Analysis of the Aging-Related AP2/ERF Transcription Factor Gene Family in Osmanthus fragrans
Abstract
:1. Introduction
2. Results
2.1. Identification Results for AP2/ERF Transcription Factor Family Members in O. fragrans
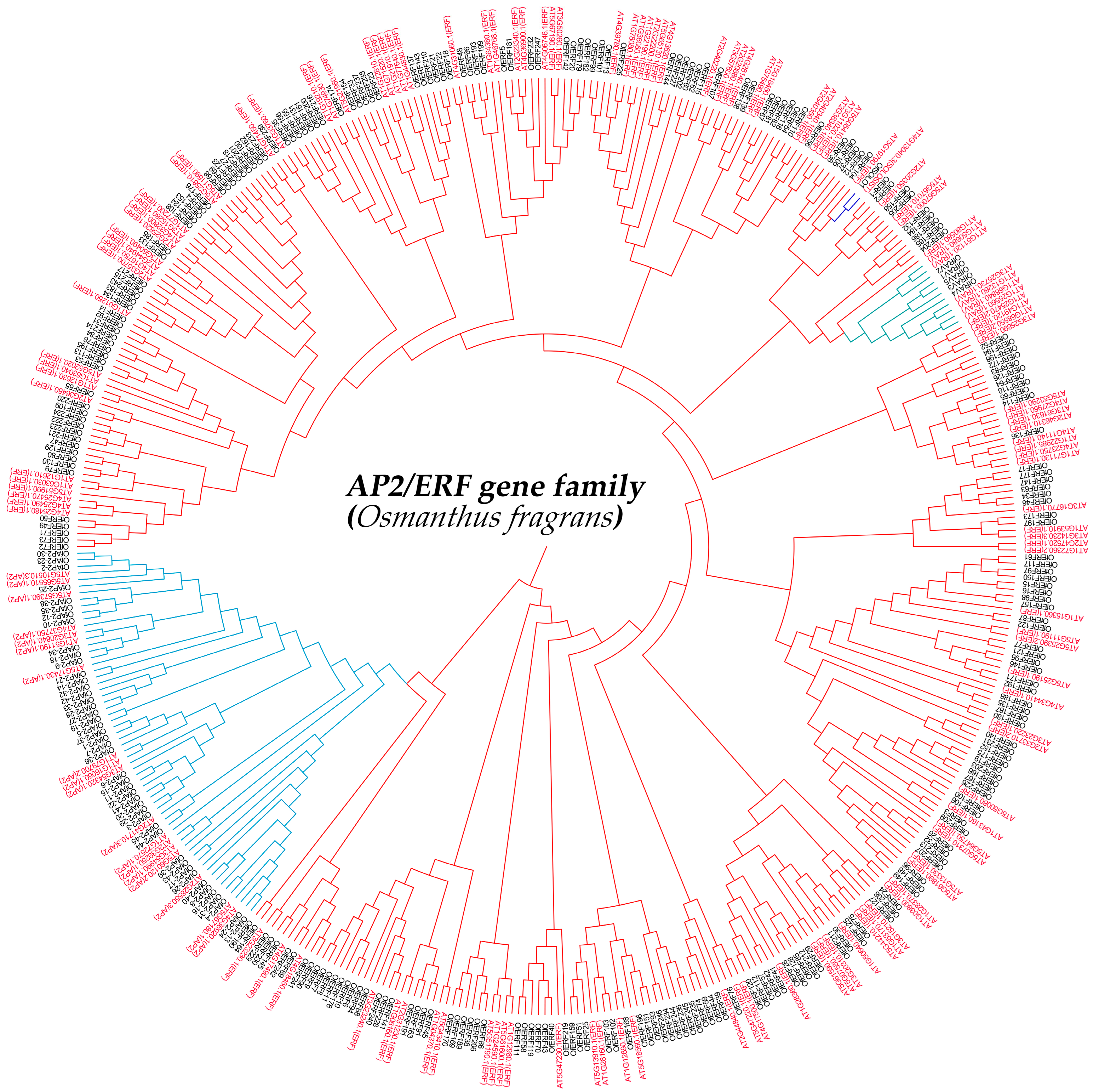
2.2. Phylogenetic Analysis of AP2/ERF Transcription Factor Family Members in O. fragrans
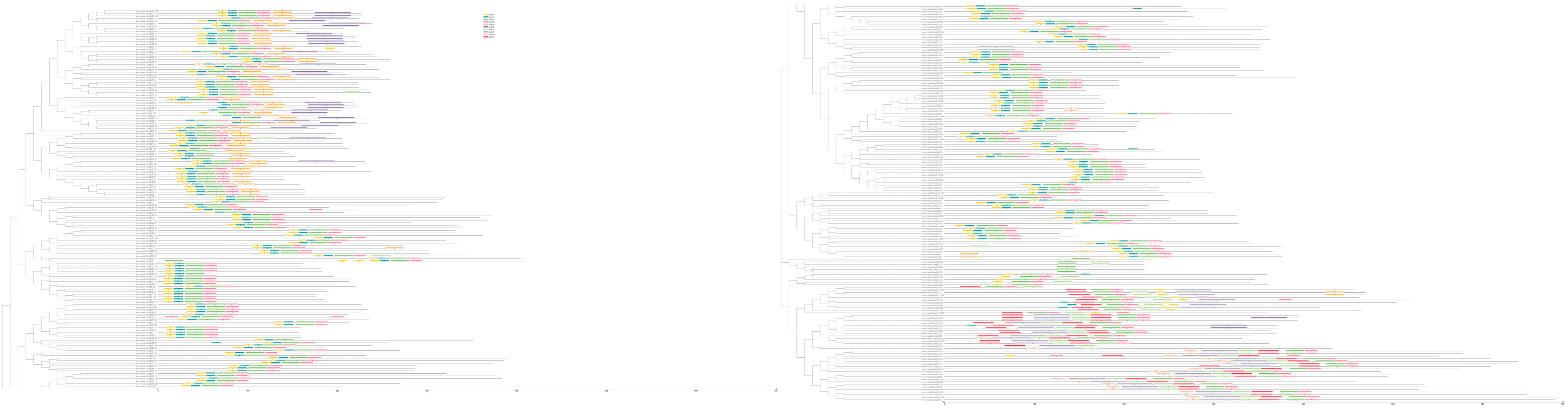
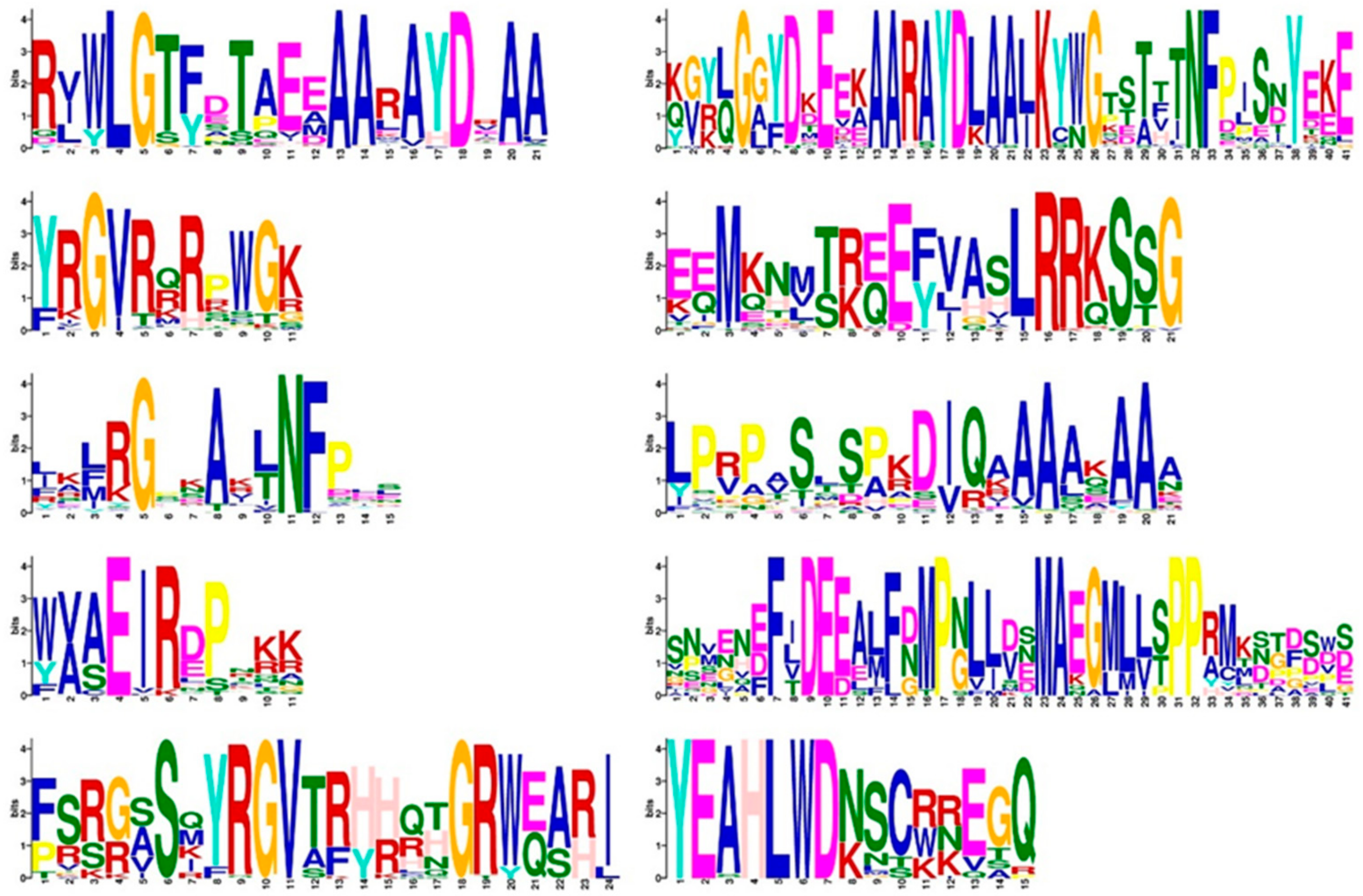
2.3. Analysis of the Gene Structure, Conserved Domain, and Subcellular Localization of AP2/ERF Transcription Factors in O. fragrans
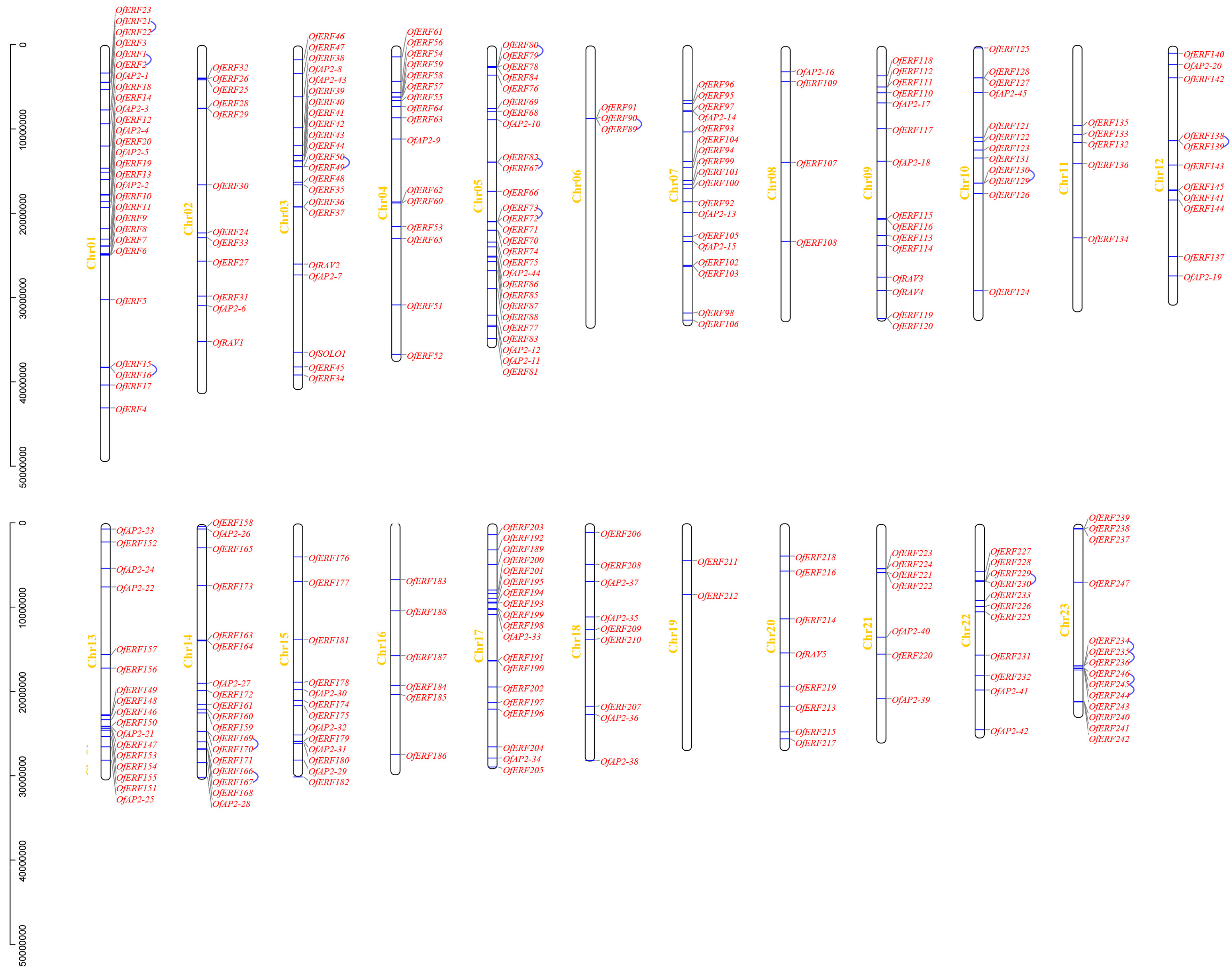
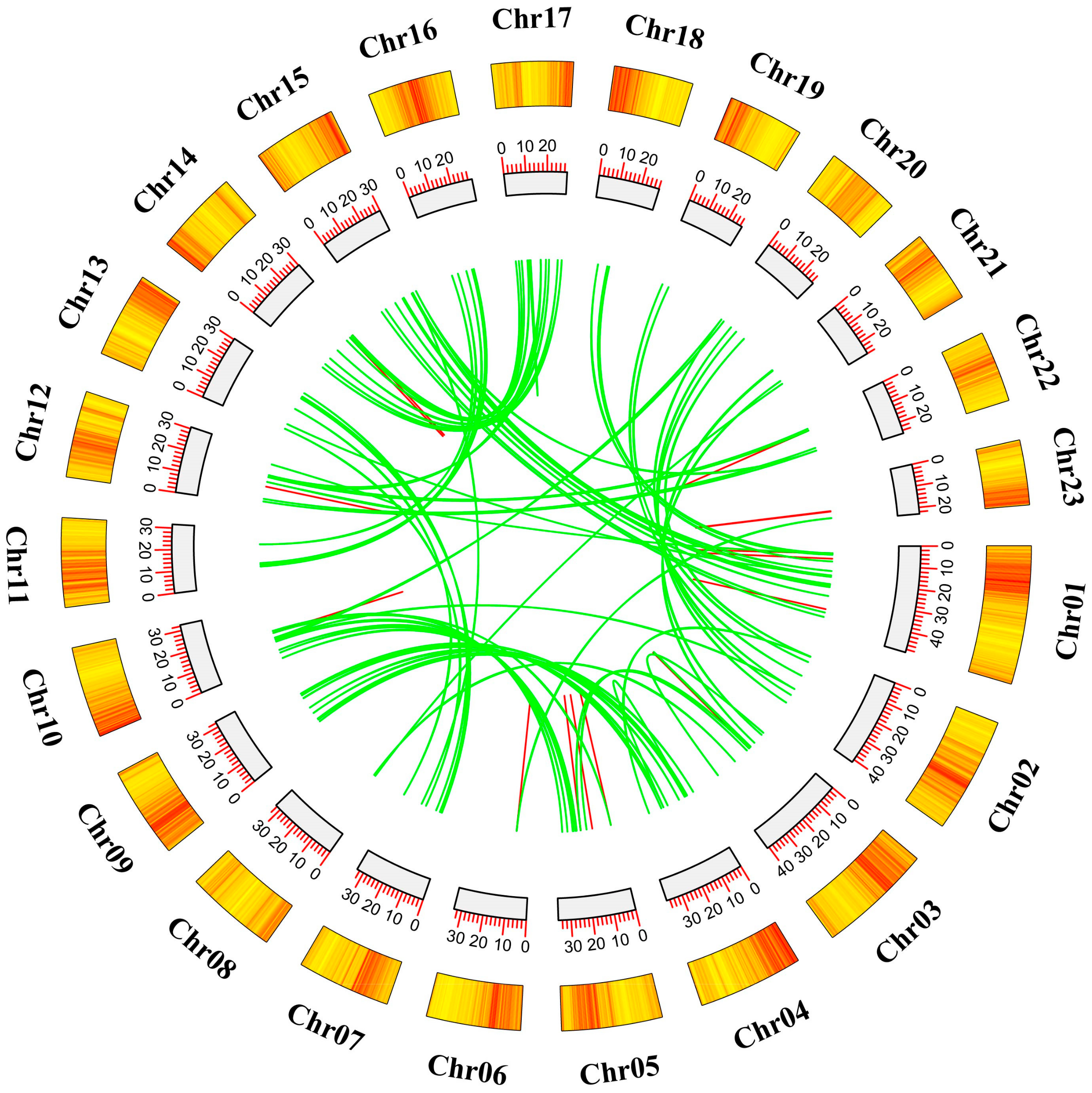
2.4. Analysis of Chromosomal Localization and Duplication Events within AP2/ERF Transcription Factors in O. fragrans
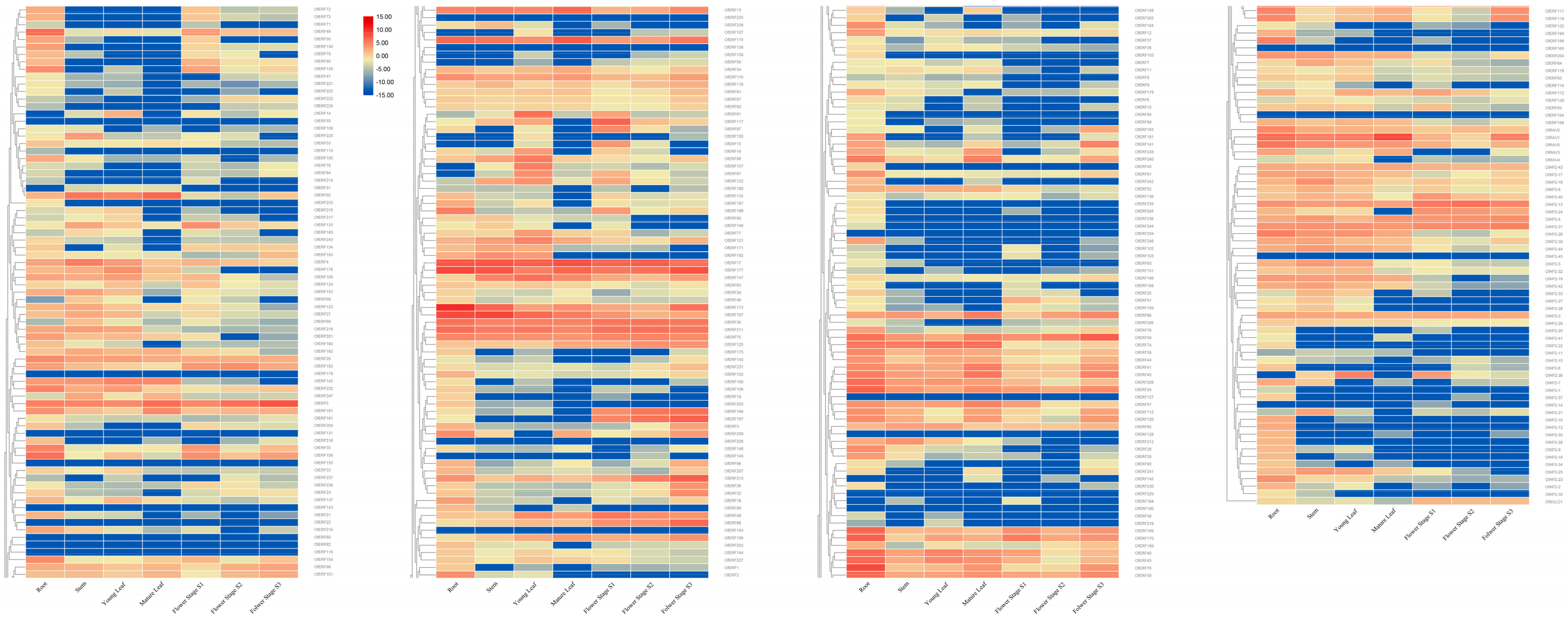
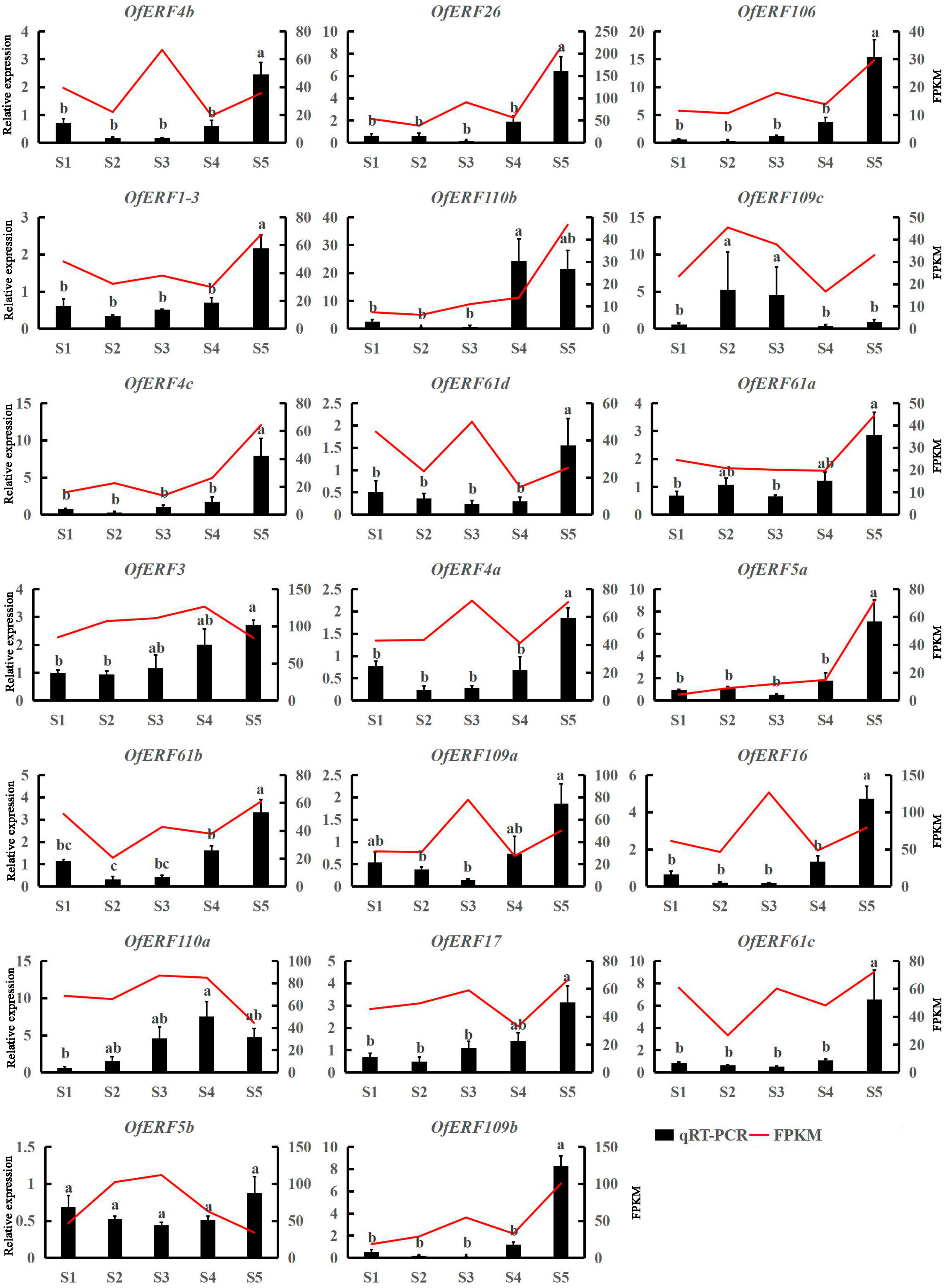
2.5. Expression Analysis Results for the AP2/ERF Gene in O. fragrans
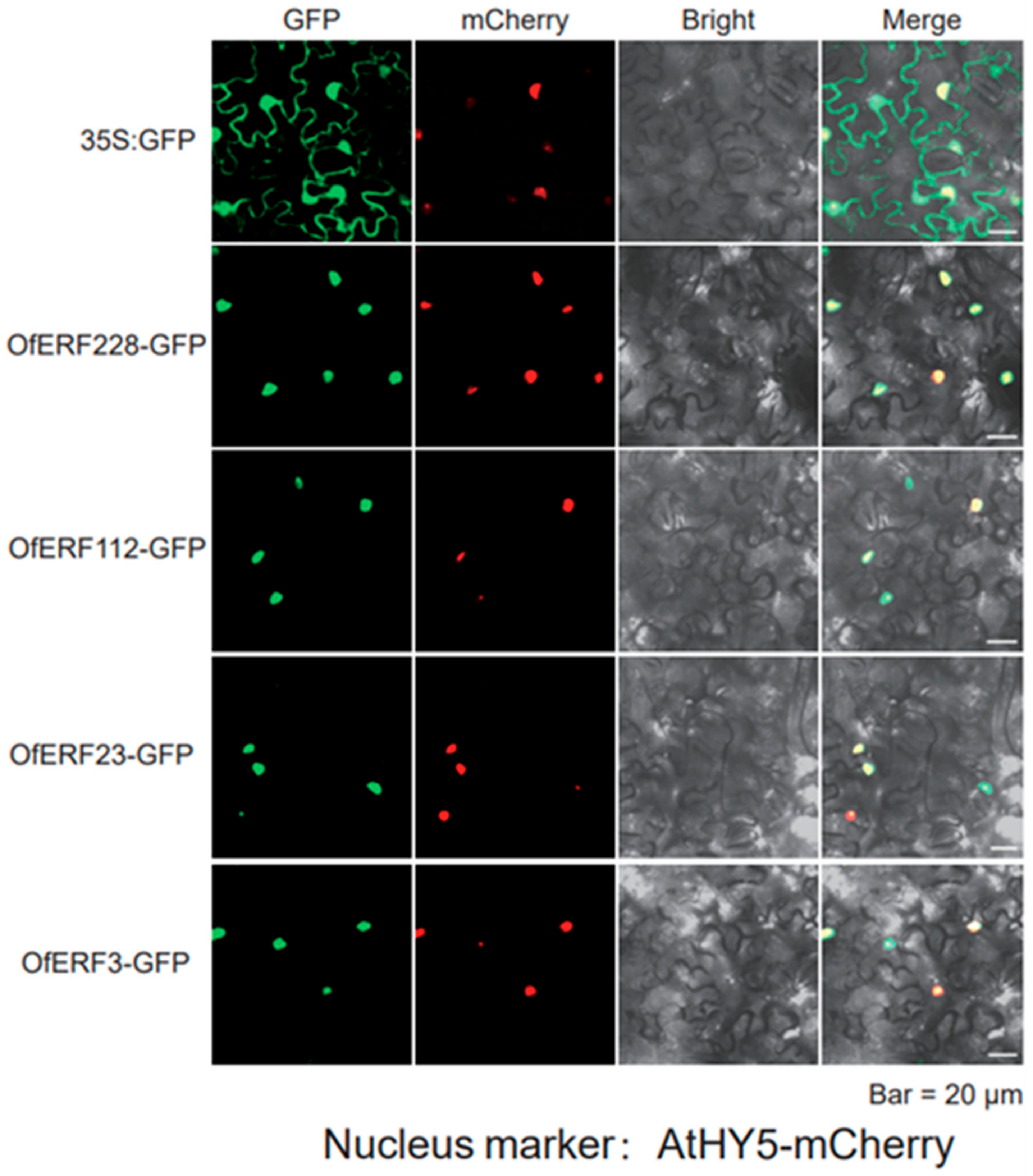
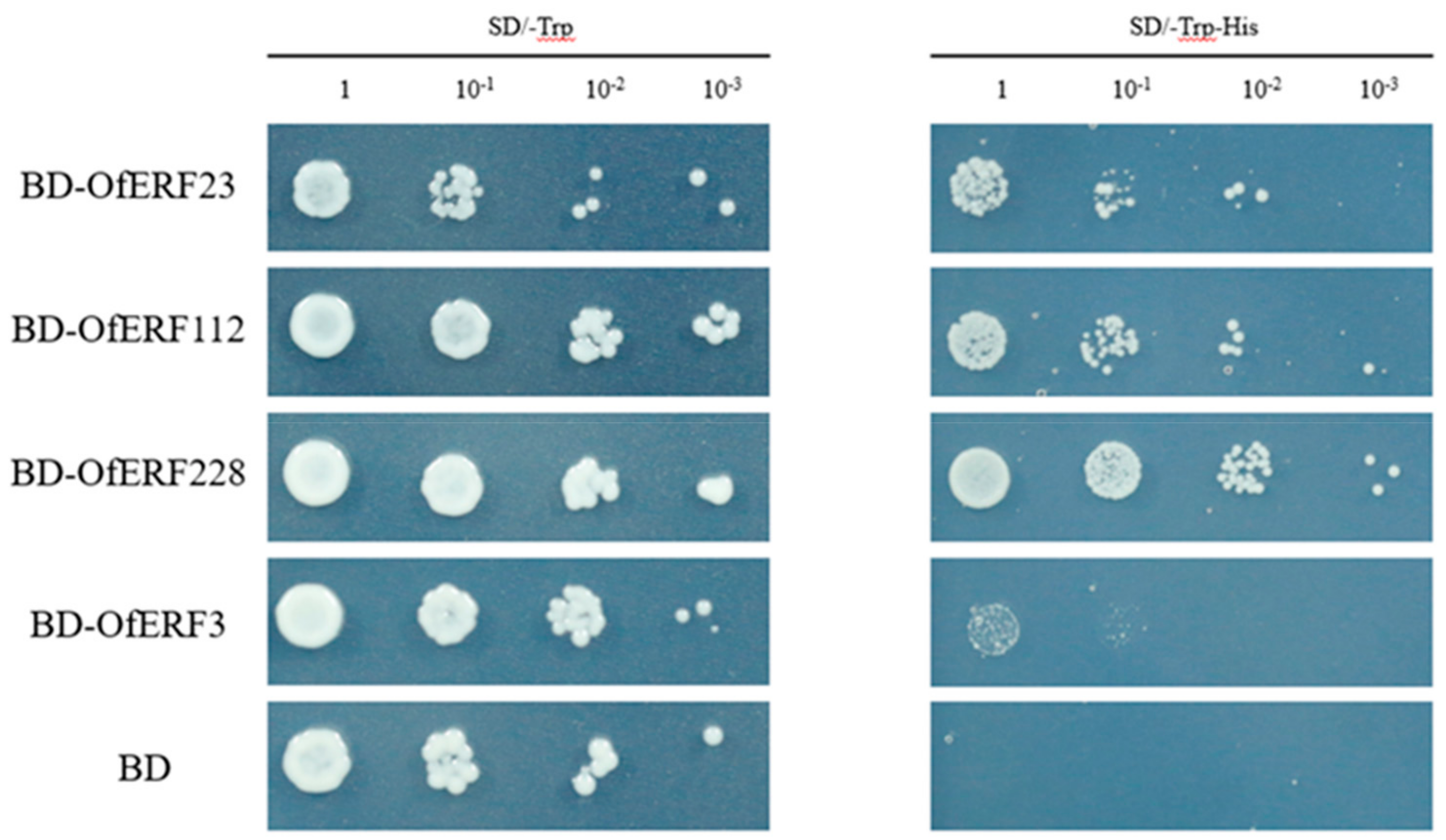
2.6. The Result of Yeast Self-Activation Verification and Subcellular Localization
3. Discussion
3.1. Expression Analysis Results for the AP2/ERF Gene in O. fragrans
3.2. OfERF qRT-PCR and Tissue Specificity
3.3. Gene Function of OfERFs
4. Materials and Methods
4.1. Experimental Materials
4.2. Identification of AP2/ERF Transcription Factor Family Members in the O. fragrans Genome
4.3. Phylogenetic Analysis of AP2/ERF Family Members in O. fragrans
4.4. Gene Structure, Conserved Domain, Conserved Motif, and Subcellular Localization Analysis
4.5. Chromosomal Localization, Gene Replication, and Evolutionary Analysis
4.6. De Novo Assembly of RNA-Seq Reads and Quantification of Gene Expression
4.7. AP2/ERF Gene Expression Analysis
4.8. Subcellular Localization and Yeast Self-Activation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abeles, F.B. Chapter 4—Hormonal and Herbicidal Regulation of Ethylene Production. In Ethylene in Plant Biology; Abeles, F.B., Ed.; Academic Press: San Diego, CA, USA, 1973; pp. 58–86. [Google Scholar]
- Wagstaff, C.; Chanasut, U.; Harren, F.J.M.; Laarhoven, L.-J.; Thomas, B.; Rogers, H.J.; Stead, A.D. Ethylene and flower longevity in Alstroemeria: Relationship between tepal senescence, abscission and ethylene biosynthesis. J. Exp. Bot. 2005, 56, 1007–1016. [Google Scholar] [CrossRef] [PubMed]
- Mizoi, J.; Yamaguchi-Shinozaki, K. Molecular approaches to improve rice abiotic stress tolerance. Methods Mol. Biol. 2013, 956, 269–283. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef] [PubMed]
- Sharoni, A.M.; Nuruzzaman, M.; Satoh, K.; Shimizu, T.; Kondoh, H.; Sasaya, T.; Choi, I.R.; Omura, T.; Kikuchi, S. Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant Cell Physiol. 2011, 52, 344–360. [Google Scholar] [CrossRef]
- Licausi, F.; Giorgi, F.M.; Zenoni, S.; Osti, F.; Pezzotti, M.; Perata, P. Genomic and transcriptomic analysis of the AP2/ERF superfamily in Vitis vinifera. BMC Genom. 2010, 11, 719. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Cai, B.; Peng, R.H.; Zhu, B.; Jin, X.F.; Xue, Y.; Gao, F.; Fu, X.Y.; Tian, Y.S.; Zhao, W.; et al. Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa. Biochem. Biophys. Res. Commun. 2008, 371, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Riaz, M.W.; Lu, J.; Shah, L.; Yang, L.; Chen, C.; Mei, X.D.; Xue, L.; Manzoor, M.A.; Abdullah, M.; Rehman, S.; et al. Expansion and Molecular Characterization of AP2/ERF Gene Family in Wheat (Triticum aestivum L.). Front. Genet. 2021, 12, 632155. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Liu, S. Genome-wide identification and phylogenetic analysis of the ERF gene family in cucumbers. Genet. Mol. Biol. 2011, 34, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Wei, Y.; Xu, R.; Lin, S.; Luan, H.; Lv, C.; Zhang, X.; Song, X.; Xu, R. Genome-Wide Analysis of APETALA2/Ethylene-Responsive Factor (AP2/ERF) Gene Family in Barley (Hordeum vulgare L.). PLoS ONE 2016, 11, e0161322. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ni, D.; Shen, J.; Deng, S.; Xuan, H.; Wang, C.; Xu, J.; Zhou, L.; Guo, N.; Zhao, J.; et al. Genome-Wide Identification of the AP2/ERF Gene Family and Functional Analysis of GmAP2/ERF144 for Drought Tolerance in Soybean. Front. Plant Sci. 2022, 13, 848766. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Wang, Y.; Li, X.; Gao, F.; Feng, J.; Zhou, Y. Characterization of the AP2/ERF Transcription Factor Family and Expression Profiling of DREB Subfamily under Cold and Osmotic Stresses in Ammopiptanthus nanus. Plants 2020, 9, 455. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.; Dai, L.J.; Li, X.; Li, W.; Qu, G.Z.; Li, S. Genome-Wide Identification of the ERF Transcription Factor Family for Structure Analysis, Expression Pattern, and Response to Drought Stress in Populus alba × Populus glandulosa. Int. J. Mol. Sci. 2023, 24, 3697. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Hou, X.L.; Xing, G.M.; Liu, J.X.; Duan, A.Q.; Xu, Z.S.; Li, M.Y.; Zhuang, J.; Xiong, A.S. Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Meyerowitz, E.M. The AP2/EREBP family of plant transcription factors. Biol. Chem. 1998, 379, 633–646. [Google Scholar] [CrossRef] [PubMed]
- Scarpeci, T.E.; Frea, V.S.; Zanor, M.I.; Valle, E.M. Overexpression of AtERF019 delays plant growth and senescence, and improves drought tolerance in Arabidopsis. J. Exp. Bot. 2017, 68, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, J.; Wang, H.; Fu, Z.; Liu, J.; Yu, Y. Identification and expression analysis of ERF transcription factor genes in petunia during flower senescence and in response to hormone treatments. J. Exp. Bot. 2011, 62, 825–840. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Liu, Y.; Liang, Y.; Lu, J.; Jiang, C.; Fei, Z.; Jiang, C.Z.; Ma, C.; Gao, J. Rosa hybrida RhERF1 and RhERF4 mediate ethylene- and auxin-regulated petal abscission by influencing pectin degradation. Plant J. 2019, 99, 1159–1171. [Google Scholar] [CrossRef] [PubMed]
- Jofuku, K.D.; den Boer, B.G.; Van Montagu, M.; Okamuro, J.K. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 1994, 6, 1211–1225. [Google Scholar] [CrossRef] [PubMed]
- Kim, J. CYTOKININ RESPONSE FACTORs Gating Environmental Signals and Hormones. Trends Plant Sci. 2016, 21, 993–996. [Google Scholar] [CrossRef]
- Zhuang, J.; Deng, D.X.; Yao, Q.H.; Zhang, J.; Xiong, F.; Chen, J.M.; Xiong, A.S. Discovery, phylogeny and expression patterns of AP2-like genes in maize. Plant Growth Regul. 2010, 62, 51–58. [Google Scholar] [CrossRef]
- Li, M.Y.; Liu, J.X.; Hao, J.N.; Feng, K.; Duan, A.Q.; Yang, Q.Q.; Xu, Z.S.; Xiong, A.S. Genomic identification of AP2/ERF transcription factors and functional characterization of two cold resistance-related AP2/ERF genes in celery (Apium graveolens L.). Planta 2019, 250, 1265–1280. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; White, M.J.; MacRae, T.H. Transcription factors and their genes in higher plants functional domains, evolution and regulation. Eur. J. Biochem. 1999, 262, 247–257. [Google Scholar] [CrossRef]
- Zhang, J.; Liao, J.; Ling, Q.; Xi, Y.; Qian, Y. Genome-wide identification and expression profiling analysis of maize AP2/ERF superfamily genes reveal essential roles in abiotic stress tolerance. BMC Genom. 2022, 23, 125. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Tang, W.; Zhou, Y.; Chen, J.; Xu, Z.; Ma, R.; Dong, Y.; Ma, Y.; Chen, M. AP2/ERF transcription factor GmDREB1 confers drought tolerance in transgenic soybean by interacting with GmERFs. Plant Physiol. Biochem. 2022, 170, 287–295. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, X. Genome-wide identification of AP2/ERF superfamily genes and their expression during fruit ripening of Chinese jujube. Sci. Rep. 2018, 8, 15612. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Peng, R.-H.; Cheng, Z.-M.; Zhang, J.; Cai, B.; Zhang, Z.; Gao, F.; Zhu, B.; Fu, X.-Y.; Jin, X.-F.; et al. Genome-wide analysis of the putative AP2/ERF family genes in Vitis vinifera. Sci. Hortic. 2009, 123, 73–81. [Google Scholar] [CrossRef]
- Cho, Y.; Qiu, Y.L.; Kuhlman, P.; Palmer, J.D. Explosive invasion of plant mitochondria by a group I intron. Proc. Natl. Acad. Sci. USA 1998, 95, 14244–14249. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Zhu, W.; Silva, J.C.; Gu, X.; Buell, C.R. Intron gain and loss in segmentally duplicated genes in rice. Genome Biol. 2006, 7, R41. [Google Scholar] [CrossRef]
- Song, X.; Li, Y.; Hou, X. Genome-wide analysis of the AP2/ERF transcription factor superfamily in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genom. 2013, 14, 573. [Google Scholar] [CrossRef]
- Xu, W.; Li, F.; Ling, L.; Liu, A. Genome-wide survey and expression profiles of the AP2/ERF family in castor bean (Ricinus communis L.). BMC Genom. 2013, 14, 785. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.W.; Werr, W. A phylogenetically conserved APETALA2/ETHYLENE RESPONSE FACTOR, ERF12, regulates Arabidopsis floral development. Plant Mol. Biol. 2020, 102, 39–54. [Google Scholar] [CrossRef]
- Chen, W.H.; Li, P.F.; Chen, M.K.; Lee, Y.I.; Yang, C.H. FOREVER YOUNG FLOWER Negatively Regulates Ethylene Response DNA-Binding Factors by Activating an Ethylene-Responsive Factor to Control Arabidopsis Floral Organ Senescence and Abscission. Plant Physiol. 2015, 168, 1666–1683. [Google Scholar] [CrossRef] [PubMed]
- Petsch, K.A.; Mylne, J.; Botella, J.R. Cosuppression of eukaryotic release factor 1-1 in Arabidopsis affects cell elongation and radial cell division. Plant Physiol. 2005, 139, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhou, W.; Wang, P.; Chen, Y.; Huo, S.; Wang, J.; Tian, D.; Niu, J.; Zhao, Y.; Song, X. Transcriptome Analysis Reveals the Senescence Process Controlling the Flower Opening and Closure Rhythm in the Waterlilies (Nymphaea L.). Front. Plant Sci. 2021, 12, 701633. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF Transcription Factor Regulatory Networks in Hormone and Abiotic Stress Responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef] [PubMed]
- El-Esawi, M.A.; Al-Ghamdi, A.A.; Ali, H.M.; Ahmad, M. Overexpression of AtWRKY30 Transcription Factor Enhances Heat and Drought Stress Tolerance in Wheat (Triticum aestivum L.). Genes 2019, 10, 163. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Wei, C.; Ma, Q.; Dong, H.; Shi, K.; Zhou, Y.; Foyer, C.H.; Yu, J. Ethylene response factors 15 and 16 trigger jasmonate biosynthesis in tomato during herbivore resistance. Plant Physiol. 2021, 185, 1182–1197. [Google Scholar] [CrossRef]
- Sun, H.Y.; Zhang, W.W.; Qu, H.Y.; Gou, S.S.; Li, L.X.; Song, H.H.; Yang, H.Q.; Li, W.J.; Zhang, H.; Hu, K.D.; et al. Transcriptomics Reveals the ERF2-bHLH2-CML5 Module Responses to H(2)S and ROS in Postharvest Calcium Deficiency Apples. Int. J. Mol. Sci. 2021, 22, 13013. [Google Scholar] [CrossRef]
- Wan, R.; Song, J.; Lv, Z.; Qi, X.; Han, X.; Guo, Q.; Wang, S.; Shi, J.; Jian, Z.; Hu, Q.; et al. Genome-Wide Identification and Comprehensive Analysis of the AP2/ERF Gene Family in Pomegranate Fruit Development and Postharvest Preservation. Genes 2022, 13, 895. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, Y.; Wu, M.; Li, L.; Li, C.; Han, Z.; Yuan, J.; Chen, C.; Song, W.; Wang, C. Genome-Wide Identification of AP2/ERF Transcription Factors in Cauliflower and Expression Profiling of the ERF Family under Salt and Drought Stresses. Front. Plant Sci. 2017, 8, 946. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Song, G.; Zhang, F.; Shu, X.; Wang, N. Functional Characterization of AP2/ERF Transcription Factors during Flower Development and Anthocyanin Biosynthesis Related Candidate Genes in Lycoris. Int. J. Mol. Sci. 2023, 24, 14464. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhu, M.; Liu, W.; Li, J.; Liao, Y.; Liu, D.; Jin, M.; Fu, C.; Wang, F. Bulk segregant analysis coupled with transcriptomics and metabolomics revealed key regulators of bacterial leaf blight resistance in rice. BMC Plant Biol. 2023, 23, 332. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Cheng, S.; Song, Y.; Huang, Y.; Zhou, S.; Liu, X.; Zhou, D.X. The Interaction between Rice ERF3 and WOX11 Promotes Crown Root Development by Regulating Gene Expression Involved in Cytokinin Signaling. Plant Cell 2015, 27, 2469–2483. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yue, Y.; Li, H.; Ding, W.; Chen, G.; Shi, T.; Chen, J.; Park, M.S.; Chen, F.; Wang, L. The chromosome-level quality genome provides insights into the evolution of the biosynthesis genes for aroma compounds of Osmanthus fragrans. Hortic. Res. 2018, 5, 72. [Google Scholar] [CrossRef]
- Finn, R.D.; Mistry, J.; Tate, J.; Coggill, P.; Heger, A.; Pollington, J.E.; Gavin, O.L.; Gunasekaran, P.; Ceric, G.; Forslund, K.; et al. The Pfam protein families database. Nucleic Acids Res. 2010, 38, D211–D222. [Google Scholar] [CrossRef] [PubMed]
- Faraji, S.; Filiz, E.; Kazemitabar, S.K.; Vannozzi, A.; Palumbo, F.; Barcaccia, G.; Heidari, P. The AP2/ERF Gene Family in Triticum durum: Genome-Wide Identification and Expression Analysis under Drought and Salinity Stresses. Genes 2020, 11, 1464. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Ding, W.; Wu, X.; Wang, L.; Yang, X.; Yue, Y. Insights Into the MYB-Related Transcription Factors Involved in Regulating Floral Aroma Synthesis in Sweet Osmanthus. Front. Plant Sci. 2022, 13, 765213. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hu, H.; Shen, H.; Tian, Q.; Ding, W.; Yang, X.; Wang, L.; Yue, Y. Insights into the Cytochrome P450 Monooxygenase Superfamily in Osmanthus fragrans and the Role of OfCYP142 in Linalool Synthesis. Int. J. Mol. Sci. 2022, 23, 12150. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, G.; Shao, T.; Zhou, Y.; Chen, F.; Zhang, D.; Gu, H.; Yue, Y.; Wang, L.; Yang, X. Analysis of the Aging-Related AP2/ERF Transcription Factor Gene Family in Osmanthus fragrans. Int. J. Mol. Sci. 2024, 25, 8025. https://doi.org/10.3390/ijms25158025
Chen G, Shao T, Zhou Y, Chen F, Zhang D, Gu H, Yue Y, Wang L, Yang X. Analysis of the Aging-Related AP2/ERF Transcription Factor Gene Family in Osmanthus fragrans. International Journal of Molecular Sciences. 2024; 25(15):8025. https://doi.org/10.3390/ijms25158025
Chicago/Turabian StyleChen, Gongwei, Tianqi Shao, Yixiao Zhou, Fengyuan Chen, Dandan Zhang, Heng Gu, Yuanzheng Yue, Lianggui Wang, and Xiulian Yang. 2024. "Analysis of the Aging-Related AP2/ERF Transcription Factor Gene Family in Osmanthus fragrans" International Journal of Molecular Sciences 25, no. 15: 8025. https://doi.org/10.3390/ijms25158025
APA StyleChen, G., Shao, T., Zhou, Y., Chen, F., Zhang, D., Gu, H., Yue, Y., Wang, L., & Yang, X. (2024). Analysis of the Aging-Related AP2/ERF Transcription Factor Gene Family in Osmanthus fragrans. International Journal of Molecular Sciences, 25(15), 8025. https://doi.org/10.3390/ijms25158025





