.76 | Chloroplast | 55.78 | 92.2 | −0.237 | 533 | 5363 | 9 |
| Phvul.010G029900.1.v2.1 | PvPUB53 | Class 2 | 5.71 | 31,658 | Cytoplasm | 36.64 | 85.72 | −0.49 | 278 | 10,297 | 10 |
| Phvul.010G099400.2.v2.1 | PvPUB54 | Class 3 | 6.73 | 99,946.7 | Nucleus | 49.48 | 82.61 | −0.438 | 890 | 6083 | 10 |
| Phvul.010G100901.1.v2.1 | PvPUB55 | Class 3 | 6.41 | 51,899.13 | Nucleus | 39.83 | 87.13 | −0.361 | 460 | 2059 | 10 |
| Phvul.010G116400.1.v2.1 | PvPUB56 | Class 2 | 5.65 | 111,409.7 | Cytoplasm | 46.67 | 108.79 | −0.087 | 997 | 6901 | 10 |
| Phvul.011G064400.1.v2.1 | PvPUB57 | Class 3 | 6.12 | 90,165.7 | Nucleus | 48.52 | 83.89 | −0.3 | 800 | 7113 | 11 |
| Phvul.011G064700.1.v2.1 | PvPUB58 | Class 2 | 5.44 | 72,823.32 | Endoplasmic Reticulum | 36.28 | 99.45 | −0.217 | 670 | 4996 | 11 |
| Phvul.011G099300.1.v2.1 | PvPUB59 | Class 1 | 9.31 | 46,252.98 | Cytoplasm | 33.11 | 100.59 | −0.069 | 410 | 1232 | 11 |
| Phvul.011G099400.1.v2.1 | PvPUB60 | Class 1 | 8.51 | 44,272.07 | Cytoplasm | 42.33 | 110.43 | 0.182 | 399 | 1505 | 11 |
| Phvul.L001610.1.v2.1 | PvPUB61 | Class 2 | 7.02 | 74,759.11 | Chloroplast | 43.97 | 103.61 | 0.072 | 689 | 2069 | scaffold_1097 |
| Phvul.L001616.1.v2.1 | PvPUB62 | Class 1 | 7.22 | 61,447.63 | Nucleus | 57.38 | 91.74 | −0.26 | 562 | 1688 | scaffold_1123 |
| Phvul.L005143.1.v2.1 | PvPUB63 | Class 3 | 6.42 | 91,771 | Chloroplast | 50.92 | 86.97 | −0.27 | 819 | 4320 | scaffold_15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Liu, Z.; She, H.; Xu, Z.; Zhang, H.; Fang, Z.; Qian, W. Genome-Wide Identification and Characterization of U-Box Gene Family Members and Analysis of Their Expression Patterns in Phaseolus vulgaris L. under Cold Stress. Int. J. Mol. Sci. 2024, 25, 7968. https://doi.org/10.3390/ijms25147968
Wang J, Liu Z, She H, Xu Z, Zhang H, Fang Z, Qian W. Genome-Wide Identification and Characterization of U-Box Gene Family Members and Analysis of Their Expression Patterns in Phaseolus vulgaris L. under Cold Stress. International Journal of Molecular Sciences. 2024; 25(14):7968. https://doi.org/10.3390/ijms25147968
Chicago/Turabian StyleWang, Jiawei, Zhiyuan Liu, Hongbing She, Zhaosheng Xu, Helong Zhang, Zhengwu Fang, and Wei Qian. 2024. "Genome-Wide Identification and Characterization of U-Box Gene Family Members and Analysis of Their Expression Patterns in Phaseolus vulgaris L. under Cold Stress" International Journal of Molecular Sciences 25, no. 14: 7968. https://doi.org/10.3390/ijms25147968
APA StyleWang, J., Liu, Z., She, H., Xu, Z., Zhang, H., Fang, Z., & Qian, W. (2024). Genome-Wide Identification and Characterization of U-Box Gene Family Members and Analysis of Their Expression Patterns in Phaseolus vulgaris L. under Cold Stress. International Journal of Molecular Sciences, 25(14), 7968. https://doi.org/10.3390/ijms25147968






