Integrated Bioinformatics-Based Identification and Validation of Neuroinflammation-Related Hub Genes in Primary Open-Angle Glaucoma
Abstract
:1. Introduction
2. Results
2.1. Identification of DEGs and DENIGs in POAG
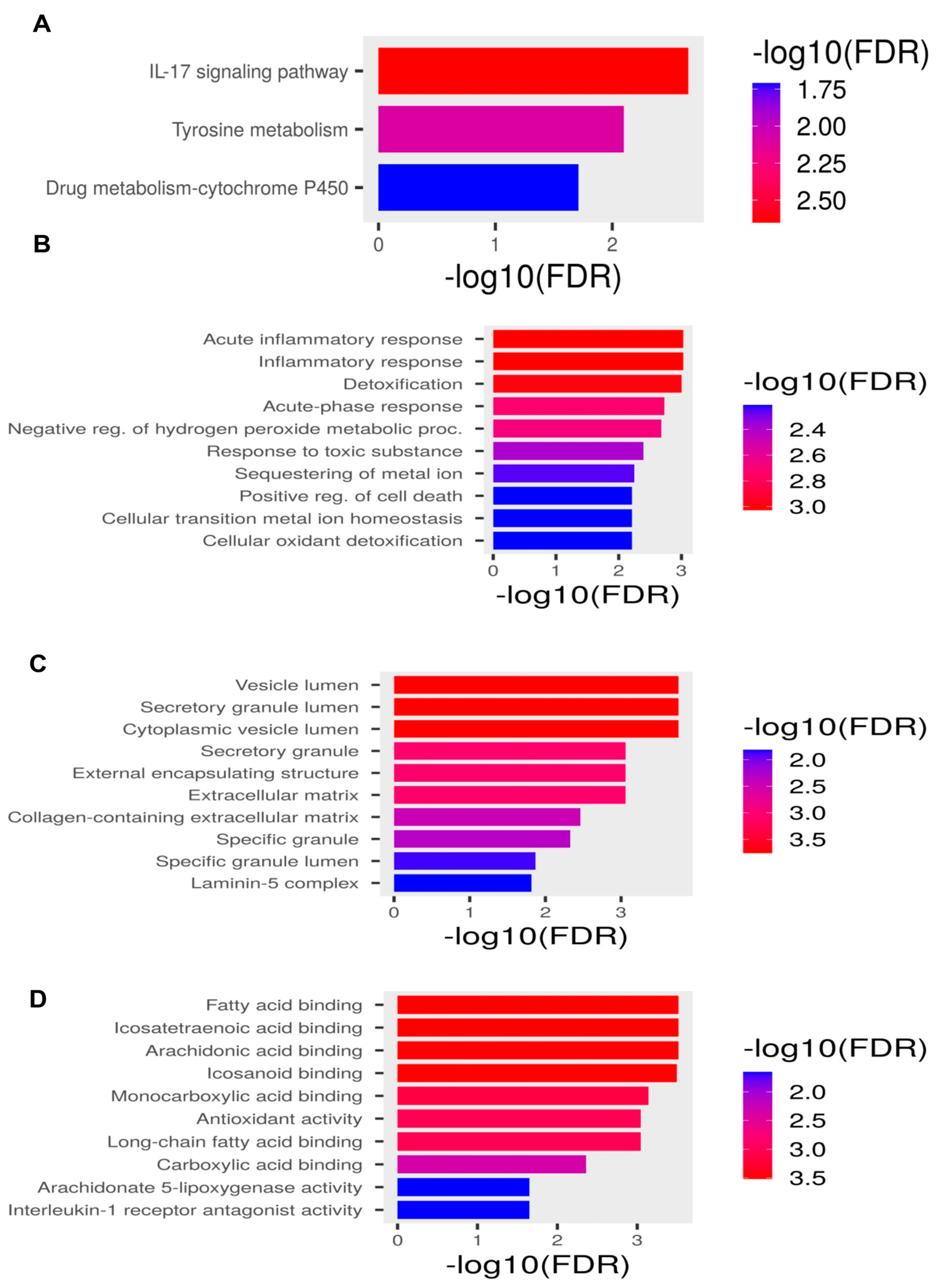
2.2. Pathway Enrichment Analysis
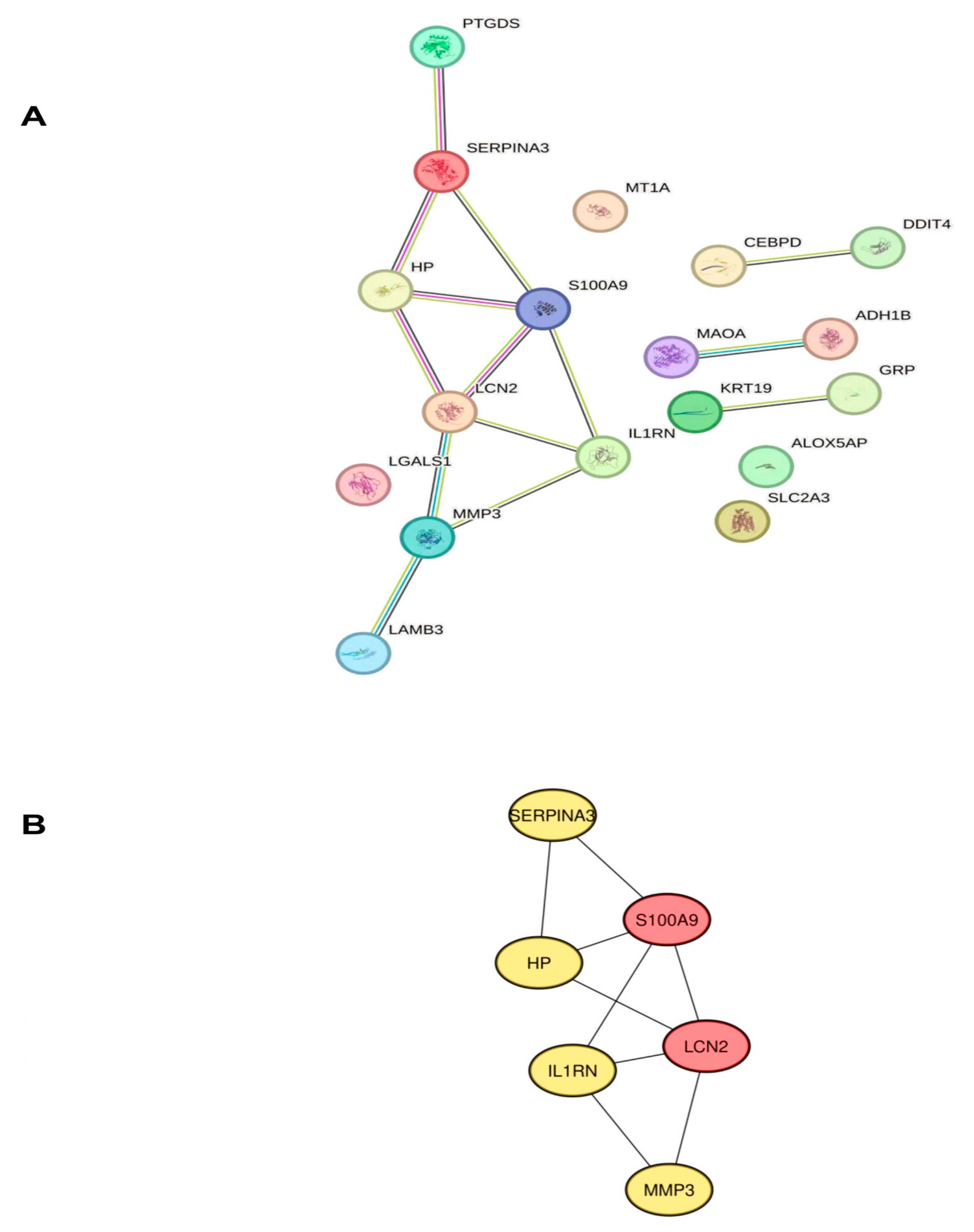
2.3. Identification of Hub Genes Associated with Neuroinflammation in POAG
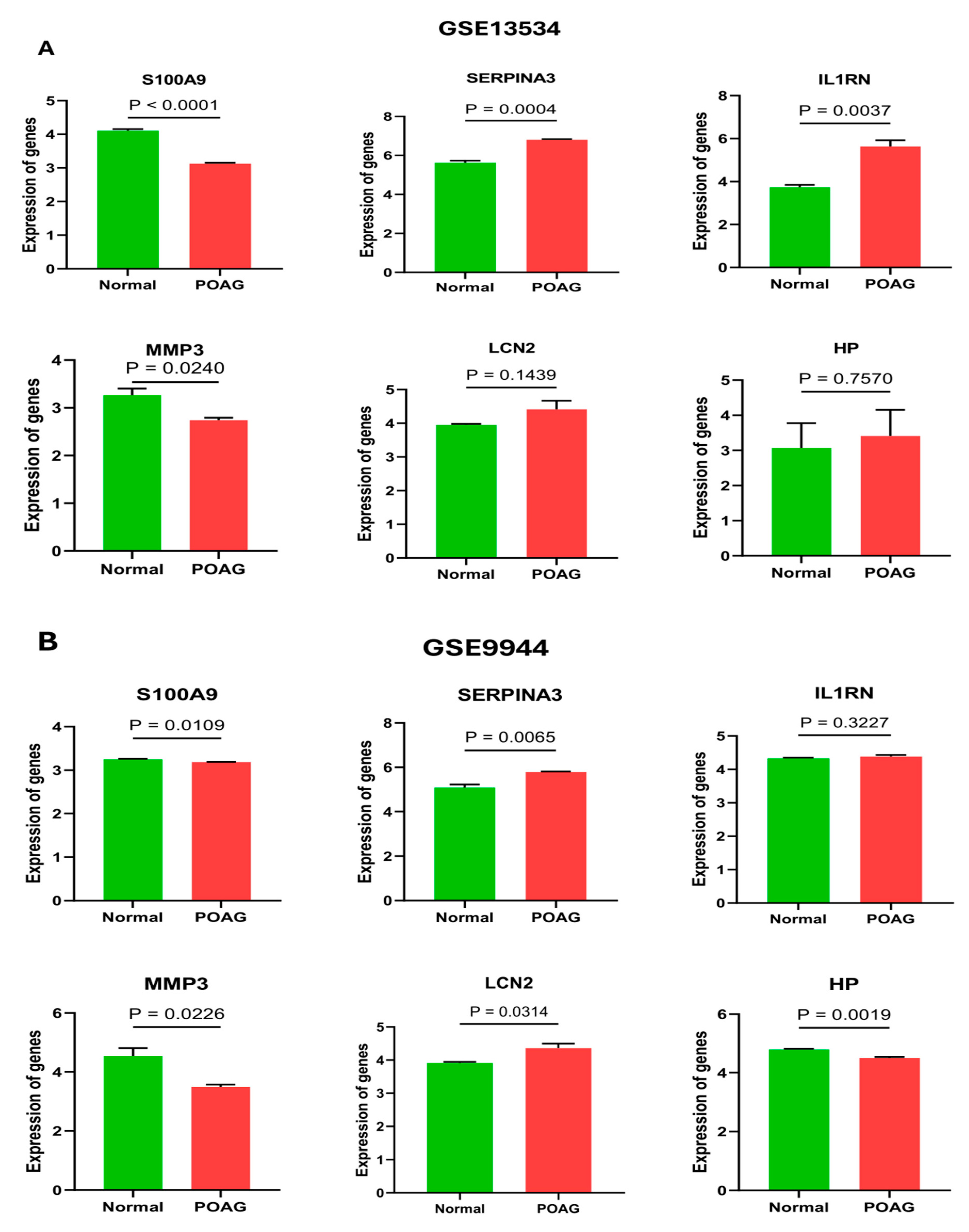
2.4. Validation of Hub Genes
2.5. Estimation of Possible miRNA Regulatory Networks of Hub Genes
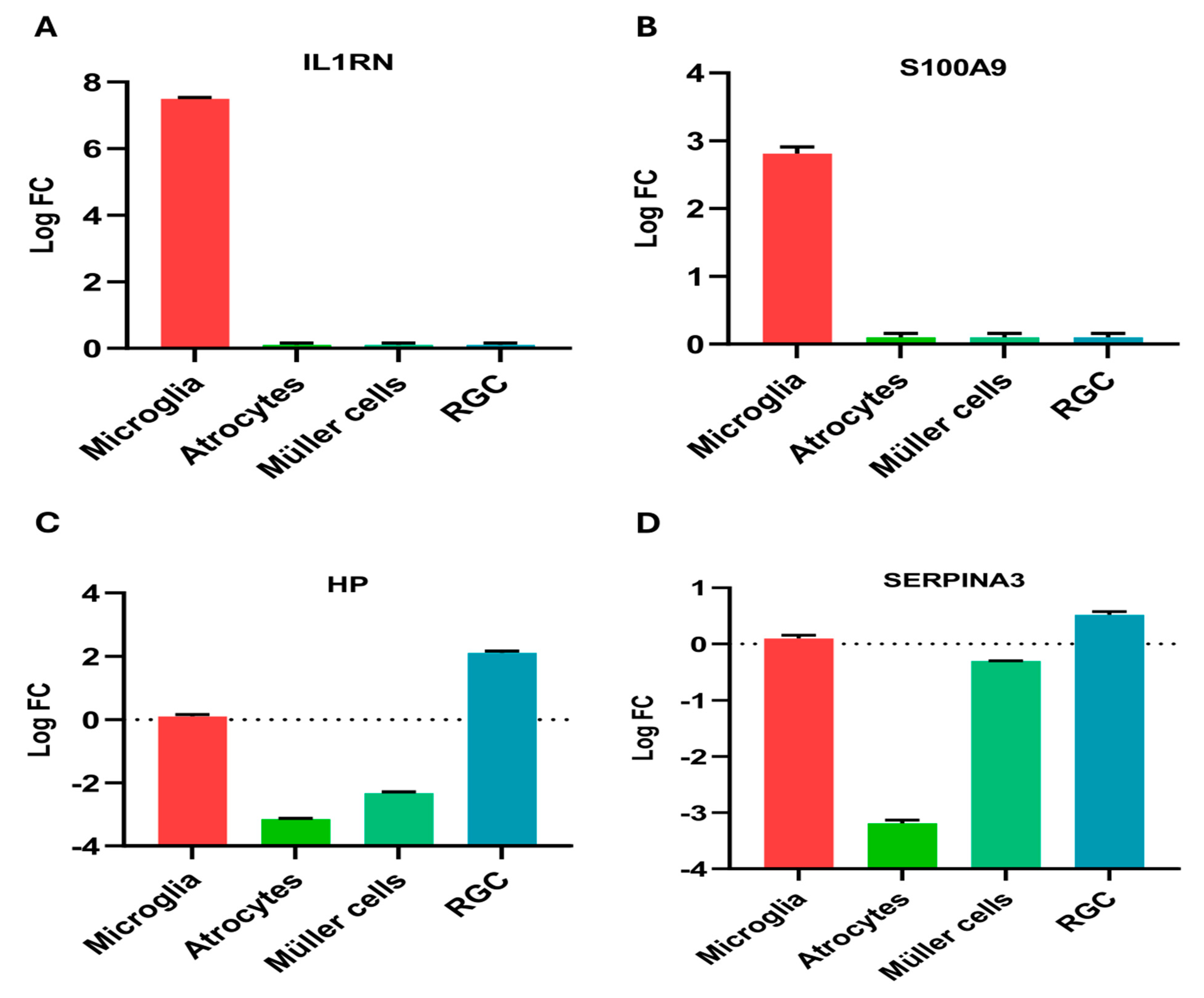
2.6. Regional Expression
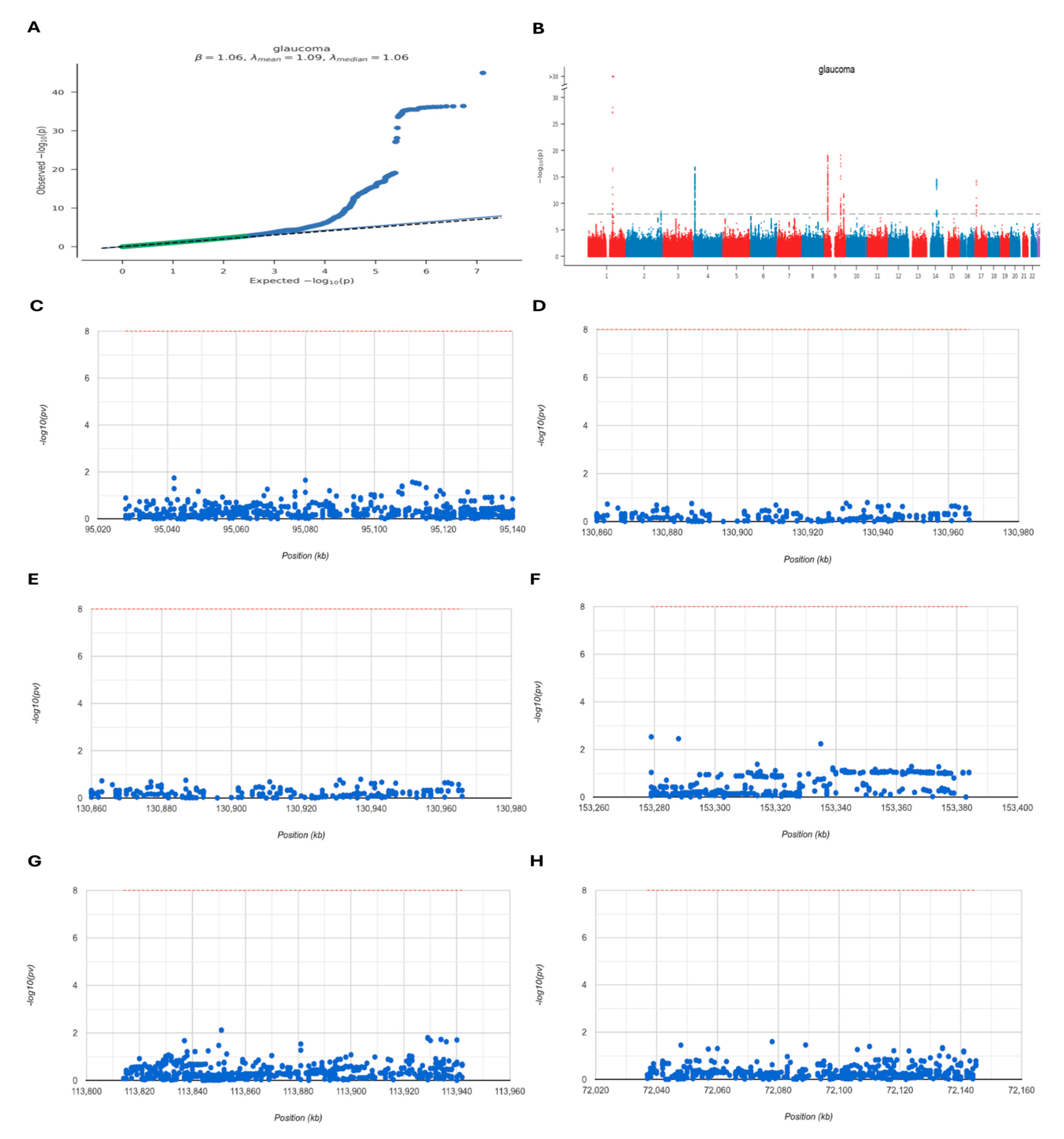
2.7. GWAS Analysis
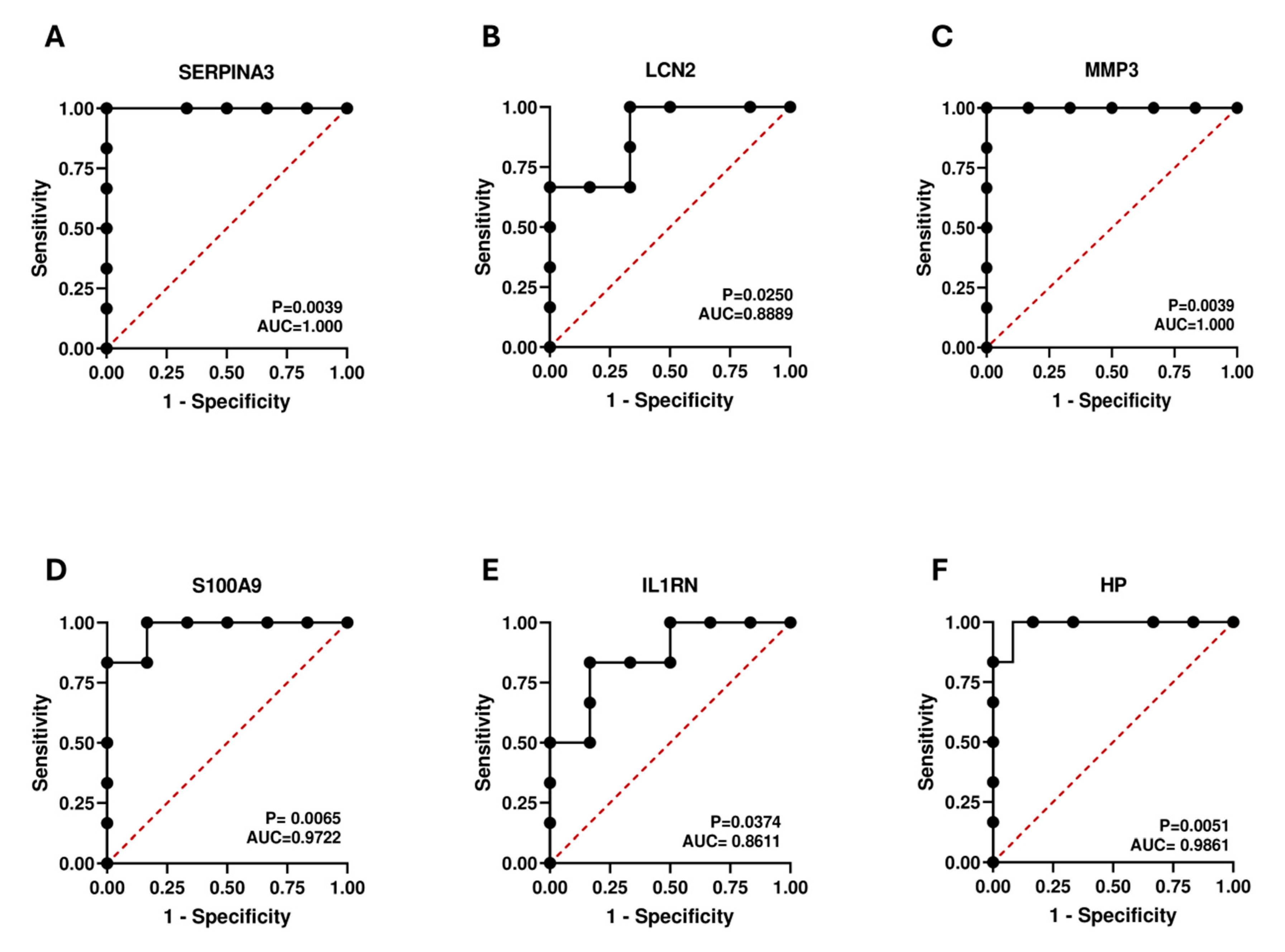
2.8. Diagnostic Value Validation
2.9. Validation of Hub Genes Using a Glaucoma Model
3. Discussion
Therapeutic Potential of Hub Genes
4. Methods
4.1. Data Sources
4.2. Data Processing and DEGs Analysis
4.3. Pathway Enrichment Analyses
4.4. Protein–Protein Interaction (PPI), Network Construction, and Gene Identification
4.5. External Validation of Hub Genes
4.6. Estimation of the Gene–miRNA Regulatory Network
4.7. Regional Expression
4.8. Genome-Wide Association Study (GWAS) Analysis
4.9. Receiver Operating Characteristic (ROC) Curve Analysis
4.10. Cell Culture
4.11. RNA Extraction and Real-Time PCR
4.12. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PPI | Protein-protein interactions |
| ECM | Extracellular matrix |
| OGD/R | Oxygen and glucose deprivation/reoxygenation |
| DMEM | Dulbecco’s Modified Eagle’s Medium |
| MF | Molecular function |
| BP | Biological process |
| CC | Cellular component |
| GO | Gene Ontology |
| DENIGs | Differentially expressed neuroinflammation-related genes |
| DEGs | Differentially expressed genes |
| IOP | Intraocular pressure |
| RGCs | Retinal ganglion cells |
| SNPs | Single nucleotide polymorphisms |
| GWAS | Genome-wide association study |
| ROC | Receiver operating characteristic |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| POAG | Primary open-angle glaucoma |
| SERPINA3 | serine protease inhibitor, clade A, member 3 |
| LCN2 | Lipocalin 2 |
| MMP3 | Matrix Metalloproteinase-3 |
| S100A9 | S100 Calcium-Binding Protein A9 |
| IL1RN | Interleukin-1 Receptor Antagonist |
| HP | Haptoglobin |
| GEO | Gene Expression Omnibus |
References
- Wu, Y.; Szymanska, M.; Hu, Y.; Fazal, M.I.; Jiang, N.; Yetisen, A.K.; Cordeiro, M.F. Measures of disease activity in glaucoma. Biosens. Bioelectron. 2022, 196, 113700. [Google Scholar] [CrossRef] [PubMed]
- Allison, K.; Patel, D.; Alabi, O. Epidemiology of Glaucoma: The Past, Present, and Predictions for the Future. Cureus 2020, 12, e11686. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Wang, J.; Li, Y.; Jiang, B. Prevalence of primary open angle glaucoma in the last 20 years: A meta-analysis and systematic review. Sci. Rep. 2021, 11, 13762. [Google Scholar] [CrossRef] [PubMed]
- Macanian, J.; Sharma, S.C. Pathogenesis of Glaucoma. Encyclopedia 2022, 2, 1803–1810. [Google Scholar] [CrossRef]
- Williams, P.A.; Marsh-Armstrong, N.; Howell, G.R.; Bosco, A.; Danias, J.; Simon, J.; Di Polo, A.; Kuehn, M.H.; Przedborski, S.; Raff, M.; et al. Neuroinflammation in glaucoma: A new opportunity. Exp. Eye Res. 2017, 157, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Baudouin, C.; Kolko, M.; Melik-Parsadaniantz, S.; Messmer, E.M. Inflammation in Glaucoma: From the back to the front of the eye, and beyond. Prog. Retin. Eye Res. 2021, 83, 100916. [Google Scholar] [CrossRef] [PubMed]
- Schuster, A.K.; Erb, C.; Hoffmann, E.M.; Dietlein, T.; Pfeiffer, N. The Diagnosis and Treatment of Glaucoma. Dtsch. Arzteblatt Int. 2020, 117, 225–234. [Google Scholar] [CrossRef]
- Loftsson, T.; Stefánsson, E. Aqueous eye drops containing drug/cyclodextrin nanoparticles deliver therapeutic drug concentrations to both anterior and posterior segment. Acta Ophthalmol. 2022, 100, 7–25. [Google Scholar] [CrossRef] [PubMed]
- Hakim, A.; Guido, B.; Narsineni, L.; Chen, D.-W.; Foldvari, M. Gene therapy strategies for glaucoma from IOP reduction to retinal neuroprotection: Progress towards non-viral systems. Adv. Drug Deliv. Rev. 2023, 196, 114781. [Google Scholar] [CrossRef]
- Tao, Y.; Zhang, Q.; Meng, M.; Huang, J. A bibliometric analysis of the application of stem cells in glaucoma research from 1999 to 2022. Front. Cell Dev. Biol. 2023, 11, 1081898. [Google Scholar] [CrossRef]
- Choi, S.; Choi, S.-H.; Bastola, T.; Park, Y.; Oh, J.; Kim, K.-Y.; Hwang, S.; Miller, Y.I.; Ju, W.-K. AIBP: A New Safeguard against Glaucomatous Neuroinflammation. Cells 2024, 13, 198. [Google Scholar] [CrossRef] [PubMed]
- Duarte, J.N. Neuroinflammatory Mechanisms of Mitochondrial Dysfunction and Neurodegeneration in Glaucoma. J. Ophthalmol. 2021, 2021, 4581909. [Google Scholar] [CrossRef] [PubMed]
- Tezel, G. Immune regulation toward immunomodulation for neuroprotection in glaucoma. Curr. Opin. Pharmacol. 2013, 13, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, A.I.; de Hoz, R.; Salobrar-Garcia, E.; Salazar, J.J.; Rojas, B.; Ajoy, D.; López-Cuenca, I.; Rojas, P.; Triviño, A.; Ramírez, J.M. The Role of Microglia in Retinal Neurodegeneration: Alzheimer’s Disease, Parkinson, and Glaucoma. Front. Aging Neurosci. 2017, 9, 214. [Google Scholar] [CrossRef]
- Bordone, M.P.; González Fleitas, M.F.; Pasquini, L.A.; Bosco, A.; Sande, P.H.; Rosenstein, R.E.; Dorfman, D. Involvement of microglia in early axoglial alterations of the optic nerve induced by experimental glaucoma. J. Neurochem. 2017, 142, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Evangelho, K.; Mogilevskaya, M.; Losada-Barragan, M.; Vargas-Sanchez, J.K. Pathophysiology of primary open-angle glaucoma from a neuroinflammatory and neurotoxicity perspective: A review of the literature. Int. Ophthalmol. 2019, 39, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Adornetto, A.; Russo, R.; Parisi, V. Neuroinflammation as a target for glaucoma therapy. Neural Regen. Res. 2019, 14, 391. [Google Scholar] [CrossRef]
- Fernández-Albarral, J.A.; Salazar, J.J.; De Hoz, R.; Marco, E.M.; Martín-Sánchez, B.; Flores-Salguero, E.; Salobrar-García, E.; López-Cuenca, I.; Barrios-Sabador, V.; Avilés-Trigueros, M.; et al. Retinal Molecular Changes Are Associated with Neuroinflammation and Loss of RGCs in an Experimental Model of Glaucoma. Int. J. Mol. Sci. 2021, 22, 2066. [Google Scholar] [CrossRef]
- Zhao, X.; Sun, R.; Luo, X.; Wang, F.; Sun, X. The Interaction Between Microglia and Macroglia in Glaucoma. Front. Neurosci. 2021, 15, 610788. [Google Scholar] [CrossRef]
- Vernazza, S.; Tirendi, S.; Bassi, A.M.; Traverso, C.E.; Saccà, S.C. Neuroinflammation in Primary Open-Angle Glaucoma. J. Clin. Med. 2020, 9, 3172. [Google Scholar] [CrossRef]
- Wang, H.; Sun, P.; Chen, Y.; Jiang, L.; Wu, H.; Zhang, W.; Gao, F. Research progress on human genes involved in the pathogenesis of glaucoma (Review). Mol. Med. Rep. 2018, 18, 656–674. [Google Scholar] [CrossRef]
- Yin, J.; Qian, Z.; Chen, Y.; Li, Y.; Zhou, X. MicroRNA regulatory networks in the pathogenesis of sarcopenia. J. Cell. Mol. Med. 2020, 24, 4900–4912. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Xiao, Z.; Shen, Y.; Yang, W.; Wang, P.; Li, P.; Wang, Z.; Pu, M.; Zhao, L.; Xie, P.; et al. SERPINA3: A novel inflammatory biomarker associated with cerebral small vessel disease burden in ischemic stroke. CNS Neurosci. Ther. 2023, 30, e14472. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Cho, K.-S.; Vu, T.H.K.; Shen, C.-H.; Kaur, M.; Chen, G.; Mathew, R.; McHam, M.L.; Fazelat, A.; Lashkari, K.; et al. Commensal microflora-induced T cell responses mediate progressive neurodegeneration in glaucoma. Nat. Commun. 2018, 9, 3209. [Google Scholar] [CrossRef]
- Zattoni, M.; Mearelli, M.; Vanni, S.; Colini Baldeschi, A.; Tran, T.H.; Ferracin, C.; Catania, M.; Moda, F.; Di Fede, G.; Giaccone, G.; et al. Serpin Signatures in Prion and Alzheimer’s Diseases. Mol. Neurobiol. 2022, 59, 3778–3799. [Google Scholar] [CrossRef] [PubMed]
- Aslam, M.S.; Yuan, L. Serpina3n: Potential drug and challenges, mini review. J. Drug Target. 2020, 28, 368–378. [Google Scholar] [CrossRef] [PubMed]
- Li, M.D.; Burns, T.C.; Morgan, A.A.; Khatri, P. Integrated multi-cohort transcriptional meta-analysis of neurodegenerative diseases. Acta Neuropathol. Commun. 2014, 2, 93. [Google Scholar] [CrossRef]
- Schrötter, A.; Park, Y.M.; Marcus, K.; Martins-de-Souza, D.; Nilsson, P.; Magraoui, F.E.; Meyer, H.E.; Grinberg, L.T. Key players in neurodegenerative disorders in focus—New insights into the proteomic profile of Alzheimer’s disease, schizophrenia, ALS, and multiple sclerosis—24th HUPO BPP Workshop: September 29, 2015, Vancouver, Canada. Proteomics 2016, 16, 1047–1050. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Wang, S.; Zhang, G.; He, J.; Liu, Z.; Wang, M.; Cai, H.; Wan, J. Highly expressed of SERPINA3 indicated poor prognosis and involved in immune suppression in glioma. Immun. Inflamm. Dis. 2021, 9, 1618–1630. [Google Scholar] [CrossRef]
- Liu, C.; Zhao, X.-M.; Wang, Q.; Du, T.-T.; Zhang, M.-X.; Wang, H.-Z.; Li, R.-P.; Liang, K.; Gao, Y.; Zhou, S.-Y.; et al. Astrocyte-derived SerpinA3N promotes neuroinflammation and epileptic seizures by activating the NF-κB signaling pathway in mice with temporal lobe epilepsy. J. Neuroinflammation 2023, 20, 161. [Google Scholar] [CrossRef]
- Yamanaka, G.; Ishida, Y.; Kanou, K.; Suzuki, S.; Watanabe, Y.; Takamatsu, T.; Morichi, S.; Go, S.; Oana, S.; Yamazaki, T.; et al. Towards a Treatment for Neuroinflammation in Epilepsy: Interleukin-1 Receptor Antagonist, Anakinra, as a Potential Treatment in Intractable Epilepsy. Int. J. Mol. Sci. 2021, 22, 6282. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Klechikov, A.G.; Gharibyan, A.L.; Wärmländer, S.K.T.S.; Jarvet, J.; Zhao, L.; Jia, X.; Shankar, S.K.; Olofsson, A.; Brännström, T.; et al. The role of pro-inflammatory S100A9 in Alzheimer’s disease amyloid-neuroinflammatory cascade. Acta Neuropathol. 2014, 127, 507–522. [Google Scholar] [CrossRef] [PubMed]
- Horvath, I.; Iashchishyn, I.A.; Moskalenko, R.A.; Wang, C.; Wärmländer, S.K.T.S.; Wallin, C.; Gräslund, A.; Kovacs, G.G.; Morozova-Roche, L.A. Co-aggregation of pro-inflammatory S100A9 with α-synuclein in Parkinson’s disease: Ex vivo and in vitro studies. J. Neuroinflammation 2018, 15, 172. [Google Scholar] [CrossRef] [PubMed]
- Xia, P.; Ji, X.; Yan, L.; Lian, S.; Chen, Z.; Luo, Y. Roles of S100A8, S100A9 and S100A12 in infection, inflammation and immunity. Immunology 2023, 171, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-Q.; Dai, X.-D.; Ran, Y.; Cao, Y.; Lan, C.-L.; Guan, J.-T.; Liu, C.; Yang, F.-M.; Gan, Y.-J.; Liu, B.-J.; et al. Circulating S100A8/A9 Levels Reflect Intraocular Inflammation in Uveitis Patients. Ocul. Immunol. Inflamm. 2020, 28, 133–141. [Google Scholar] [CrossRef]
- Tong, L.; Lan, W.; Lim, R.R.; Chaurasia, S.S. S100A Proteins as Molecular Targets in the Ocular Surface Inflammatory Diseases. Ocul. Surf. 2014, 12, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Ali, S.A. Multifunctional Role of S100 Protein Family in the Immune System: An Update. Cells 2022, 11, 2274. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.T.; Zhou, L.; Li, J.; Tong, L.; Zhao, S.Z.; Li, X.R.; Yu, S.J.; Koh, S.K.; Beuerman, R.W. Proteomic Profiling of Inflammatory Signaling Molecules in the Tears of Patients on Chronic Glaucoma Medication. Investig. Opthalmol. Vis. Sci. 2011, 52, 7385. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.C.; Dá Mesquita, S.; Sousa, J.C.; Correia-Neves, M.; Sousa, N.; Palha, J.A.; Marques, F. From the periphery to the brain: Lipocalin-2, a friend or foe? Prog. Neurobiol. 2015, 131, 120–136. [Google Scholar] [CrossRef]
- Dekens, D.W.; Naudé, P.J.W.; Engelborghs, S.; Vermeiren, Y.; Van Dam, D.; Oude Voshaar, R.C.; Eisel, U.L.M.; De Deyn, P.P. Neutrophil Gelatinase-Associated Lipocalin and its Receptors in Alzheimer’s Disease (AD) Brain Regions: Differential Findings in AD with and without Depression. J. Alzheimers Dis. 2016, 55, 763–776. [Google Scholar] [CrossRef]
- Llorens, F.; Hermann, P.; Villar-Piqué, A.; Diaz-Lucena, D.; Nägga, K.; Hansson, O.; Santana, I.; Schmitz, M.; Schmidt, C.; Varges, D.; et al. Cerebrospinal fluid lipocalin 2 as a novel biomarker for the differential diagnosis of vascular dementia. Nat. Commun. 2020, 11, 619. [Google Scholar] [CrossRef]
- Kim, B.-W.; Jeong, K.H.; Kim, J.-H.; Jin, M.; Kim, J.-H.; Lee, M.-G.; Choi, D.-K.; Won, S.-Y.; McLean, C.; Jeon, M.-T.; et al. Pathogenic Upregulation of Glial Lipocalin-2 in the Parkinsonian Dopaminergic System. J. Neurosci. 2016, 36, 5608–5622. [Google Scholar] [CrossRef] [PubMed]
- Al Nimer, F.; Elliott, C.; Bergman, J.; Khademi, M.; Dring, A.M.; Aeinehband, S.; Bergenheim, T.; Romme Christensen, J.; Sellebjerg, F.; Svenningsson, A.; et al. Lipocalin-2 is increased in progressive multiple sclerosis and inhibits remyelination. Neurol. Neuroimmunol. Neuroinflammation 2016, 3, e191. [Google Scholar] [CrossRef]
- Mesquita, S.D.; Ferreira, A.C.; Falcao, A.M.; Sousa, J.C.; Oliveira, T.G.; Correia-Neves, M.; Sousa, N.; Marques, F.; Palha, J.A. Lipocalin 2 modulates the cellular response to amyloid beta. Cell Death Differ. 2014, 21, 1588–1599. [Google Scholar] [CrossRef]
- Yasuda, M.; Tanaka, Y.; Omodaka, K.; Nishiguchi, K.M.; Nakamura, O.; Tsuda, S.; Nakazawa, T. Transcriptome profiling of the rat retina after optic nerve transection. Sci. Rep. 2016, 6, 28736. [Google Scholar] [CrossRef] [PubMed]
- Ueno, S.; Yoneshige, A.; Koriyama, Y.; Hagiyama, M.; Shimomura, Y.; Ito, A. Early Gene Expression Profile in Retinal Ganglion Cell Layer After Optic Nerve Crush in Mice. Investig. Opthalmol. Vis. Sci. 2018, 59, 370. [Google Scholar] [CrossRef]
- Yoneshige, A.; Hagiyama, M.; Takashima, Y.; Ueno, S.; Inoue, T.; Kimura, R.; Koriyama, Y.; Ito, A. Elevated Hydrostatic Pressure Causes Retinal Degeneration Through Upregulating Lipocalin-2. Front. Cell Dev. Biol. 2021, 9, 664327. [Google Scholar] [CrossRef]
- Tokito, A.; Jougasaki, M. Matrix Metalloproteinases in Non-Neoplastic Disorders. Int. J. Mol. Sci. 2016, 17, 1178. [Google Scholar] [CrossRef] [PubMed]
- O’Callaghan, J.; Crosbie, D.E.; Cassidy, P.S.; Sherwood, J.M.; Flügel-Koch, C.; Lütjen-Drecoll, E.; Humphries, M.M.; Reina-Torres, E.; Wallace, D.; Kiang, A.-S.; et al. Therapeutic potential of AAV-mediated MMP-3 secretion from corneal endothelium in treating glaucoma. Hum. Mol. Genet. 2017, 26, 1230–1246. [Google Scholar] [CrossRef]
- Weinreb, R.N.; Robinson, M.R.; Dibas, M.; Stamer, W.D. Matrix Metalloproteinases and Glaucoma Treatment. J. Ocul. Pharmacol. Ther. 2020, 36, 208–228. [Google Scholar] [CrossRef]
- O’Callaghan, J.; Delaney, C.; O’Connor, M.; Van Batenburg-Sherwood, J.; Schicht, M.; Lütjen-Drecoll, E.; Hudson, N.; Ni Dhubhghaill, S.; Humphries, P.; Stanley, C.; et al. Matrix metalloproteinase-3 (MMP-3)–mediated gene therapy for glaucoma. Sci. Adv. 2023, 9, eadf6537. [Google Scholar] [CrossRef] [PubMed]
- Zenobia, C.; Hajishengallis, G. Basic biology and role of interleukin-17 in immunity and inflammation. Periodontol. 2000 2015, 69, 142–159. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, X.; Zhong, Y. Interleukin-17A: The Key Cytokine in Neurodegenerative Diseases. Front. Aging Neurosci. 2020, 12, 566922. [Google Scholar] [CrossRef] [PubMed]
- Milovanovic, J.; Arsenijevic, A.; Stojanovic, B.; Kanjevac, T.; Arsenijevic, D.; Radosavljevic, G.; Milovanovic, M.; Arsenijevic, N. Interleukin-17 in Chronic Inflammatory Neurological Diseases. Front. Immunol. 2020, 11, 947. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhong, H.; Yu, H.; Sun, J.; Shen, B.; Xu, X.; Huang, S.; Huang, P.; Zhong, Y. Interleukin-17A modulates retinal inflammation by regulating microglial activation via the p38 MAPK pathway in experimental glaucoma neuropathy. FASEB J. 2023, 37, e22945. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.S.; Koh, S.-H. Neuroinflammation in neurodegenerative disorders: The roles of microglia and astrocytes. Transl. Neurodegener. 2020, 9, 42. [Google Scholar] [CrossRef] [PubMed]
- Scimone, C.; Granata, F.; Longo, M.; Mormina, E.; Turiaco, C.; Caragliano, A.A.; Donato, L.; Sidoti, A.; D’Angelo, R. Germline Mutation Enrichment in Pathways Controlling Endothelial Cell Homeostasis in Patients with Brain Arteriovenous Malformation: Implication for Molecular Diagnosis. Int. J. Mol. Sci. 2020, 21, 4321. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.; Sun, X. Contribution of Interleukin-17A to Retinal Degenerative Diseases. Front. Immunol. 2022, 13, 847937. [Google Scholar] [CrossRef] [PubMed]
- Dobrzycka, M.; Sulewska, A.; Biecek, P.; Charkiewicz, R.; Karabowicz, P.; Charkiewicz, A.; Golaszewska, K.; Milewska, P.; Michalska-Falkowska, A.; Nowak, K.; et al. miRNA Studies in Glaucoma: A Comprehensive Review of Current Knowledge and Future Perspectives. Int. J. Mol. Sci. 2023, 24, 14699. [Google Scholar] [CrossRef]
- Wang, Y.; Niu, L.; Zhao, J.; Wang, M.; Li, K.; Zheng, Y. An update: Mechanisms of microRNA in primary open-angle glaucoma. Brief. Funct. Genomics 2021, 20, 19–27. [Google Scholar] [CrossRef]
- Kondkar, A.A.; Azad, T.A.; Sultan, T.; Radhakrishnan, R.; Osman, E.A.; Almobarak, F.A.; Lobo, G.P.; Al-Obeidan, S.A. Polymorphism rs3742330 in microRNA Biogenesis Gene DICER1 Is Associated with Pseudoexfoliation Glaucoma in Saudi Cohort. Genes 2022, 13, 489. [Google Scholar] [CrossRef]
- Rao, A.; Chakraborty, M.; Roy, A.; Sahay, P.; Pradhan, A.; Raj, N. Differential miRNA Expression: Signature for Glaucoma in Pseudoexfoliation. Clin. Ophthalmol. 2020, 14, 3025–3038. [Google Scholar] [CrossRef]
- Kathirvel, K.; Fan, X.; Haribalaganesh, R.; Bharanidharan, D.; Sharmila, R.; Krishnadas, R.; Muthukkaruppan, V.; Willoughby, C.E.; Senthilkumari, S. Small RNA Sequencing Reveals a Distinct MicroRNA Signature between Glucocorticoid Responder and Glucocorticoid Non-Responder Primary Human Trabecular Meshwork Cells after Dexamethasone Treatment. Genes 2023, 14, 2012. [Google Scholar] [CrossRef]
- Zingale, V.D.; Gugliandolo, A.; Mazzon, E. MiR-155: An Important Regulator of Neuroinflammation. Int. J. Mol. Sci. 2021, 23, 90. [Google Scholar] [CrossRef]
- Yan, L.; Lee, S.; Lazzaro, D.R.; Aranda, J.; Grant, M.B.; Chaqour, B. Single and Compound Knock-outs of MicroRNA (miRNA)-155 and Its Angiogenic Gene Target CCN1 in Mice Alter Vascular and Neovascular Growth in the Retina via Resident Microglia. J. Biol. Chem. 2015, 290, 23264–23281. [Google Scholar] [CrossRef]
- Yin, H.; Song, S.; Pan, X. Knockdown of miR-155 protects microglia against LPS-induced inflammatory injury via targeting RACK1: A novel research for intracranial infection. J. Inflamm. 2017, 14, 17. [Google Scholar] [CrossRef]
- Wu, D.; Cerutti, C.; Lopez-Ramirez, M.A.; Pryce, G.; King-Robson, J.; Simpson, J.E.; Van Der Pol, S.M.; Hirst, M.C.; De Vries, H.E.; Sharrack, B.; et al. Brain Endothelial miR-146a Negatively Modulates T-Cell Adhesion through Repressing Multiple Targets to Inhibit NF-κB Activation. J. Cereb. Blood Flow Metab. 2015, 35, 412–423. [Google Scholar] [CrossRef]
- Chen, L.; Dong, R.; Lu, Y.; Zhou, Y.; Li, K.; Zhang, Z.; Peng, M. MicroRNA-146a protects against cognitive decline induced by surgical trauma by suppressing hippocampal neuroinflammation in mice. Brain. Behav. Immun. 2019, 78, 188–201. [Google Scholar] [CrossRef]
- Ishikawa, M.; Izumi, Y.; Sato, K.; Sato, T.; Zorumski, C.F.; Kunikata, H.; Nakazawa, T. Glaucoma and microglia-induced neuroinflammation. Front. Ophthalmol. 2023, 3, 1132011. [Google Scholar] [CrossRef]
- Tezel, G. Molecular regulation of neuroinflammation in glaucoma: Current knowledge and the ongoing search for new treatment targets. Prog. Retin. Eye Res. 2022, 87, 100998. [Google Scholar] [CrossRef] [PubMed]
- Howell, G.R.; Macalinao, D.G.; Sousa, G.L.; Walden, M.; Soto, I.; Kneeland, S.C.; Barbay, J.M.; King, B.L.; Marchant, J.K.; Hibbs, M.; et al. Molecular clustering identifies complement and endothelin induction as early events in a mouse model of glaucoma. J. Clin. Investig. 2011, 121, 1429–1444. [Google Scholar] [CrossRef]
- Tribble, J.R.; Kokkali, E.; Otmani, A.; Plastino, F.; Lardner, E.; Vohra, R.; Kolko, M.; André, H.; Morgan, J.E.; Williams, P.A. When Is a Control Not a Control? Reactive Microglia Occur Throughout the Control Contralateral Pathway of Retinal Ganglion Cell Projections in Experimental Glaucoma. Transl. Vis. Sci. Technol. 2021, 10, 22. [Google Scholar] [CrossRef]
- Rutigliani, C.; Tribble, J.R.; Hagström, A.; Lardner, E.; Jóhannesson, G.; Stålhammar, G.; Williams, P.A. Widespread retina and optic nerve neuroinflammation in enucleated eyes from glaucoma patients. Acta Neuropathol. Commun. 2022, 10, 118. [Google Scholar] [CrossRef]
- Williams, P.A.; Tribble, J.R.; Pepper, K.W.; Cross, S.D.; Morgan, B.P.; Morgan, J.E.; John, S.W.M.; Howell, G.R. Inhibition of the classical pathway of the complement cascade prevents early dendritic and synaptic degeneration in glaucoma. Mol. Neurodegener. 2016, 11, 26. [Google Scholar] [CrossRef]
- Wu, M.; Xu, L.; Wang, Y.; Zhou, N.; Zhen, F.; Zhang, Y.; Qu, X.; Fan, H.; Liu, S.; Chen, Y.; et al. S100A8/A9 induces microglia activation and promotes the apoptosis of oligodendrocyte precursor cells by activating the NF-κB signaling pathway. Brain Res. Bull. 2018, 143, 234–245. [Google Scholar] [CrossRef]
- Bai, Q.; Sun, D.; Zeng, Y.; Zhu, J.; Zhang, C.; Zhang, X.; Chen, L.; Zhou, X.; Ye, L.; Tang, Y.; et al. Effect of Proinflammatory S100A9 Protein on Migration and Proliferation of Microglial Cells. J. Mol. Neurosci. 2023, 73, 983–995. [Google Scholar] [CrossRef]
- Matt, S.M. Epigenetic Regulation of Neuroinflammation in the Aged Mouse Brain; University of Illinois: Champaign, IL, USA, 2018. [Google Scholar]
- Retinasamy, T.; Shaikh, M.F. Interleukin 1 receptor antagonist and neurodegenerative diseases: The future treatment strategy. Neurosci. Res. Notes 2023, 6, 164. [Google Scholar] [CrossRef]
- Naaman, E.; Qarawani, A.; Ben-Zvi Elimelech, R.; Harel, M.; Sigal-Dror, S.; Safuri, S.; Smirnovas, V.; Baronaite, I.; Romanova, N.V.; Morozova-Roche, L.A.; et al. The Surprising Nonlinear Effects of S100A9 Proteins in the Retina. ACS Chem. Neurosci. 2024, 15, 735–744. [Google Scholar] [CrossRef]
- Zhang, X.; Sun, D.; Zhou, X.; Zhang, C.; Yin, Q.; Chen, L.; Tang, Y.; Liu, Y.; Morozova-Roche, L.A. Proinflammatory S100A9 stimulates TLR4/NF-κB signaling pathways causing enhanced phagocytic capacity of microglial cells. Immunol. Lett. 2023, 255, 54–61. [Google Scholar] [CrossRef]
- Wang, G.; Huang, K.; Tian, Q.; Guo, Y.; Liu, C.; Li, Z.; Yu, Z.; Zhang, Z.; Li, M. S100A9 aggravates early brain injury after subarachnoid hemorrhage via inducing neuroinflammation and inflammasome activation. iScience 2024, 27, 109165. [Google Scholar] [CrossRef]
- Liao, Y.-L.; Zhou, X.-Y.; Ji, M.-H.; Qiu, L.-C.; Chen, X.-H.; Gong, C.-S.; Lin, Y.; Guo, Y.-H.; Yang, J.-J. S100A9 Upregulation Contributes to Learning and Memory Impairments by Promoting Microglia M1 Polarization in Sepsis Survivor Mice. Inflammation 2021, 44, 307–320. [Google Scholar] [CrossRef]
- Shi, M.; Cao, Y.; Zhang, X.; Zhang, Y.; Zhu, Y.; Liu, L.; Mi, L.; Liu, X.; Zhou, Y.; Li, S.; et al. Inhibition of S100a8/A9 Ameliorates Neuroinflammation by Blocking NET Formation Via AMPK/Nrf2/HO-1 Pathway after Traumatic Brain Injury in Mice; SSRN: Rochester, NY, USA, 2023. [Google Scholar]
- Sun, F.; Zhang, H.; Huang, T.; Shi, J.; Wei, T.; Wang, Y. S100A9 blockade improves the functional recovery after spinal cord injury via mediating neutrophil infiltration. Exp. Ther. Med. 2022, 23, 291. [Google Scholar] [CrossRef]
- Rolle, T.; Ponzetto, A.; Malinverni, L. The Role of Neuroinflammation in Glaucoma: An Update on Molecular Mechanisms and New Therapeutic Options. Front. Neurol. 2021, 11, 612422. [Google Scholar] [CrossRef]
- Silva, C.R.; Melo, B.M.S.; Silva, J.R.; Lopes, A.H.; Pereira, J.A.; Cecilio, N.T.; Berlink, J.; Souza, G.G.; Lucas, G.; Vogl, T.; et al. S100A9 plays a pivotal role in a mouse model of herpetic neuralgia via TLR4/TNF pathway. Brain. Behav. Immun. 2020, 88, 353–362. [Google Scholar] [CrossRef]
- Jing, Y.; Yang, D.; Fu, Y.; Wang, W.; Yang, G.; Yuan, F.; Chen, H.; Ding, J.; Chen, S.; Tian, H. Neuroprotective Effects of Serpina3k in Traumatic Brain Injury. Front. Neurol. 2019, 10, 1215. [Google Scholar] [CrossRef]
- Zhou, T.; Chen, L.; Huang, C.; Lin, Z.; Zong, R.; Zhu, C.; Pan, F.; Ma, J.; Liu, Z.; Zhou, Y. Serine Proteinase Inhibitor SERPINA3K Suppresses Corneal Neovascularization Via Inhibiting Wnt Signaling and VEGF. Investig. Opthalmol. Vis. Sci. 2014, 55, 4863. [Google Scholar] [CrossRef]
- Sanchez-Navarro, A.; González-Soria, I.; Caldiño-Bohn, R.; Bobadilla, N.A. Integrative view of serpins in health and disease: The contribution of serpinA3. Am. J. Physiol.-Cell Physiol. 2020, 320, C106–C118. [Google Scholar] [CrossRef]
- De Mezer, M.; Rogaliński, J.; Przewoźny, S.; Chojnicki, M.; Niepolski, L.; Sobieska, M.; Przystańska, A. SERPINA3: Stimulator or Inhibitor of Pathological Changes. Biomedicines 2023, 11, 156. [Google Scholar] [CrossRef]
- Jung, B.-K.; Ryu, K.-Y. Lipocalin-2: A therapeutic target to overcome neurodegenerative diseases by regulating reactive astrogliosis. Exp. Mol. Med. 2023, 55, 2138–2146. [Google Scholar] [CrossRef]
- Jung, B.-K.; Park, Y.; Yoon, B.; Bae, J.-S.; Han, S.-W.; Heo, J.-E.; Kim, D.-E.; Ryu, K.-Y. Reduced secretion of LCN2 (lipocalin 2) from reactive astrocytes through autophagic and proteasomal regulation alleviates inflammatory stress and neuronal damage. Autophagy 2023, 19, 2296–2317. [Google Scholar] [CrossRef]
- Raeeszadeh-Sarmazdeh, M.; Do, L.; Hritz, B. Metalloproteinases and Their Inhibitors: Potential for the Development of New Therapeutics. Cells 2020, 9, 1313. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinforma. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef] [PubMed]
- Hunt, G.P.; Grassi, L.; Henkin, R.; Smeraldi, F.; Spargo, T.P.; Kabiljo, R.; Koks, S.; Ibrahim, Z.; Dobson, R.J.B.; Al-Chalabi, A.; et al. GEOexplorer: A webserver for gene expression analysis and visualisation. Nucleic Acids Res. 2022, 50, W367–W374. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Manzoni, C.; Kia, D.A.; Vandrovcova, J.; Hardy, J.; Wood, N.W.; Lewis, P.A.; Ferrari, R. Genome, transcriptome and proteome: The rise of omics data and their integration in biomedical sciences. Brief. Bioinform. 2018, 19, 286–302. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J. miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [CrossRef]
- Voigt, A.P.; Whitmore, S.S.; Lessing, N.D.; DeLuca, A.P.; Tucker, B.A.; Stone, E.M.; Mullins, R.F.; Scheetz, T.E. Spectacle: An interactive resource for ocular single-cell RNA sequencing data analysis. Exp. Eye Res. 2020, 200, 108204. [Google Scholar] [CrossRef]
- Li, S.; Xiao, J.; Huang, C.; Sun, J. Identification and validation of oxidative stress and immune-related hub genes in Alzheimer’s disease through bioinformatics analysis. Sci. Rep. 2023, 13, 657. [Google Scholar] [CrossRef] [PubMed]
- Yao, F.; Peng, J.; Zhang, E.; Ji, D.; Gao, Z.; Tang, Y.; Yao, X.; Xia, X. Pathologically high intraocular pressure disturbs normal iron homeostasis and leads to retinal ganglion cell ferroptosis in glaucoma. Cell Death Differ. 2023, 30, 69–81. [Google Scholar] [CrossRef] [PubMed]
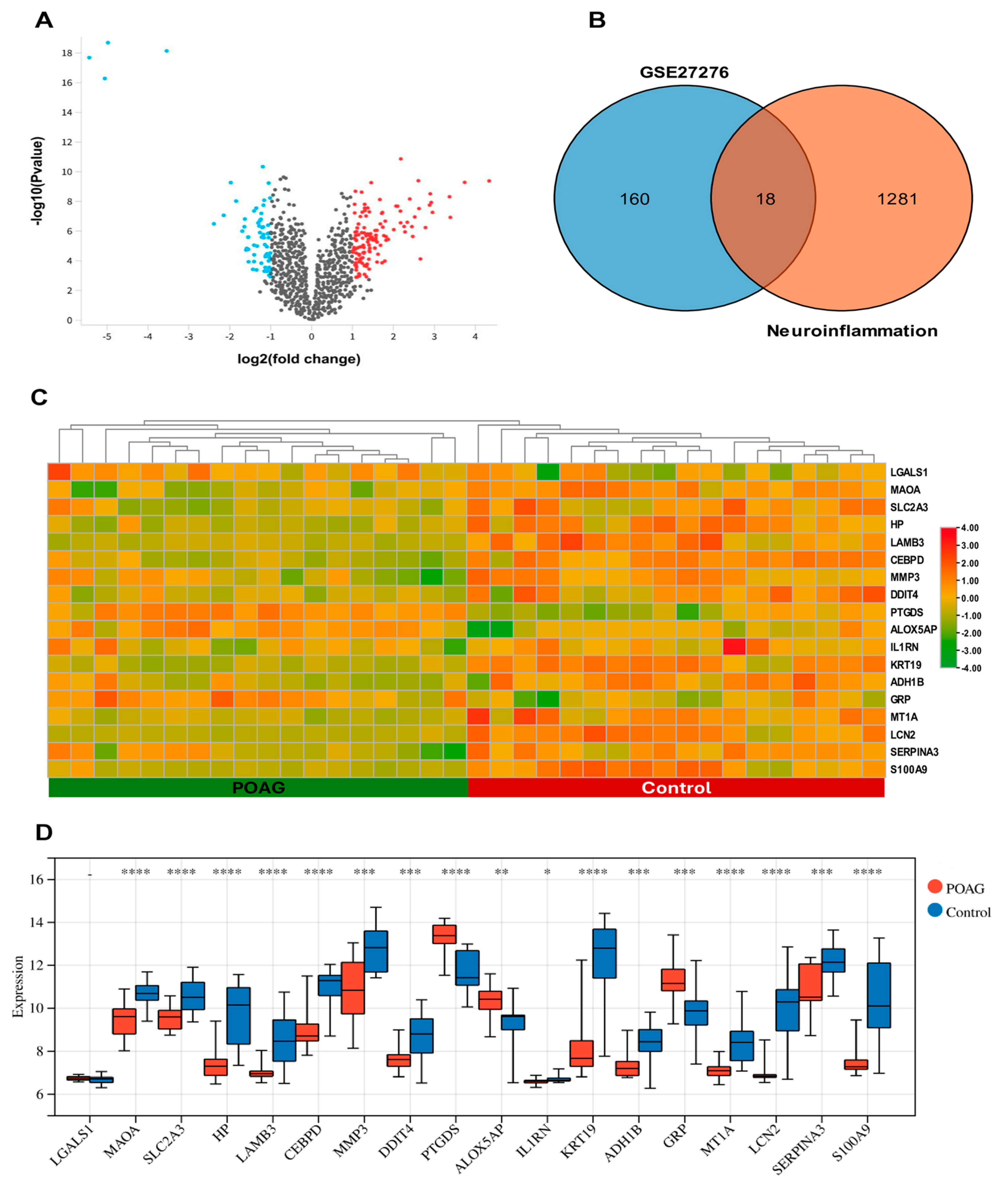



| ID | Gene Symbol | Gene Title | Change | log2 (Fold Change) | log10 (p Value) |
|---|---|---|---|---|---|
| GI_13027803-S | MMP3 | matrix metallopeptidase 3 | down | −1.898 | 4.308 |
| GI_15718674-S | ALOX5AP | arachidonate 5-lipoxygenase activating protein | up | 1.175 | 3.247 |
| GI_27894318-A | IL1RN | interleukin 1 receptor antagonist | down | −1.535 | 4.606 |
| GI_28866959-S | MT1A | metallothionein 1A | down | −1.414 | 5.486 |
| GI_28872797-S | CEBPD | CCAAT/enhancer binding protein delta | down | −2.256 | 11.354 |
| GI_33469954-S | MAOA | monoamine oxidase A | down | −1.452 | 7.383 |
| GI_34222290-S | GRP | gastrin releasing peptide | up | 1.4 | 3.175 |
| GI_34577060-S | ADH1B | alcohol dehydrogenase 1B (class I), beta polypeptide | down | −1.102 | 4.372 |
| GI_38455401-S | LCN2 | lipocalin 2 | down | −2.901 | 7.469 |
| GI_38505192-S | PTGDS | prostaglandin D2 synthase | up | 1.636 | 6.538 |
| GI_40217850-S | KRT19 | keratin 19 | down | −4.316 | 8.929 |
| GI_4557712-S | LAMB3 | laminin subunit beta 3 | down | −1.554 | 5.392 |
| GI_4826761-S | HP | haptoglobin | down | −2.46 | 7.126 |
| GI_5902089-S | SLC2A3 | solute carrier family 2 member 3 | down | −1.021 | 4.429 |
| GI_6006015-S | LGALS1 | galectin 1 | up | 1.598 | 4.509 |
| GI_9506686-S | DDIT4 | DNA damage inducible transcript 4 | down | −1.206 | 4.006 |
| GI_9665246-S | SERPINA3 | serpin family A member 3 | down | −1.474 | 4.358 |
| GI_9845520-S | S100A9 | S100 calcium binding protein A9 | down | −2.799 | 6.001 |
| Term | Description | Enrichment FDR (p-Value) | Gene Count | Fold Enrichment | Genes |
|---|---|---|---|---|---|
| KEGG pathway | |||||
| Path:hsa04657 | IL-17 signaling pathway | 0.0022836 | 3 | 41.00537634 | LCN2 MMP3 S100A9 |
| Path:hsa00350 | Tyrosine metabolism | 0.008155124 | 2 | 70.62037037 | ADH1B MAOA |
| Path:hsa00982 | Drug metabolism-cytochrome P450 | 0.019936646 | 2 | 36.84541063 | ADH1B MAOA |
| Path:hsa04151 | PI3K-Akt signaling pathway | 0.132326939 | 2 | 7.18173258 | LAMB3 DDIT4 |
| Biological processes | |||||
| GO:0010727 | negative reg. of hydrogen peroxide metabolic proc. | 0.000161944 | 2 | 953.375 | MMP3 HP |
| GO:0006953 | acute-phase response | 7.84 × 10−5 | 3 | 200.7105263 | IL1RN SERPINA3 HP |
| GO:0002526 | acute inflammatory response | 0.000268135 | 3 | 88.00384615 | HP IL1RN SERPINA3 |
| GO:0098869 | cellular oxidant detoxification | 0.009726848 | 2 | 71.28037383 | S100A9 HP |
| GO:0046916 | cellular transition metal ion homeostasis | 0.009977053 | 2 | 65.18803419 | LCN2 S100A9 |
| GO:1990748 | cellular detoxification | 0.009977053 | 2 | 62.51639344 | S100A9 HP |
| GO:0097237 | cellular response to a toxic substance | 0.010245208 | 2 | 58.66923077 | S100A9 HP |
| GO:0098754 | detoxification | 0.011070075 | 2 | 51.88435374 | S100A9 HP |
| GO:0042742 | defense response to bacterium | 0.002732388 | 3 | 32.13623596 | LCN2 HP S100A9 |
| GO:0006954 | inflammatory response | 7.84 × 10−5 | 5 | 21.37612108 | IL1RN S100A9 HP SERPINA3 MMP3 |
| Cellular components | |||||
| GO:0031838 | haptoglobin-hemoglobin complex | 0.006548498 | 1 | 346.6818182 | HP |
| GO:0071682 | endocytic vesicle lumen | 0.012072376 | 1 | 158.8958333 | HP |
| GO:0035580 | specific granule lumen | 0.000785589 | 2 | 93.01219512 | LCN2 HP |
| GO:1904724 | tertiary granule lumen | 0.030363329 | 1 | 56.91791045 | HP |
| GO:0031093 | platelet alpha granule lumen | 0.030363329 | 1 | 54.47857143 | SERPINA3 |
| GO:0072562 | blood microparticle | 0.001545822 | 2 | 54.09219858 | SERPINA3 HP |
| GO:0034774 | secretory granule lumen | 8.47 × 10−6 | 4 | 41.45108696 | LCN2 S100A9 SERPINA3 HP |
| GO:0060205 | cytoplasmic vesicle lumen | 8.47 × 10−6 | 4 | 41.11590296 | LCN2 S100A9 SERPINA3 HP |
| GO:0031983 | vesicle lumen | 8.47 × 10−6 | 4 | 40.89544236 | LCN2 S100A9 SERPINA3 HP |
| GO:0035578 | azurophil granule lumen | 0.040033456 | 1 | 36.31904762 | SERPINA3 |
| GO:0042581 | specific granule | 0.003126567 | 2 | 35.97641509 | LCN2 HP |
| GO:0031012 | extracellular matrix | 0.001072635 | 3 | 19.00415282 | S100A9 SERPINA3 MMP3 |
| GO:0030312 | external encapsulating structure | 0.001072635 | 3 | 18.97263682 | S100A9 SERPINA3 MMP3 |
| GO:0062023 | collagen-containing extracellular matrix | 0.011593959 | 2 | 16.83664459 | S100A9 SERPINA3 |
| Molecular functions | |||||
| GO:0005152 | interleukin-1 receptor antagonist activity | 0.010024678 | 1 | 1271.166667 | IL1RN |
| GO:0035662 | Toll-like receptor 4 binding | 0.010024678 | 1 | 953.375 | S100A9 |
| GO:0050543 | icosatetraenoic acid binding | 0.010024678 | 1 | 635.5833333 | S100A9 |
| GO:0050544 | arachidonic acid binding | 0.010024678 | 1 | 635.5833333 | S100A9 |
| GO:0050542 | icosanoid binding | 0.010394826 | 1 | 544.7857143 | S100A9 |
| GO:0035325 | Toll-like receptor binding | 0.011449266 | 1 | 317.7916667 | S100A9 |
| GO:0036041 | long-chain fatty acid binding | 0.013350181 | 1 | 254.2333333 | S100A9 |
| GO:0005149 | interleukin-1 receptor binding | 0.013350181 | 1 | 224.3235294 | IL1RN |
| GO:0019966 | interleukin-1 binding | 0.013350181 | 1 | 224.3235294 | IL1RN |
| GO:0048019 | receptor antagonist activity | 0.015568406 | 1 | 181.5952381 | IL1RN |
| GO:0030547 | signaling receptor inhibitor activity | 0.027296208 | 1 | 93.01219512 | IL1RN |
| GO:0016209 | antioxidant activity | 0.010024678 | 2 | 79.44791667 | S100A9 HP |
| GO:0017171 | serine hydrolase activity | 0.010024678 | 2 | 33.01731602 | HP MMP3 |
| GO:0004175 | endopeptidase activity | 0.017891408 | 2 | 15.34607646 | MMP3 HP |
| Gene | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| LCN2 | CGTCCTAAATGGCCAACCCT | TAGGAAGAGGGGGAGAAGCC |
| S100A9 | TGGCTGCCAAAACAGGATCT | GCCCCAGAACCAAGGTCATT |
| SERPINA3 | GCTGAACTGCACTGTTGTGG | ATGCACACAGAGACCCACAG |
| MMP3 | ATCCCTTTTGATGGGCCTGG | GGATGGAAGAGACGGCCAAA |
| IL1R1 | CTGATCATCCCGTGAGCCTC | GTCTGGACTGTGGACATGCA |
| HP | AGTGAGAATGCGACAGCCAA | TCAGTTGCCCTCACGTACAC |
| GAPDH | AGGTCGGTGTGAACGGATTTG | GGGGTCGTTGATGGCAACA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ullah, Z.; Tao, Y.; Huang, J. Integrated Bioinformatics-Based Identification and Validation of Neuroinflammation-Related Hub Genes in Primary Open-Angle Glaucoma. Int. J. Mol. Sci. 2024, 25, 8193. https://doi.org/10.3390/ijms25158193
Ullah Z, Tao Y, Huang J. Integrated Bioinformatics-Based Identification and Validation of Neuroinflammation-Related Hub Genes in Primary Open-Angle Glaucoma. International Journal of Molecular Sciences. 2024; 25(15):8193. https://doi.org/10.3390/ijms25158193
Chicago/Turabian StyleUllah, Zakir, Yuanyuan Tao, and Jufang Huang. 2024. "Integrated Bioinformatics-Based Identification and Validation of Neuroinflammation-Related Hub Genes in Primary Open-Angle Glaucoma" International Journal of Molecular Sciences 25, no. 15: 8193. https://doi.org/10.3390/ijms25158193






