Alterations of the Fatty Acid Profile and the Expression of Genes Related to FA Metabolism in Cirrhotic Liver Tissue
Abstract
:1. Introduction
2. Results
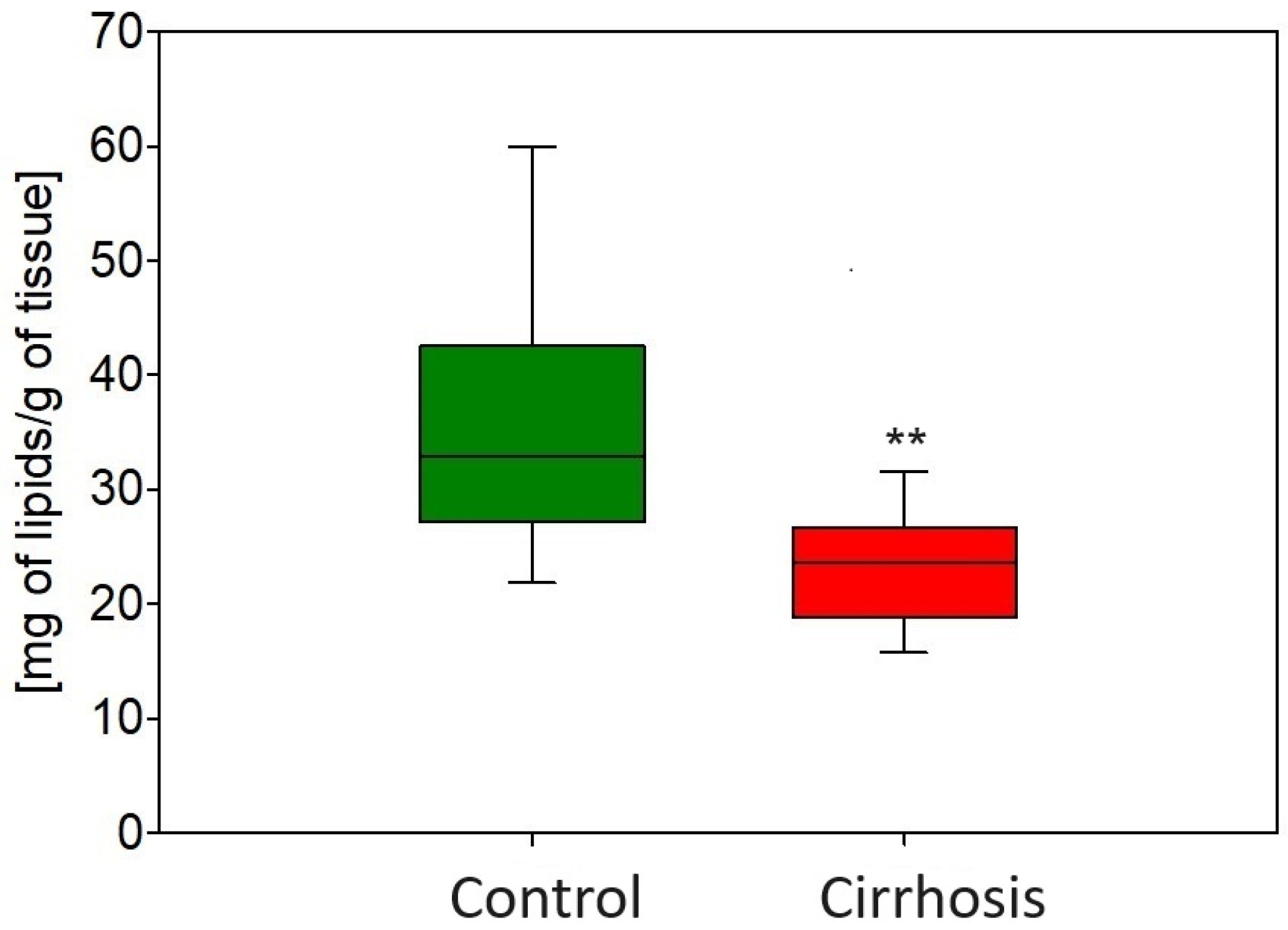
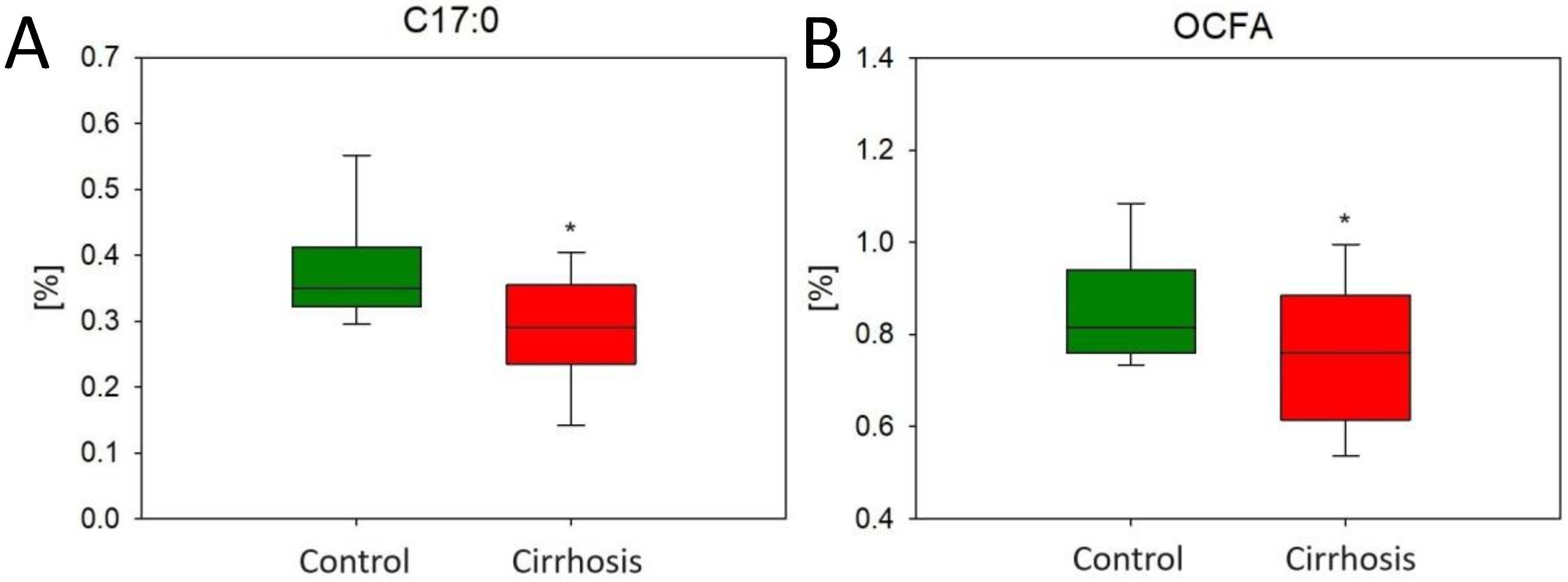
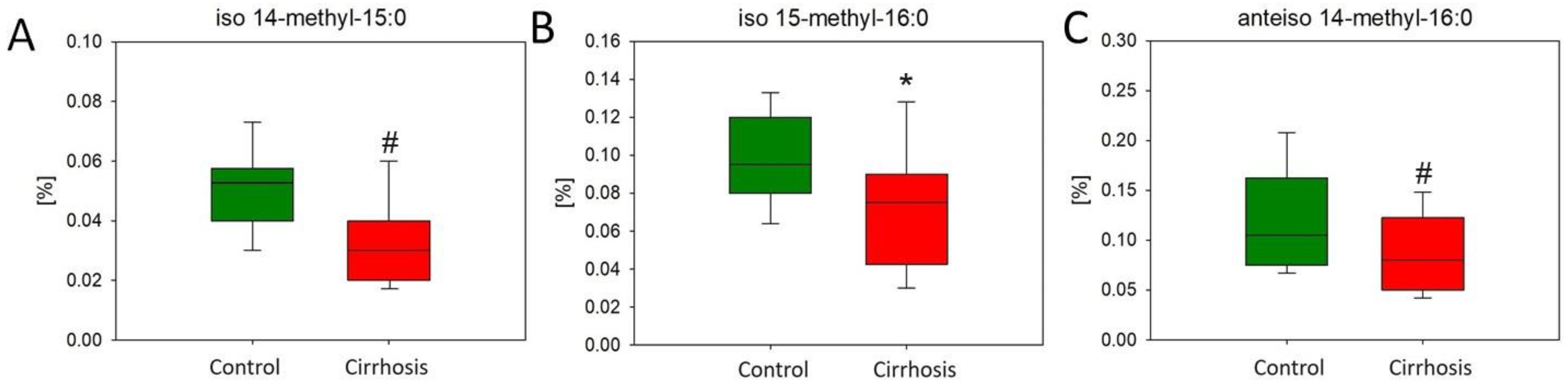
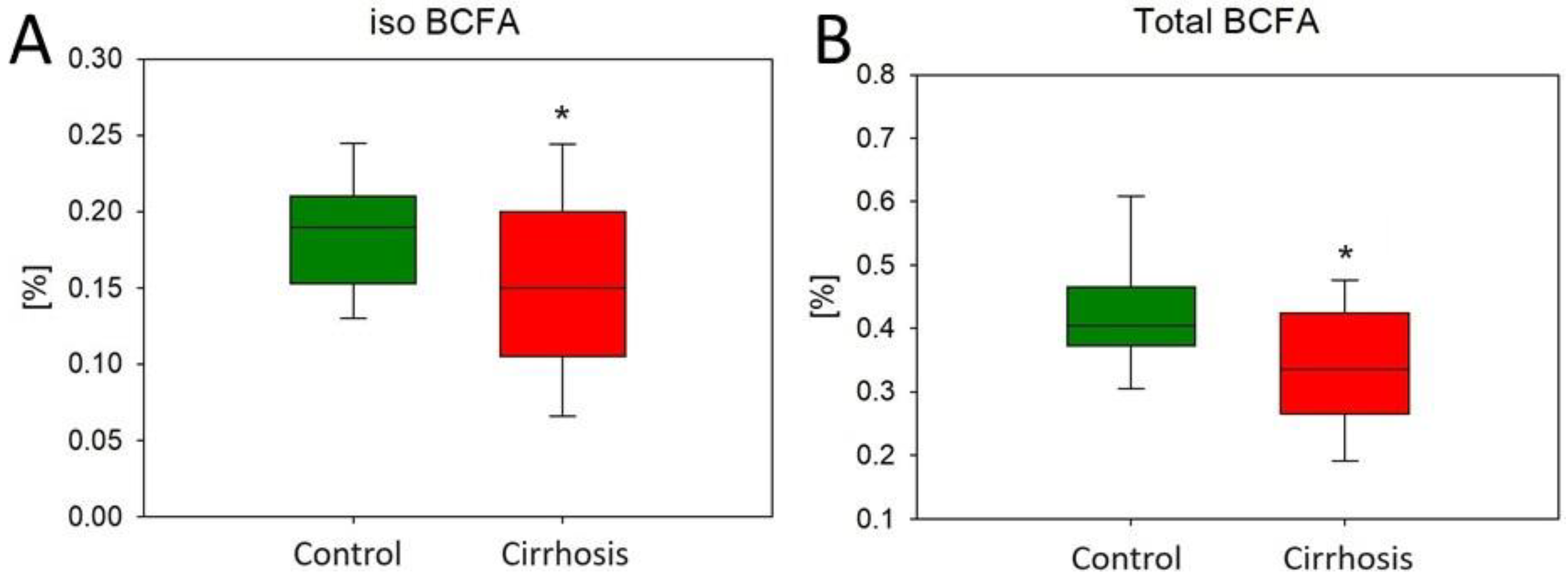
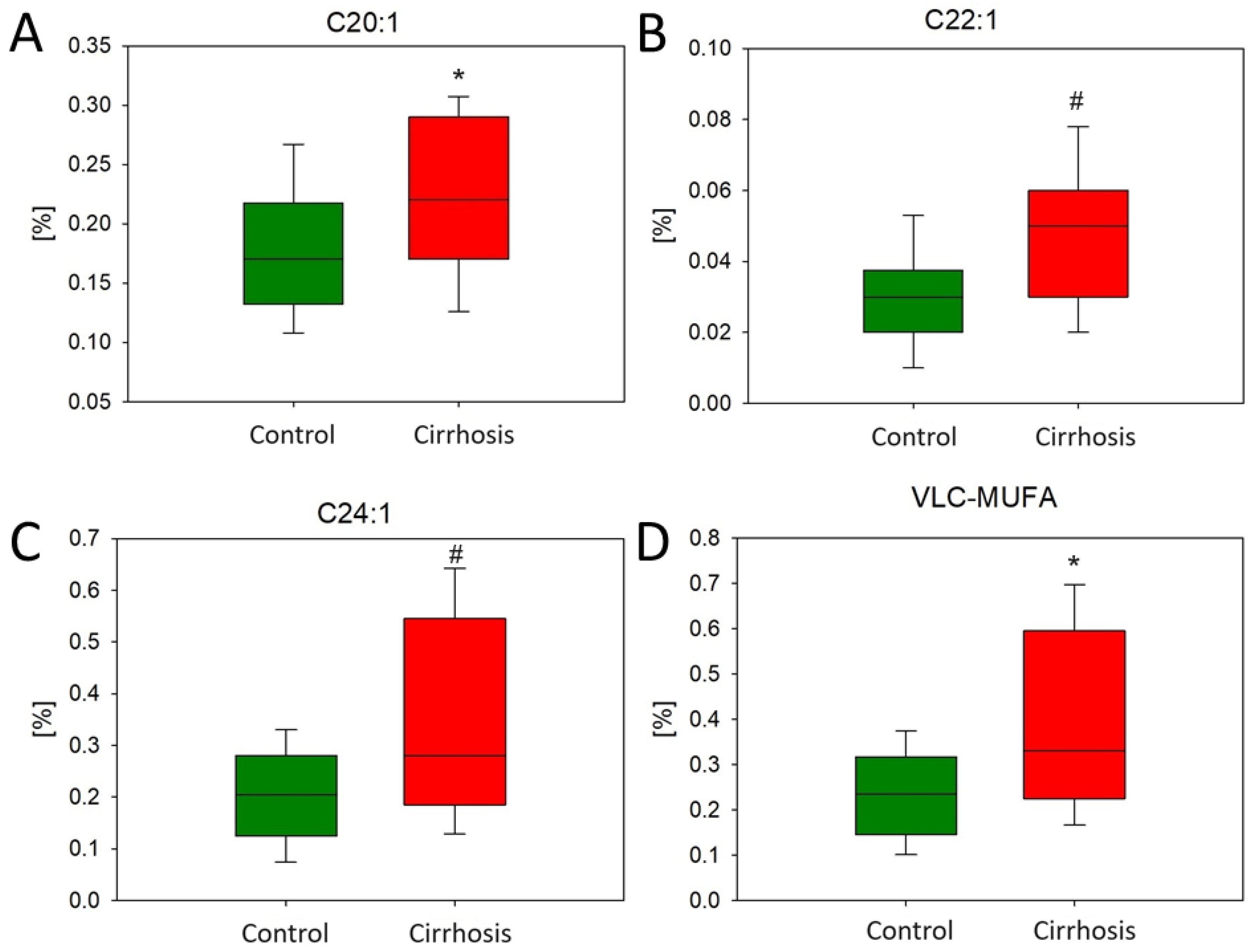
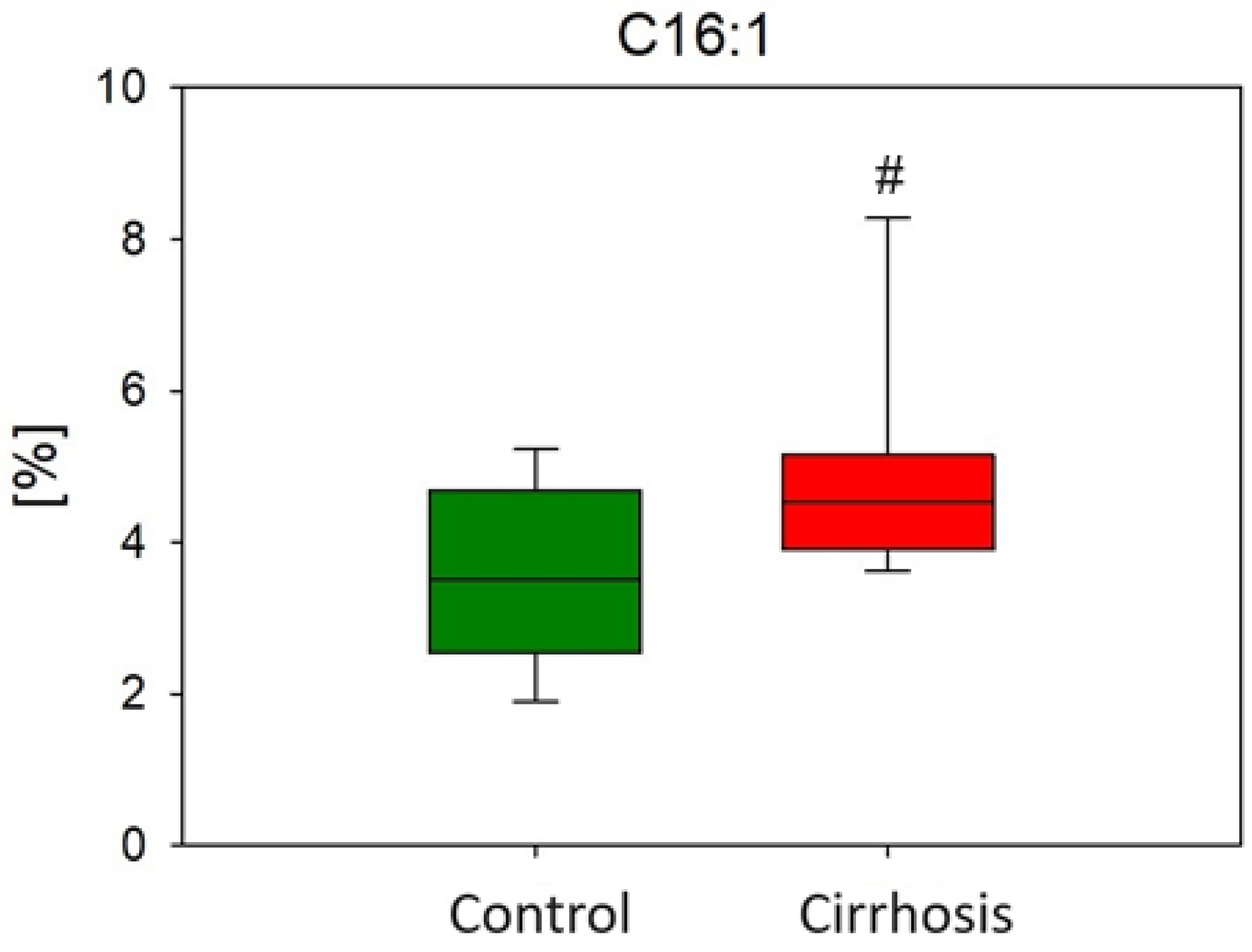
2.1. Effect of Cirrhosis on Hepatic FA Profile
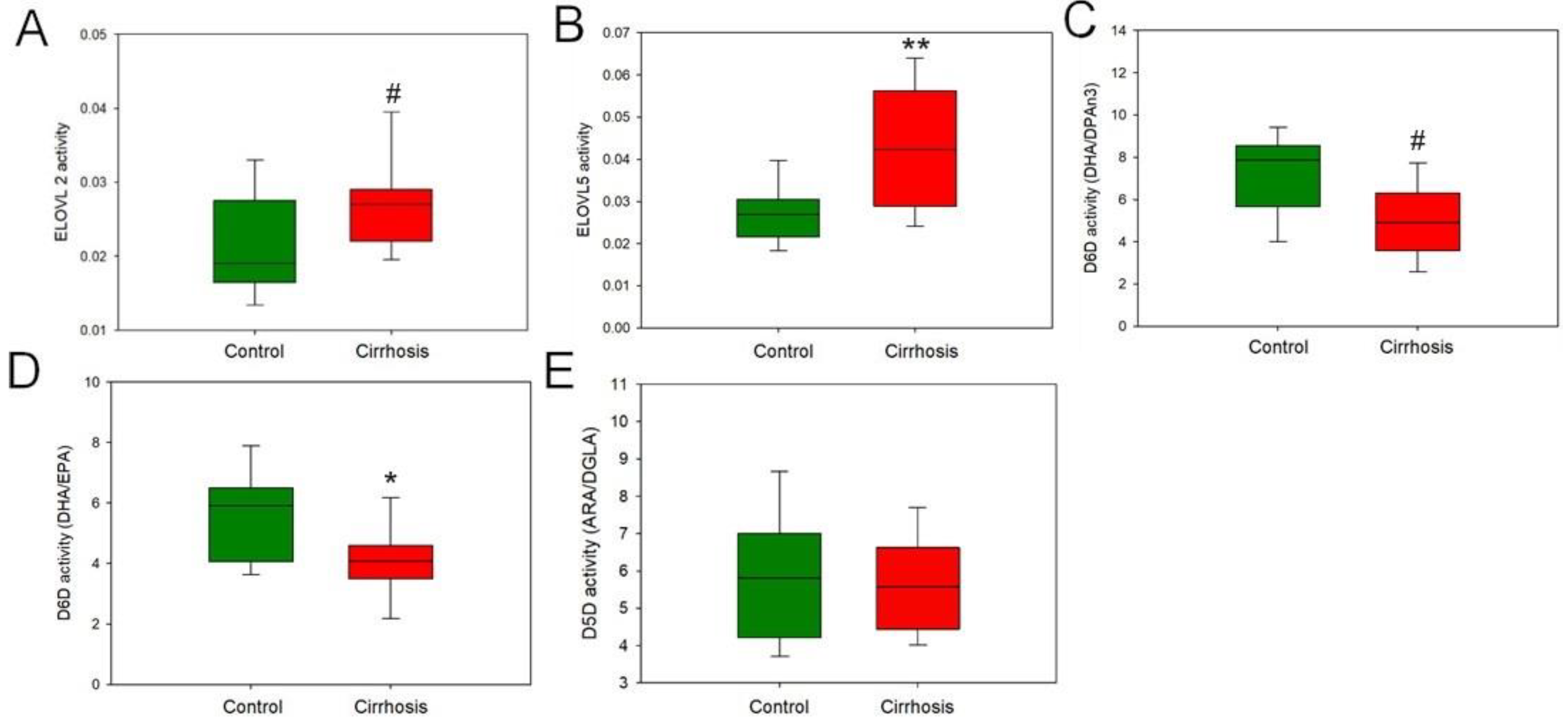
2.2. Effect of Cirrhosis on Hepatic FA Metabolism
3. Discussion
4. Materials and Methods
4.1. Subjects
4.2. Sample Collection
4.3. Sample Preparation
4.4. Gas Chromatography–Mass Spectrometry Analysis
4.5. Analysis of mRNA Level
4.6. Analysis of Hydroxyproline Content
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ginès, P.; Krag, A.; Abraldes, J.G.; Solà, E.; Fabrellas, N.; Kamath, P.S. Liver cirrhosis. Lancet 2021, 398, 1359–1376. [Google Scholar] [CrossRef]
- Singal, A.K.; Bataller, R.; Ahn, J.; Kamath, P.S.; Shah, V.H. ACG Clinical Guideline: Alcoholic Liver Disease. Am. J. Gastroenterol. 2018, 113, 175–194. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.Y.; Tsai, I.T.; Hsu, Y.C. Alcohol-related liver disease: Basic mechanisms and clinical perspectives. Int. J. Mol. Sci. 2021, 22, 5170. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Hu, W.; Tu, J.; Li, J.; Liang, Q.; Han, S. Pathogenic mechanisms and regulatory factors involved in alcoholic liver disease. J. Transl. Med. 2023, 21, 300. [Google Scholar] [CrossRef]
- Hliwa, A.; Ramos-Molina, B.; Laski, D.; Mika, A.; Sledzinski, T. The role of fatty acids in non-alcoholic fatty liver disease progression: An update. Int. J. Mol. Sci. 2021, 22, 6900. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M.; Golabi, P.; Paik, J.M.; Henry, A.; Van Dongen, C.; Henry, L. The global epidemiology of nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH): A systematic review. Hepatology 2023, 77, 1335–1347. [Google Scholar] [CrossRef] [PubMed]
- Eslam, M.; Newsome, P.N.; Sarin, S.K.; Anstee, Q.M.; Targher, G.; Romero-Gomez, M.; Zelber-Sagi, S.; Wai-Sun Wong, V.; Dufour, J.F.; Schattenberg, J.M.; et al. A new definition for metabolic dysfunction-associated fatty liver disease: An international expert consensus statement. J. Hepatol. 2020, 73, 202–209. [Google Scholar] [CrossRef]
- Nguyen, P.; Leray, V.; Diez, M.; Serisier, S.; Le Bloc’H, J.; Siliart, B.; Dumon, H. Liver lipid metabolism. J. Anim. Physiol. Anim. Nutr. 2008, 92, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Puri, P.; Baillie, R.A.; Wiest, M.M.; Mirshahi, F.; Choudhury, J.; Cheung, O.; Sargeant, C.; Contos, M.J.; Sanyal, A.J. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology 2007, 46, 1081–1090. [Google Scholar] [CrossRef]
- Yamada, K.; Mizukoshi, E.; Sunagozaka, H.; Arai, K.; Yamashita, T.; Takeshita, Y.; Misu, H.; Takamura, T.; Kitamura, S.; Zen, Y.; et al. Characteristics of hepatic fatty acid compositions in patients with nonalcoholic steatohepatitis. Liver Int. 2015, 35, 582–590. [Google Scholar] [CrossRef]
- Pawlosky, R.J.; Salem, N. Perspectives on alcohol consumption: Liver polyunsaturated fatty acids and essential fatty acid metabolism. Alcohol 2004, 34, 27–33. [Google Scholar] [CrossRef]
- Gabr, S.A.; Alghadir, A.H.; Sherif, Y.E.; Ghfar, A.A. Hydroxyproline as a Biomarker in Liver Disease. Expo. Health 2016, 2016, 1–21. [Google Scholar] [CrossRef]
- Mozaffarian, D.; Wu, J.H.Y. Omega-3 fatty acids and cardiovascular disease: Effects on risk factors, molecular pathways, and clinical events. J. Am. Coll. Cardiol. 2011, 58, 2047–2067. [Google Scholar] [CrossRef]
- Sehgal, R.; Perfilyev, A.; Männistö, V.; Ågren, J.; Nilsson, E.; Käkelä, P.; Ling, C.; de Mello, V.D.; Pihlajamäki, J. Liver saturated fat content associates with hepatic DNA methylation in obese individuals. Clin. Epigenet. 2023, 15, 21. [Google Scholar] [CrossRef]
- Rada, P.; González-Rodríguez, Á.; García-Monzón, C.; Valverde, Á.M. Understanding lipotoxicity in NAFLD pathogenesis: Is CD36 a key driver? Cell Death Dis. 2020, 11, 802. [Google Scholar] [CrossRef]
- Lundbom, J.; Hakkarainen, A.; Söderlund, S.; Westerbacka, J.; Lundbom, N.; Taskinen, M.R. Long-TE 1H MRS suggests that liver fat is more saturated than subcutaneous and visceral fat. NMR Biomed. 2011, 24, 238–245. [Google Scholar] [CrossRef]
- Yan, P.; Luo, Y.; Huang, Z.; Mou, T.; Yang, H.; Peng, D.; Wu, Z. Establishment of a prognostic signature based on fatty acid metabolism genes in HCC associated with hepatitis B. BMC Gastroenterol. 2023, 23, 390. [Google Scholar] [CrossRef]
- Zhang, H.; Axinbai, M.; Zhao, Y.; Wei, J.; Qu, T.; Kong, J.; He, Y.; Zhang, L. Bioinformatics analysis of ferroptosis-related genes and immune cell infiltration in non-alcoholic fatty liver disease. Eur. J. Med. Res. 2023, 28, 605. [Google Scholar] [CrossRef]
- Kris-Etherton, P.M. Monounsaturated Fatty Acids and Risk of Cardiovascular Disease AHA Science Advisory. Circulation 1999, 100, 1253–1258. [Google Scholar] [CrossRef]
- Sarabhai, T.; Kahl, S.; Szendroedi, J.; Markgraf, D.F.; Zaharia, O.P.; Barosa, C.; Herder, C.; Wickrath, F.; Bobrov, P.; Hwang, J.H.; et al. Monounsaturated fat rapidly induces hepatic gluconeogenesis and whole-body insulin resistance. JCI Insight 2020, 5, e134520. [Google Scholar] [CrossRef]
- Imamura, F.; Lemaitre, R.N.; King, I.B.; Song, X.; Steffen, L.M.; Folsom, A.R.; Siscovick, D.S.; Mozaffarian, D. Long-chain monounsaturated fatty acids and incidence of congestive heart failure in 2 prospective cohorts. Circulation 2013, 127, 1512–1521. [Google Scholar] [CrossRef]
- Gillingham, L.G.; Harris-Janz, S.; Jones, P.J.H. Dietary monounsaturated fatty acids are protective against metabolic syndrome and cardiovascular disease risk factors. Lipids 2011, 46, 209–228. [Google Scholar] [CrossRef]
- Jiang, S.; Yang, W.; Li, Y.; Feng, J.; Miao, J.; Shi, H.; Xue, H. Monounsaturated and polyunsaturated fatty acids concerning prediabetes and type 2 diabetes mellitus risk among participants in the National Health and Nutrition Examination Surveys (NHANES) from 2005 to March 2020. Front. Nutr. 2023, 10, 1284800. [Google Scholar] [CrossRef]
- Schwingshackl, L.; Hoffmann, G. Monounsaturated fatty acids, olive oil and health status: A systematic review and meta-analysis of cohort studies. Lipids Health Dis. 2014, 13, 154. [Google Scholar] [CrossRef]
- Yang, Z.H.; Miyahara, H.; Iwasaki, Y.; Takeo, J.; Katayama, M. Dietary supplementation with long-chain monounsaturated fatty acids attenuates obesity-related metabolic dysfunction and increases expression of PPAR gamma in adipose tissue in type 2 diabetic KK-Ay mice. Nutr. Metab. 2013, 10, 16. [Google Scholar] [CrossRef]
- Bozzetto, L.; Prinster, A.; Annuzzi, G.; Costagliola, L.; Mangione, A.; Vitelli, A.; Mazzarella, R.; Longobardo, M.; Mancini, M.; Vigorito, C.; et al. Liver Fat Is Reduced by an Isoenergetic MUFA Diet in a Controlled Randomized Study in Type 2 Diabetic Patients. Diabetes Care 2012, 35, 1429–1435. [Google Scholar] [CrossRef]
- Gozdzik, P.; Magkos, F.; Sledzinski, T.; Mika, A. Monomethyl branched-chain fatty acids: Health effects and biological mechanisms. Prog. Lipid Res. 2023, 90, 101226. [Google Scholar] [CrossRef]
- Yan, Y.; Wang, Z.; Greenwald, J.; Kothapalli, K.S.D.; Park, H.G.; Liu, R.; Mendralla, E.; Lawrence, P.; Wang, X.; Brenna, J.T. BCFA suppresses LPS induced IL-8 mRNA expression in human intestinal epithelial cells. Prostaglandins. Leukot. Essent. Fatty Acids 2017, 116, 27–31. [Google Scholar] [CrossRef]
- Jeyakumar, S.M.; Vajreswari, A. Stearoyl-CoA desaturase 1: A potential target for non-alcoholic fatty liver disease?—Perspective on emerging experimental evidence. World J. Hepatol. 2022, 14, 168–179. [Google Scholar] [CrossRef]
- Du, T.Y.; Gao, Y.X.; Zheng, Y.S. Identification of key genes related to immune infiltration in cirrhosis via bioinformatics analysis. Sci. Rep. 2023, 13, 1876. [Google Scholar] [CrossRef]
- Paul, B.; Lewinska, M.; Andersen, J.B. Lipid alterations in chronic liver disease and liver cancer. JHEP Rep. 2022, 4, 100479. [Google Scholar] [CrossRef]
- You, M.; Fischer, M.; Deeg, M.A.; Crabb, D.W. Ethanol Induces Fatty Acid Synthesis Pathways by Activation of Sterol Regulatory Element-binding Protein (SREBP). J. Biol. Chem. 2002, 277, 29342–29347. [Google Scholar] [CrossRef]
- Ipsen, D.H.; Skat-Rørdam, J.; Tsamouri, M.M.; Latta, M.; Lykkesfeldt, J.; Tveden-Nyborg, P. Molecular drivers of non-alcoholic steatohepatitis are sustained in mild-to-late fibrosis progression in a guinea pig model. Mol. Genet. Genom. 2019, 294, 649–661. [Google Scholar] [CrossRef]
- Samarasinghe, S.M.; Hewage, A.S.; Siriwardana, R.C.; Tennekoon, K.H.; Niriella, M.A.; De Silva, S. Genetic and metabolic aspects of non-alcoholic fatty liver disease (NAFLD) pathogenicity. Egypt. J. Med. Hum. Genet. 2023, 24, 53. [Google Scholar] [CrossRef]
- Cootway, D.; Kalyesubula, M.; Miller, J.; Huff, H.; Lefers, L.; Christofi, V.P.; Anderson, E.; Ntambi, J. SCD1 deficiency promotes hepatic fibrosis and cirrhosis under a high carbohydrate low-fat diet. J. Biol. Chem. 2024, 300, 106471. [Google Scholar] [CrossRef]
- Shimomura, Y.; Honda, T.; Goto, H.; Nonami, T.; Kurokawa, T.; Nagasaki, M.; Murakami, T. Effects of liver failure on the enzymes in the branched-chain amino acid catabolic pathway. Biochem. Biophys. Res. Commun. 2004, 313, 381–385. [Google Scholar] [CrossRef]
- Kyritsi, K.; Wu, N.; Zhou, T.; Carpino, G.; Baiocchi, L.; Kennedy, L.; Chen, L.; Ceci, L.; Meyer, A.A.; Barupala, N.; et al. Knockout of secretin ameliorates biliary and liver phenotypes during alcohol-induced hepatotoxicity. Cell Biosci. 2023, 13, 5. [Google Scholar] [CrossRef]
- Han, J.; Lin, K.; Zhang, X.; Yan, L.; Chen, Y.; Chen, H.; Liu, J.; Liu, J.; Wu, Y. PTEN-mediated AKT/β-catenin signaling enhances the proliferation and expansion of Lgr5+ hepatocytes. Int. J. Biol. Sci. 2021, 17, 861–868. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]






| Parameter | Hepatic Control | Patients with Cirrhosis | p Value |
|---|---|---|---|
| Age (years) | 57.9 ± 17.6 | 50.6 ± 11.3 | NS |
| BMI (kg/m3) | 27.5 (23.5; 32.0) | 26.0 (23.5; 32.5) | NS |
| MELD-Na | 7.00 ± 1.63 | 17.0 ± 7.27 | 0.012 * |
| Total cholesterol (mg/dL) | 151 ± 38.1 | 138 ± 50.9 | NS |
| LDL-C (mg/dL) | 80.9 ± 27.9 | 90.0 ± 40.3 | NS |
| HDL-C (mg/dL) | 51.2 ± 15.2 | 35.0 ± 13.3 | 0.017 * |
| TAG (mg/dL) | 98.0 ± 43.9 | 91.7 ± 24.7 | NS |
| Glucose (mg/dL) | 96.0 (89.5; 110) | 101 (93.0; 116) | NS |
| Bilirubin (mg/dL) | 0.58 (0.40; 0.75) | 2.0 (1.16; 4.05) | <0.001 ## |
| GGTP (IU/L) | 43.0 (24.0; 96.0) | 79.0 (47.0; 238) | NS |
| ALT (U/L) | 20.0 (17.0; 41.0) | 36.0 (13.0; 72.0) | NS |
| AST (U/L) | 25.5 (19.0; 37.8) | 51.0 (28.5; 90.5) | 0.033 # |
| WBC (109/l) | 7.37 (5.66; 8.87) | 5.46 (3.56; 7.54) | 0.022 # |
| ALP (U/L) | 76.0 (66.5; 113) | 116 (91.3; 218) | 0.034 # |
| Creatinine (mg/dL) | 0.81 (0.69; 0.94) | 0.82 (0.67; 1.25) | NS |
| Albumin (g/L) | 38.6 ± 3.75 | 33.2 ± 6.81 | 0.028 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hliwa, A.; Lange-Andrzejewska, O.; Laski, D.; Sledzinski, M.; Remiszewski, P.; Drobinska, A.; Mika, A.; Sledzinski, T. Alterations of the Fatty Acid Profile and the Expression of Genes Related to FA Metabolism in Cirrhotic Liver Tissue. Int. J. Mol. Sci. 2024, 25, 8115. https://doi.org/10.3390/ijms25158115
Hliwa A, Lange-Andrzejewska O, Laski D, Sledzinski M, Remiszewski P, Drobinska A, Mika A, Sledzinski T. Alterations of the Fatty Acid Profile and the Expression of Genes Related to FA Metabolism in Cirrhotic Liver Tissue. International Journal of Molecular Sciences. 2024; 25(15):8115. https://doi.org/10.3390/ijms25158115
Chicago/Turabian StyleHliwa, Aleksandra, Oliwia Lange-Andrzejewska, Dariusz Laski, Maciej Sledzinski, Piotr Remiszewski, Anna Drobinska, Adriana Mika, and Tomasz Sledzinski. 2024. "Alterations of the Fatty Acid Profile and the Expression of Genes Related to FA Metabolism in Cirrhotic Liver Tissue" International Journal of Molecular Sciences 25, no. 15: 8115. https://doi.org/10.3390/ijms25158115






