SingleNucleotide Polymorphisms as Biomarkers of Mepolizumab and Benralizumab Treatment Response in Severe Eosinophilic Asthma
Abstract
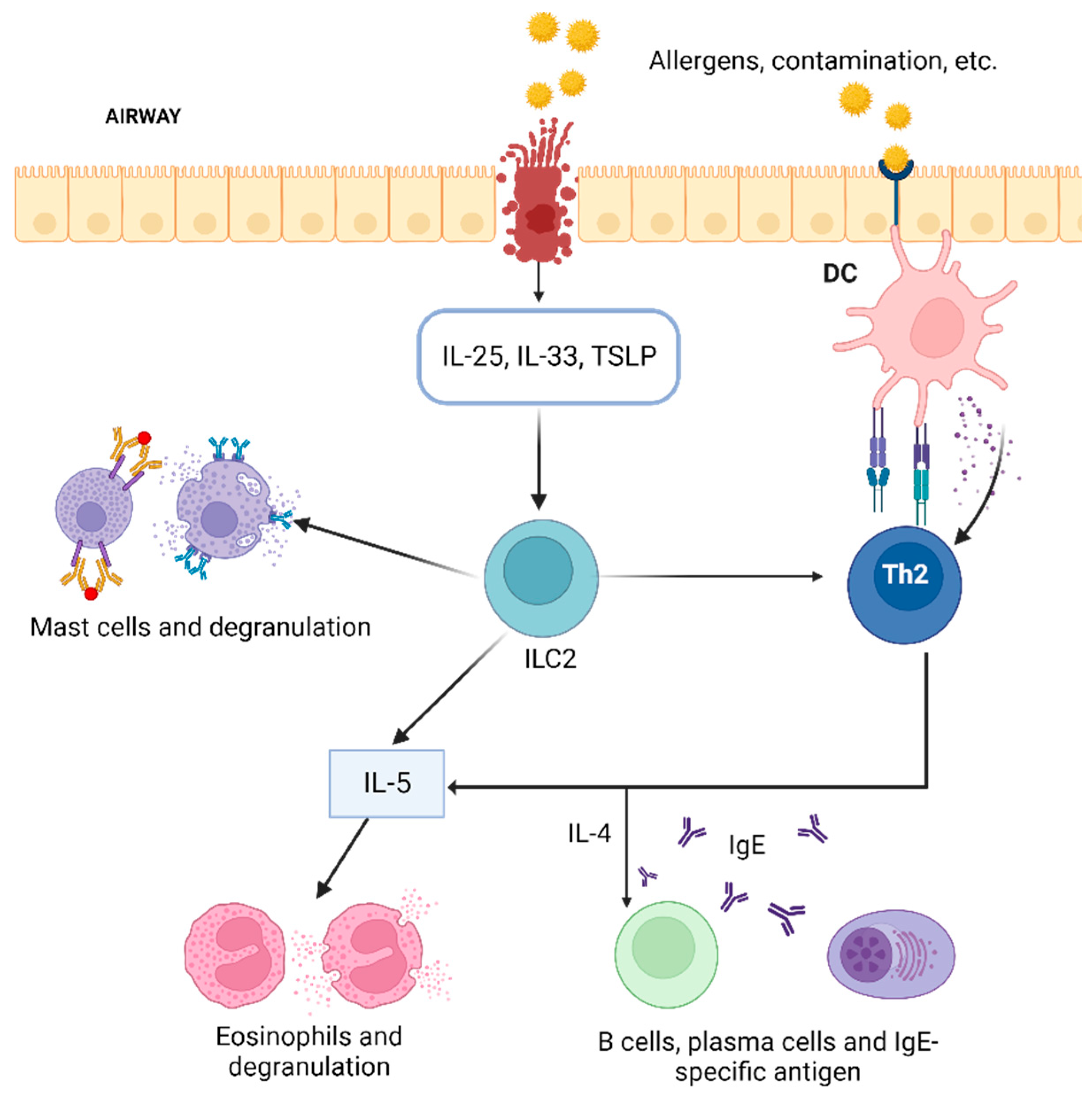
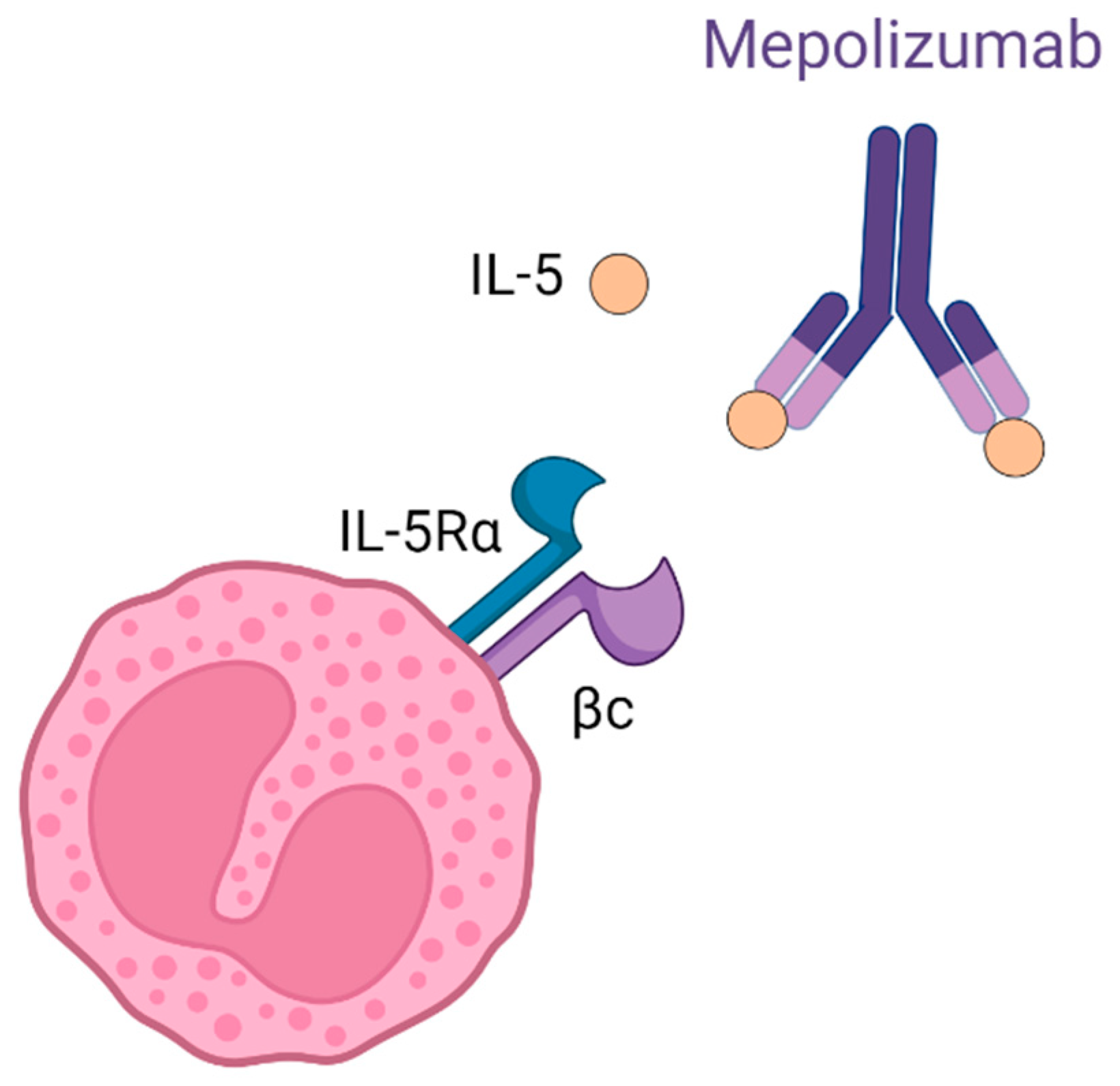
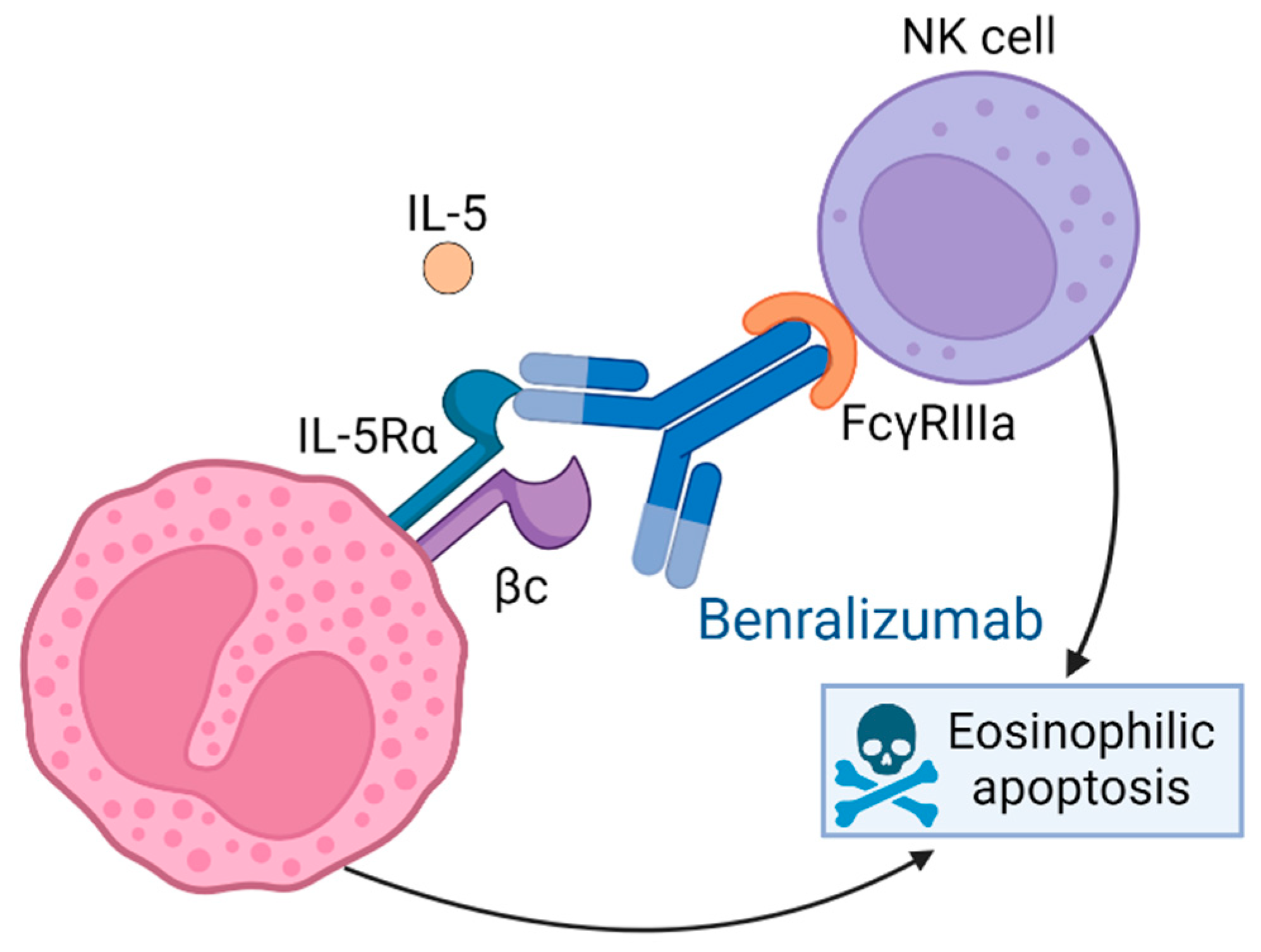
1. Introduction
2. Results
2.1. Characteristics of the Patients
2.2. Clinical Effectiveness
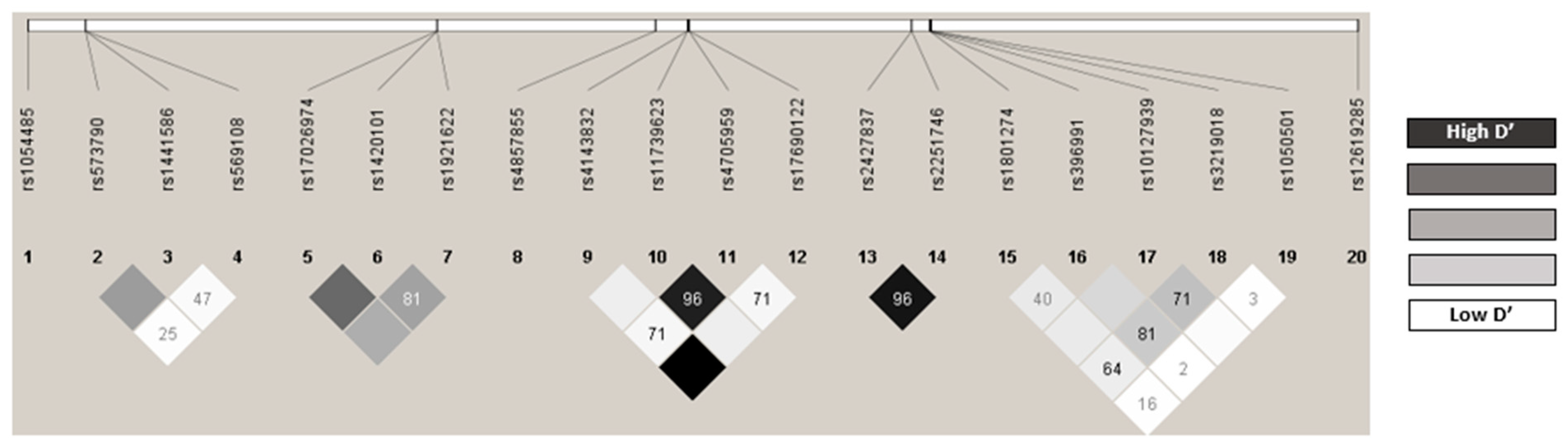
2.3. Distribution of the Genotypes Analyzed
2.4. Predictors of Mepolizumab Response at 12 Months
2.4.1. Predictors of Treatment Response Using the Exacerbation Reduction Criterion
2.4.2. Predictors of Response for OCS Reduction
2.4.3. Predictors of Response for Lung Function Improvement
2.4.4. Predictors of Meeting at Least 1 Response Criterion
2.4.5. Predictors of Meeting at Least Two Response Criteria
2.4.6. Predictors of Meeting All 3 Criteria
| OR (95% CI) | p Value | p Value a | |
|---|---|---|---|
| Response with respect to exacerbation reduction | |||
| ZNF415 rs1054485 (T vs. GG) | 5.33 (1.06–30.02) | 0.042 | 0.042 |
| Response with respect to reduction in OCS | |||
| Exacerbation in previous year (No) | 3.89 (1.24–14.92) | 0.029 | 0.029 |
| Response with respect to improved lung function | |||
| Age upon starting mepolizumab (years) | 1.10 (1.04–1.18) | 0.002 | 0.003 |
| FCER1B rs569108 (AA vs. G) | 171.06 (12.94–6264.11) | <0.001 | 0.003 |
| FCER1A rs2427837 (A vs. GG) | 8.61 (1.71–76.62) | 0.021 | 0.21 |
| Meeting at least 1 criterion | |||
| GATA2 rs4857855 (C vs. TT) | 34 (1.1–1136.7) | 0.026 | 0.026 |
| Meeting at least 2 criteria | |||
| Exacerbation in previous year (No) | 13.79 (2.12–296.89) | 0.024 | 0.036 |
| FCER1A rs2427837 (A vs. GG) | 4.99 (1.18–30.1) | 0.044 | 0.044 |
| FCER1B rs569108 (AA vs. G) | 11.86 (1.92–109.31) | 0.013 | 0.036 |
| Meeting all 3 criteria | |||
| IL1RL1 rs1921622 (AA vs. G) | 3.44 (1.18–11.09) | 0.028 | 0.028 |
2.5. Predictors of Benralizumab Response at 12 Months
2.5.1. Predictors of Treatment Response Using the Exacerbation Reduction Criterion
2.5.2. Predictors of Response for OCS Reduction
2.5.3. Predictors of Response for Lung Function Improvement
2.5.4. Predictors of Meeting at Least 1 Response Criterion
2.5.5. Predictors of Meeting at Least Two Response Criteria
2.5.6. Predictors of Meeting All Three Criteria
| OR (95% CI) | p Value | p Value a | |
|---|---|---|---|
| Response with respect to exacerbation reduction | |||
| Exacerbation in previous year (No) | 1.3 × 108 (1.8 × 10−19–NA) | 0.073 | 0.073 |
| Response with respect to reduction in OCS | |||
| Sex (Female) | 4.78 (1.22–20.63) | 0.028 | 0.050 |
| Allergies (Yes) | 4.02 (1.05–16.74) | 0.045 | 0.050 |
| FCER1B rs569108 (AA vs. G) | 11.51 (1.19–269.78) | 0.050 | 0.050 |
| Response with respect to improved lung function | |||
| IL5 rs4143832 (T vs. GG) | 11.1 (1.9–112.17) | 0.017 | 0.035 |
| IKZF2 rs12619285 (AA vs. G) | 9.1 (1.7–75.78) | 0.019 | 0.035 |
| FCER1B rs1441586 (C vs. TT) | 7.81 (1.16–73.45) | 0.045 | 0.045 |
| Polyposis (No) | 9.16 (1.58–91.4) | 0.027 | 0.035 |
| Meeting at least 1 criterion | |||
| - | - | - | - |
| Meeting at least 2 criteria | |||
| IKZF2 rs12619285 (AA vs. G) | 9.68 (1.58–188.03) | 0.039 | 0.039 |
| Meeting all 3 criteria | |||
| IL5 rs4143832 (T vs. GG) | 4.55 (1.36–17.22) | 0.014 | 0.014 |
3. Discussion
4. Materials and Methods
4.1. Ethics Statements
4.2. Study Design
4.3. Study Population
4.4. Socio-Demographic and Clinical Variables
4.5. Genetic Variables
4.5.1. DNA Isolation
4.5.2. Detection of Gene Polymorphisms and Quality Control
4.6. Response Variables
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACT | Asthma Control Test |
| BMI | Body Mass Index |
| CI | Confidence Interval |
| COPD | Chronic Obstructive Pulmonary Disease |
| FEV1 | Forced Expiratory Volume in one second |
| GEMA | Guía Española para el Manejo del Asma |
| GERD | Gastroesophageal Reflux Disease |
| GINA | Global Initiative for Asthma |
| HWE | Hardy-Weinberg Equilibrium |
| ICS | Inhaled Corticosteroids |
| Ig | Immunoglobulin |
| IL | Interleukin |
| ILC2 | Type 2 Innate Lymphoid Cells |
| LD | Linkage Disequilibrium |
| MAF | Minor Allele Frequency |
| NK | Natural Killer |
| OCS | Oral Corticosteroids Systemic |
| OR | Odds Ratio |
| PCR | Polymerase Chain Reaction |
| SAHS | Sleep Apnea-Hypopnea Syndrome |
| SD | Standard Deviation |
| SNP | Single-Nucleotide Polymorphisms |
| SUA | Severe Uncontrolled asthma |
| Th2 | Type 2 Helper T |
| TSLP | thymic stromal lymphopoietin |
References
- Sociedad Española de Neumología y Cirugía Torácica. Gema 5.2. Guía Española Para El Manejo Del Asma. 2022. Available online: https://se-fc.org/wp-content/uploads/2022/05/GEMA-5.2-Final.pdf (accessed on 3 April 2024).
- Global Initiative for Asthma. GINA 2022. Global Strategy for Asthma Management and Prevention; Global Initiative for Asthma: Fontana, WI, USA, 2022. [Google Scholar]
- Caminati, M.; Le Pham, D.; Bagnasco, D.; Canonica, G.W. Type 2 Immunity in Asthma. World Allergy Organ. J. 2018, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Pelaia, C.; Paoletti, G.; Puggioni, F.; Racca, F.; Pelaia, G.; Canonica, G.W.; Heffler, E. Interleukin-5 in the Pathophysiology of Severe Asthma. Front. Physiol. 2019, 10, 1514. [Google Scholar] [CrossRef] [PubMed]
- Bobolea, D.I. Fenotipos Del Asma Grave Del Adulto: Las Claves Para La Medicina Personalizada. Available online: http://www.alergoaragon.org/2017/0103.pdf (accessed on 3 April 2024).
- Agencia Española del Medicamento y Productos Sanitarios. Ficha Técnica Nucala 100 Mg Polvo Para Solucion Inyectable. Available online: https://cima.aemps.es/cima/dochtml/ft/1151043001/FT_1151043001.html (accessed on 5 April 2022).
- Agencia Española del Medicamento y Productos Sanitarios. Ficha Tecnica Fasenra 30 Mg Solucion Inyectable En Jeringa Precargada. Available online: https://cima.aemps.es/cima/dochtml/ft/1171252001/FT_1171252001.html (accessed on 5 April 2022).
- Lobo, E.D.; Hansen, R.J.; Balthasar, J.P. Antibody Pharmacokinetics and Pharmacodynamics. J. Pharm. Sci. 2004, 93, 2645–2668. [Google Scholar] [CrossRef] [PubMed]
- Nimmerjahn, F.; Ravetch, J.V. Fcγ Receptors: Old Friends and New Family Members. Immunity 2006, 24, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Tracey, D.; Klareskog, L.; Sasso, E.H.; Salfeld, J.G.; Tak, P.P. Tumor Necrosis Factor Antagonist Mechanisms of Action: A Comprehensive Review. Pharmacol. Ther. 2008, 117, 244–279. [Google Scholar] [CrossRef] [PubMed]
- March, M.E.; Sleiman, P.M.; Hakonarson, H. Genetic Polymorphisms and Associated Susceptibility to Asthma. Int. J. Gen. Med. 2013, 6, 253. [Google Scholar] [CrossRef] [PubMed]
- Kabesch, M.; Depner, M.; Dahmen, I.; Weiland, S.K.; Vogelberg, C.; Niggemann, B.; Lau, S.; Illig, T.; Klopp, N.; Wahn, U.; et al. Polymorphisms in Eosinophil Pathway Genes, Asthma and Atopy. Allergy 2007, 62, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, C.; Rothenberg, M.E. Chapter 3 Biology of the Eosinophil. Adv. Immunol. 2009, 101, 81–121. [Google Scholar] [CrossRef]
- Melo, R.C.N.; Spencer, L.A.; Dvorak, A.M.; Weller, P.F. Mechanisms of Eosinophil Secretion: Large Vesiculotubular Carriers Mediate Transport and Release of Granule-Derived Cytokines and Other Proteins. J. Leukoc. Biol. 2008, 83, 229–236. [Google Scholar] [CrossRef]
- McLeod, O.; Silveira, A.; Valdes-Marquez, E.; Björkbacka, H.; Almgren, P.; Gertow, K.; Gådin, J.R.; Bäcklund, A.; Sennblad, B.; Baldassarre, D.; et al. Genetic Loci on Chromosome 5 Are Associated with Circulating Levels of Interleukin-5 and Eosinophil Count in a European Population with High Risk for Cardiovascular Disease. Cytokine 2016, 81, 1–9. [Google Scholar] [CrossRef]
- Morales, P.; Reyes, J. Estudio de Polimorfismos e Interacciones Génicas de los Genes IL1A, IL1B, IL1R, IL1RA, IL4R A, IL12, IFNG, TGFB1, TNFA, IL2, IL4, IL6, IL10 en Pacientes con Asma; Universidad de Salamanca: Salamanca, Spain, 2010. [Google Scholar] [CrossRef]
- Zhang, G.; Lu, J.; Yin, X.; Sun, Y.; Li, S. Association of IL-33, IL1RL1 Gene Polymorphisms with Serum IL-33 Levels and Risk of Asthma in Adults and Asthmatic Bronchitis in Children (Chinese). Biotechnol. Biotechnol. Equip. 2018, 32, 1251–1256. [Google Scholar] [CrossRef]
- IKZF2 IKAROS Family Zinc Finger 2 [Homo Sapiens (Human)]—Gene—NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/22807 (accessed on 15 February 2023).
- Gudbjartsson, D.F.; Bjornsdottir, U.S.; Halapi, E.; Helgadottir, A.; Sulem, P.; Jonsdottir, G.M.; Thorleifsson, G.; Helgadottir, H.; Steinthorsdottir, V.; Stefansson, H.; et al. Sequence Variants Affecting Eosinophil Numbers Associate with Asthma and Myocardial Infarction. Nat. Genet. 2009, 41, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Akhabir, L.; Sandford, A.J. Genome-Wide Association Studies for Discovery of Genes Involved in Asthma. Respirology 2011, 16, 396–406. [Google Scholar] [CrossRef] [PubMed]
- Beaven, M.A.; Metzger, H. Signal Transduction by Fc Receptors: The Fc Epsilon RI Case. Immunol. Today 1993, 14, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Salmon, J.E.; Brogle, N.L.; Edberg, J.C.; Kimberly, R.P. Fc Gamma Receptor III Induces Actin Polymerization in Human Neutrophils and Primes Phagocytosis Mediated by Fc Gamma Receptor II. J. Immunol. 1991, 146, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Narasimhanl, V.; Holowkag, D.; Bairdqll, B. A Guanine Nucleotide-Binding Protein Participates in IgE Receptor-Mediated Activation of Endogenous and Reconstituted Phospholipase A2 in a Permeabilized Cell System*. J. Biol. Chem. 1990, 264, 1459–1464. [Google Scholar] [CrossRef]
- Ravetch, J.V.; Bolland, S. IgG Fc Receptors. Annu. Rev. Immunol. 2001, 19, 275–290. [Google Scholar] [CrossRef] [PubMed]
- van de Winkel, J.G.J.; Capel, P.J.A. Human IgG Fc Receptor Heterogeneity: Molecular Aspects and Clinical Implications. Immunol. Today 1993, 14, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Auton, A.; Abecasis, G.R.; Altshuler, D.M.; Durbin, R.M.; Bentley, D.R.; Chakravarti, A.; Clark, A.G.; Donnelly, P.; Eichler, E.E.; Flicek, P.; et al. A Global Reference for Human Genetic Variation. Nature 2015, 526, 68. [Google Scholar] [CrossRef]
- Nair, P.; Wenzel, S.; Rabe, K.F.; Bourdin, A.; Lugogo, N.L.; Kuna, P.; Barker, P.; Sproule, S.; Ponnarambil, S.; Goldman, M. Oral Glucocorticoid-Sparing Effect of Benralizumab in Severe Asthma. N. Engl. J. Med. 2017, 376, 2448–2458. [Google Scholar] [CrossRef]
- Chupp, G.L.; Bradford, E.S.; Albers, F.C.; Bratton, D.J.; Wang-Jairaj, J.; Nelsen, L.M.; Trevor, J.L.; Magnan, A.; ten Brinke, A. Efficacy of Mepolizumab Add-on Therapy on Health-Related Quality of Life and Markers of Asthma Control in Severe Eosinophilic Asthma (MUSCA): A Randomised, Double-Blind, Placebo-Controlled, Parallel-Group, Multicentre, Phase 3b Trial. Lancet Respir. Med. 2017, 5, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Ortega, H.G.; Liu, M.C.; Pavord, I.D.; Brusselle, G.G.; FitzGerald, J.M.; Chetta, A.; Humbert, M.; Katz, L.E.; Keene, O.N.; Yancey, S.W.; et al. Mepolizumab Treatment in Patients with Severe Eosinophilic Asthma. N. Engl. J. Med. 2014, 371, 1198–1207. [Google Scholar] [CrossRef]
- Agache, I.; Rocha, C.; Beltran, J.; Song, Y.; Posso, M.; Solà, I.; Alonso-Coello, P.; Akdis, C.; Akdis, M.; Canonica, G.W.; et al. Efficacy and Safety of Treatment with Biologicals (Benralizumab, Dupilumab and Omalizumab) for Severe Allergic Asthma: A Systematic Review for the EAACI Guidelines—Recommendations on the Use of Biologicals in Severe Asthma. Allergy 2020, 75, 1043–1057. [Google Scholar] [CrossRef] [PubMed]
- FitzGerald, J.M.; Bleecker, E.R.; Nair, P.; Korn, S.; Ohta, K.; Lommatzsch, M.; Ferguson, G.T.; Busse, W.W.; Barker, P.; Sproule, S.; et al. Benralizumab, an Anti-Interleukin-5 Receptor α Monoclonal Antibody, as Add-on Treatment for Patients with Severe, Uncontrolled, Eosinophilic Asthma (CALIMA): A Randomised, Double-Blind, Placebo-Controlled Phase 3 Trial. Lancet 2016, 388, 2128–2141. [Google Scholar] [CrossRef] [PubMed]
- Agache, I.; Beltran, J.; Akdis, C.; Akdis, M.; Canelo-Aybar, C.; Canonica, G.W.; Casale, T.; Chivato, T.; Corren, J.; Del Giacco, S.; et al. Efficacy and Safety of Treatment with Biologicals (Benralizumab, Dupilumab, Mepolizumab, Omalizumab and Reslizumab) for Severe Eosinophilic Asthma. A Systematic Review for the EAACI Guidelines—Recommendations on the Use of Biologicals in Severe Asthma. Allergy 2020, 75, 1023–1042. [Google Scholar] [CrossRef] [PubMed]
- Bleecker, E.R.; FitzGerald, J.M.; Chanez, P.; Papi, A.; Weinstein, S.F.; Barker, P.; Sproule, S.; Gilmartin, G.; Aurivillius, M.; Werkström, V.; et al. Efficacy and Safety of Benralizumab for Patients with Severe Asthma Uncontrolled with High-Dosage Inhaled Corticosteroids and Long-Acting Β2-Agonists (SIROCCO): A Randomised, Multicentre, Placebo-Controlled Phase 3 Trial. Lancet 2016, 388, 2115–2127. [Google Scholar] [CrossRef] [PubMed]
- Crimi, C.; Campisi, R.; Cacopardo, G.; Intravaia, R.; Nolasco, S.; Porto, M.; Pelaia, C.; Crimi, N. Real-Life Effectiveness of Mepolizumab in Patients with Severe Refractory Eosinophilic Asthma and Multiple Comorbidities. World Allergy Organ. J. 2020, 13, 100462. [Google Scholar] [CrossRef] [PubMed]
- Maglio, A.; Vitale, C.; Pellegrino, S.; Calabrese, C.; D’amato, M.; Molino, A.; Pelaia, C.; Triggiani, M.; Pelaia, G.; Stellato, C.; et al. Real-Life Effectiveness of Mepolizumab on Forced Expiratory Flow between 25% and 75% of Forced Vital Capacity in Patients with Severe Eosinophilic Asthma. Biomedicines 2021, 9, 1550. [Google Scholar] [CrossRef] [PubMed]
- Haldar, P.; Brightling, C.E.; Hargadon, B.; Gupta, S.; Monteiro, W.; Sousa, A.; Marshall, R.P.; Bradding, P.; Green, R.H.; Wardlaw, A.J.; et al. Mepolizumab and Exacerbations of Refractory Eosinophilic Asthma. N. Engl. J. Med. 2009, 360, 973–984. [Google Scholar] [CrossRef] [PubMed]
- Renner, A.; Marth, K.; Patocka, K.; Idzko, M.; Pohl, W. Effectiveness of Mepolizumab Therapy in Patients with Severe Eosinophilic Asthma: Austrian Real-Life Data. Pulm. Pharmacol. Ther. 2020, 64, 101946. [Google Scholar] [CrossRef]
- Brás, R.; Paulino, M.; Varandas, C.; Coutinho, C.; Silva, M.I.; Limão, R.; Costa, C.; Alonso, E.; Pedro, E.; Mendes, A. Mepolizumab for Severe Eosinophilic Asthma—A One-Year Real Life Portuguese Study. Pulmonology 2021, 27, 579–581. [Google Scholar] [CrossRef] [PubMed]
- Israel, E.; Canonica, G.W.; Brusselle, G.; Yang, S.; Howarth, P.H.; Martin, A.L.; Koufopoulou, M.; Smith, S.G.; Alfonso-Cristancho, R. Real-Life Effectiveness of Mepolizumab in Severe Asthma: A Systematic Literature Review. J. Asthma 2022, 59, 2201–2217. [Google Scholar] [CrossRef] [PubMed]
- Padilla-Galo, A.; Levy-Abitbol, R.; Olveira, C.; Valencia Azcona, B.; Pérez Morales, M.; Rivas-Ruiz, F.; Tortajada-Goitia, B.; Moya-Carmona, I.; Levy-Naon, A. Real-Life Experience with Benralizumab during 6 Months. BMC Pulm. Med. 2020, 20, 184. [Google Scholar] [CrossRef] [PubMed]
- Kavanagh, J.E.; d’Ancona, G.; Elstad, M.; Green, L.; Fernandes, M.; Thomson, L.; Roxas, C.; Dhariwal, J.; Nanzer, A.M.; Kent, B.D.; et al. Real-World Effectiveness and the Characteristics of a “Super-Responder” to Mepolizumab in Severe Eosinophilic Asthma. Chest 2020, 158, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Rojo-Tolosa, S.; González-Gutiérrez, M.V.; Jiménez-Gálvez, G.; Sánchez-Martínez, J.A.; Pineda-Lancheros, L.E.; Gálvez-Navas, J.M.; Jiménez-Morales, A.; Pérez-Ramírez, C.; Morales-García, C. Impact of Anti-IL5 Therapies on Patients with Severe Uncontrolled Asthma and Possible Predictive Biomarkers of Response: A Real-Life Study. Int. J. Mol. Sci. 2023, 24, 2011. [Google Scholar] [CrossRef] [PubMed]
- Farzan, N.; Vijverberg, S.J.H.; Arets, H.G.; Raaijmakers, J.A.M.; Maitland-van der Zee, A.H. Pharmacogenomics of Inhaled Corticosteroids and Leukotriene Modifiers: A Systematic Review. Clin. Exp. Allergy 2017, 47, 271–293. [Google Scholar] [CrossRef] [PubMed]
- Vijverberg, S.J.; Raaijmakers, J.A.; Maitland-Van Der Zee, A.H. ADRB2 Arg16 and the Need for Collaboration in Childhood Asthma Pharmacogenomics. Pharmacogenomics 2013, 14, 1937–1939. [Google Scholar] [CrossRef] [PubMed]
- Pijnenburg, M.W.; Szefler, S. Personalized Medicine in Children with Asthma. Paediatr. Respir. Rev. 2015, 16, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Rojo-Tolosa, S.; Pineda-Lancheros, L.E.; Gálvez-Navas, J.M.; Sánchez-Martínez, J.A.; González-Gutiérrez, M.V.; Fernández-Alonso, A.; Morales-García, C.; Jiménez-Morales, A.; Pérez-Ramírez, C. Association between Single Nucleotide Polymorphisms Related to Vitamin D Metabolism and the Risk of Developing Asthma. Nutrients 2023, 15, 823. [Google Scholar] [CrossRef] [PubMed]
- Amo, G.; García-Menaya, J.; Campo, P.; Cordobés, C.; Serón, M.C.P.; Ayuso, P.; Esguevillas, G.; Blanca, M.; Agúndez, J.A.G.; García-Martín, E. A Nonsynonymous FCER1B SNP Is Associated with Risk of Developing Allergic Rhinitis and with IgE Levels. Sci. Rep. 2016, 6, 19724. [Google Scholar] [CrossRef]
- Dar, S.A.; Rai, G.; Ansari, M.A.; Akhter, N.; Gupta, N.; Sharma, S.; Haque, S.; Ramachandran, V.G.; Wahid, M.; Rudramurthy, S.M.; et al. FcɛR1α Gene Polymorphism Shows Association with High IgE and Anti-FcɛR1α in Chronic Rhinosinusitis with Nasal Polyposis. J. Cell. Biochem. 2018, 119, 4142–4149. [Google Scholar] [CrossRef]
- Sharma, S.; Ghosh, B. Promoter Polymorphism in the MS4A2 Gene and Asthma in the Indian Population. Int. Arch. Allergy Immunol. 2009, 149, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Dharajiya, N.; Vaidya, S.V.; Murai, H.; Cardenas, V.; Kurosky, A.; Boldogh, I.; Sur, S.A. FcgammaRIIb Inhibits Allergic Lung Inflammation in a Murine Model of Allergic Asthma. PLoS ONE 2010, 5, e9337. [Google Scholar] [CrossRef][Green Version]
- Jiang, Y.; Hirose, S.; Abe, M.; Sanokawa-Akakura, R.; Ohtsuji, M.; Mi, X.; Li, N.; Xiu, Y.; Zhang, D.; Shirai, J.; et al. Polymorphisms in IgG Fc Receptor IIB Regulatory Regions Associated with Autoimmune Susceptibility. Immunogenetics 2000, 51, 429–435. [Google Scholar] [CrossRef]
- Cañete, J.D.; Suárez, B.; Hernández, M.V.; Sanmartí, R.; Rego, I.; Celis, R.; Moll, C.; Pinto, J.A.; Blanco, F.J.; Lozano, F. Influence of Variants of Fc Gamma Receptors IIA and IIIA on the American College of Rheumatology and European League Against Rheumatism Responses to Anti-Tumour Necrosis Factor Alpha Therapy in Rheumatoid Arthritis. Ann. Rheum. Dis. 2009, 68, 1547–1552. [Google Scholar] [CrossRef]
- Dong, C.; Ptacek, T.S.; Redden, D.T.; Zhang, K.; Brown, E.E.; Edberg, J.C.; McGwin, G.J.; Alarcón, G.S.; Ramsey-Goldman, R.; Reveille, J.D.; et al. Fcγ Receptor IIIa Single-Nucleotide Polymorphisms and Haplotypes Affect Human IgG Binding and Are Associated with Lupus Nephritis in African Americans. Arthritis Rheumatol. 2014, 66, 1291–1299. [Google Scholar] [CrossRef] [PubMed]
- Liao, E.C.; Chang, C.Y.; Hsieh, C.W.; Yu, S.J.; Yin, S.C.; Tsai, J.J. An Exploratory Pilot Study of Genetic Marker for IgE-Mediated Allergic Diseases with Expressions of FcεR1α and Cε. Int. J. Mol. Sci. 2015, 16, 9504–9519. [Google Scholar] [CrossRef]
- Rebollo, A.; Schmitt, C. Ikaros, Aiolos and Helios: Transcription Regulators and Lymphoid Malignancies. Immunol. Cell Biol. 2003, 81, 171–175. [Google Scholar] [CrossRef]
- Spear, M.L.; Hu, D.; Pino-Yanes, M.; Huntsman, S.; Eng, C.; Levin, A.M.; Ortega, V.E.; White, M.J.; McGarry, M.E.; Thakur, N.; et al. A Genome-Wide Association and Admixture Mapping Study of Bronchodilator Drug Response in African Americans with Asthma. Pharmacogenom. J. 2018, 19, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.Y.; Hur, G.; Lee, S.W.; Lee, S.J.; Lee, S.; Kim, S.H.; Rho, M.C. AGK2 Ameliorates Mast Cell-Mediated Allergic Airway Inflammation and Fibrosis by Inhibiting FcεRI/TGF-β Signaling Pathway. Pharmacol. Res. 2020, 159, 105027. [Google Scholar] [CrossRef]
- Yang, H.J.; Zheng, L.; Zhang, X.F.; Yang, M.; Huang, X. Association of the MS4A2 Gene Promoter C-109T or the 7th Exon E237G Polymorphisms with Asthma Risk: A Meta-Analysis. Clin. Biochem. 2014, 47, 605–611. [Google Scholar] [CrossRef]
- Bai, S.; Hua, L.; Wang, X.; Liu, Q.; Bao, Y. Association of a 4-Locus Gene Model Including IL13, IL4, FCER1B, and ADRB2 with the Asthma Predictive Index and Atopy in Chinese Han Children. J. Investig. Allergol. Clin. Immunol. 2018, 28, 407–413. [Google Scholar] [CrossRef]
- Hua, L.; Zuo, X.B.; Bao, Y.X.; Liu, Q.H.; Li, J.Y.; Lv, J.; Fang, D.Z.; Lin, Q.; Bao, J.; Ji, R.X. Four-Locus Gene Interaction between IL13, IL4, FCER1B, and ADRB2 for Asthma in Chinese Han Children. Pediatr. Pulmonol. 2016, 51, 364–371. [Google Scholar] [CrossRef]
- Ramphul, K.; Lv, J.; Hua, L.; Liu, Q.H.; Fang, D.Z.; Ji, R.X.; Bao, Y.X. Single Nucleotide Polymorphisms Predisposing to Asthma in Children of Mauritian Indian and Chinese Han Ethnicity. Braz. J. Med. Biol. Res. = Rev. Bras. Pesqui. Med. Biol. 2014, 47, 394–397. [Google Scholar] [CrossRef]
- Green, S.L.; Gaillard, M.C.; Song, E.; Dewar, J.B.; Halkas, A. Polymorphisms of the Beta Chain of the High-Affinity Immunoglobulin E Receptor (Fc ɛ RI-β) in South African Black and White Asthmatic and Nonasthmatic Individuals. Am. J. Respir. Crit. Care Med. 2012, 158, 1487–1492. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Hopkin, J.M. Atopy Phenotype in Subjects with Variants of the Beta Subunit of the High Affinity IgE Receptor. Thorax 1997, 52, 654. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Asthma Control Test (ACT). Available online: https://www.thoracic.org/members/assemblies/assemblies/srn/questionaires/act.php (accessed on 4 May 2022).
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2013. [Google Scholar]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and Visualization of LD and Haplotype Maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Solé, X.; Guinó, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A Web Tool for the Analysis of Association Studies. Bioinformatics 2006, 22, 1928–1929. [Google Scholar] [CrossRef]
| Mepolizumab | Benralizumab | |||
|---|---|---|---|---|
| Variables | N | Cases (N = 72) | N | Cases (N = 51) |
| Sex | ||||
| Women | 48 | 66.7 | 34 | 66.7 |
| Men | 24 | 33.3 | 17 | 33.3 |
| Age upon starting biological therapy (years) | 72 | 53 ± 13 | 51 | 58 ± 15 |
| Years with asthma | 72 | 7 (3, 10) | 51 | 6 (4, 10) |
| BMI (kg/m2) | ||||
| <25 | 19 | 26.4 | 9 | 17.7 |
| >25 | 53 | 73.6 | 42 | 82.3 |
| Previous respiratory disease | ||||
| Yes | 34 | 47.2 | 24 | 47.1 |
| No | 38 | 52.8 | 27 | 52.9 |
| Smoking status | ||||
| Non-smoker | 12 | 16.7 | 39 | 76.5 |
| Active smoker | 0 | 0 | 2 | 3.9 |
| Ex-smoker | 60 | 83.3 | 10 | 19.6 |
| Polyposis | ||||
| Yes | 33 | 45.8 | 20 | 39.2 |
| No | 39 | 54.2 | 31 | 60.8 |
| Allergies | ||||
| Yes | 37 | 51.4 | 33 | 64.7 |
| No | 35 | 48.6 | 18 | 35.3 |
| GERD | ||||
| Yes | 32 | 44.4 | 22 | 43.1 |
| No | 40 | 55.6 | 29 | 56.9 |
| SAHS | ||||
| Yes | 15 | 20.8 | 10 | 19.6 |
| No | 57 | 79.2 | 41 | 80.4 |
| COPD | ||||
| Yes | 13 | 18.1 | 10 | 19.6 |
| No | 59 | 81.9 | 41 | 80.4 |
| Age at diagnosis (years) | 72 | 49 ± 15 | 51 | 52 ± 15 |
| <18 | 2 | 2.8 | 1 | 2 |
| >18 | 70 | 97.2 | 50 | 98 |
| ICS (µg/day) | 72 | 500 (500, 1000) | 51 | 1000 (500, 1000) |
| OCS courses per year | ||||
| Yes | 57 | 79.2 | 45 | 88.2 |
| No | 15 | 20.8 | 6 | 11.8 |
| Baseline %FEV1 | ||||
| <80 | 51 | 70.8 | 34 | 66.7 |
| >80 | 21 | 29.2 | 17 | 33.3 |
| Exacerbation in previous year | ||||
| Yes | 47 | 65.3 | 22 | 43.1 |
| No | 25 | 34.7 | 29 | 56.9 |
| Baseline blood eosinophils (cells/µL) | ||||
| <300 | 15 | 20.8 | 21 | 41.2 |
| >300 | 57 | 79.2 | 30 | 58.8 |
| Previous biological therapy | ||||
| Yes | 21 | 29.2 | 20 | 39.2 |
| No | 51 | 70.8 | 31 | 60.8 |
| Response Variable | Mepolizumab | Benralizumab | ||
|---|---|---|---|---|
| N | % | N | % | |
| Reduction in OCS ≥ 50% | ||||
| R | 47 | 65.3 | 32 | 62.8 |
| NR | 25 | 34.7 | 19 | 37.2 |
| Reduction in exacerbations ≥ 50% | ||||
| R | 65 | 90.3 | 48 | 94.1 |
| NR | 7 | 9.7 | 3 | 5.9 |
| Increase in %FEV1 ≥ 10% or %FEV1 ≥ 80% | ||||
| Yes | 53 | 73.6 | 37 | 72.6 |
| No | 19 | 26.4 | 14 | 27.4 |
| Responsive for 1 criterion | ||||
| R | 70 | 97.2 | 50 | 98.0 |
| NR | 2 | 2.8 | 1 | 2.0 |
| Responsive for 2 criteria | ||||
| R | 57 | 79.2 | 42 | 82.4 |
| NR | 15 | 20.8 | 9 | 17.6 |
| Responsive for 3 criteria | ||||
| R | 35 | 48.6 | 25 | 49 |
| NR | 37 | 51.4 | 26 | 51 |
| Gene | SNP | dbSNP ID | Assay ID |
|---|---|---|---|
| IL5 (5q31) | T > G | rs4143832 | C__28028637_10 |
| A > G | rs17690122 | C__33291516_10 | |
| RAD50 (5q31.1) | C > T | rs11739623 | C__31237883_10 |
| T > C | rs4705959 | C___2549990_10 | |
| FCER1A (1q23) | T > C | rs2251746 | C___1840470_20 |
| G > A | rs2427837 | C__16233438_20 | |
| FCER1B (11q12–13) | T > C | rs1441586 | C___1842226_10 |
| T > C | rs573790 | C____900105_20 | |
| A > G | rs569108 | C____900116_10 | |
| ZNF415 (19q) | T > G | rs1054485 | C___2932371_10 |
| IKZF22 (2q13) | A > G | rs12619285 | C___1861821_10 |
| FCGR2A (1q23.3) | A > G | rs1801274 | C___9077561_20 |
| FCGR2B (1q23.3) | G > C | rs3219018 | ANPRZAZ |
| T > C | rs1050501 | ANRWUVX | |
| FCGR3A (1q23.3) | A > C | rs10127939 | C__57480226_10 |
| A > C | rs396991 | C__25815666_10 | |
| IL1RL1 (2q12) | C > T | rs1420101 | C___8906009_20 |
| G > A | rs17026974 | C__33551182_10 | |
| G > A | rs1921622 | C___1226146_10 | |
| GATA2 (3q21) | C > T | rs4857855 | C__11231076_10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rojo-Tolosa, S.; Sánchez-Martínez, J.A.; Caballero-Vázquez, A.; Pineda-Lancheros, L.E.; González-Gutiérrez, M.V.; Pérez-Ramírez, C.; Jiménez-Morales, A.; Morales-García, C. SingleNucleotide Polymorphisms as Biomarkers of Mepolizumab and Benralizumab Treatment Response in Severe Eosinophilic Asthma. Int. J. Mol. Sci. 2024, 25, 8139. https://doi.org/10.3390/ijms25158139
Rojo-Tolosa S, Sánchez-Martínez JA, Caballero-Vázquez A, Pineda-Lancheros LE, González-Gutiérrez MV, Pérez-Ramírez C, Jiménez-Morales A, Morales-García C. SingleNucleotide Polymorphisms as Biomarkers of Mepolizumab and Benralizumab Treatment Response in Severe Eosinophilic Asthma. International Journal of Molecular Sciences. 2024; 25(15):8139. https://doi.org/10.3390/ijms25158139
Chicago/Turabian StyleRojo-Tolosa, Susana, José Antonio Sánchez-Martínez, Alberto Caballero-Vázquez, Laura Elena Pineda-Lancheros, María Victoria González-Gutiérrez, Cristina Pérez-Ramírez, Alberto Jiménez-Morales, and Concepción Morales-García. 2024. "SingleNucleotide Polymorphisms as Biomarkers of Mepolizumab and Benralizumab Treatment Response in Severe Eosinophilic Asthma" International Journal of Molecular Sciences 25, no. 15: 8139. https://doi.org/10.3390/ijms25158139
APA StyleRojo-Tolosa, S., Sánchez-Martínez, J. A., Caballero-Vázquez, A., Pineda-Lancheros, L. E., González-Gutiérrez, M. V., Pérez-Ramírez, C., Jiménez-Morales, A., & Morales-García, C. (2024). SingleNucleotide Polymorphisms as Biomarkers of Mepolizumab and Benralizumab Treatment Response in Severe Eosinophilic Asthma. International Journal of Molecular Sciences, 25(15), 8139. https://doi.org/10.3390/ijms25158139







