An Overview on MADS Box Members in Plants: A Meta-Review
Abstract
:1. Introduction
2. Study-Based Meta-Review: Basic Strategy
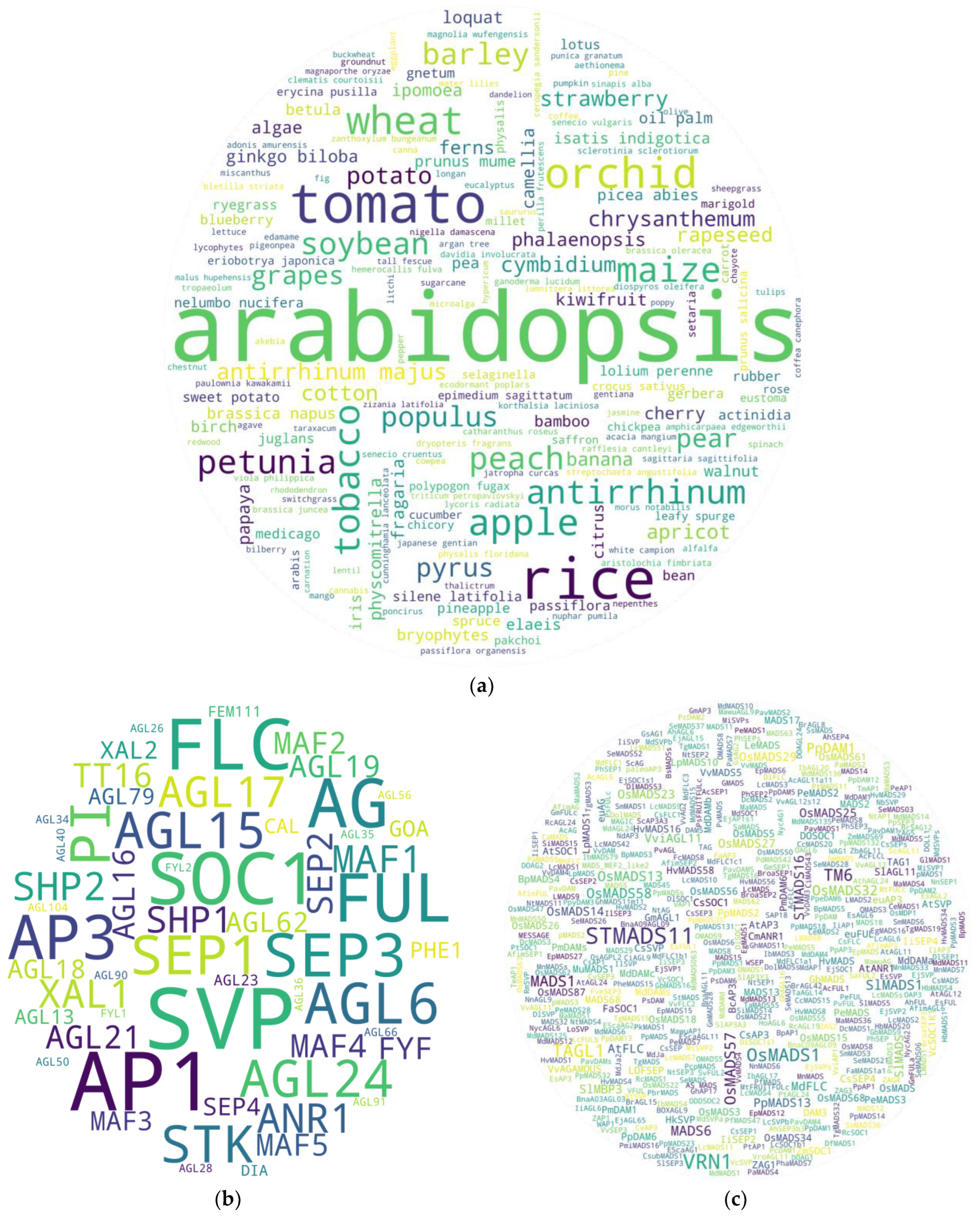
3. General Overview: Plants and Genes of Studies
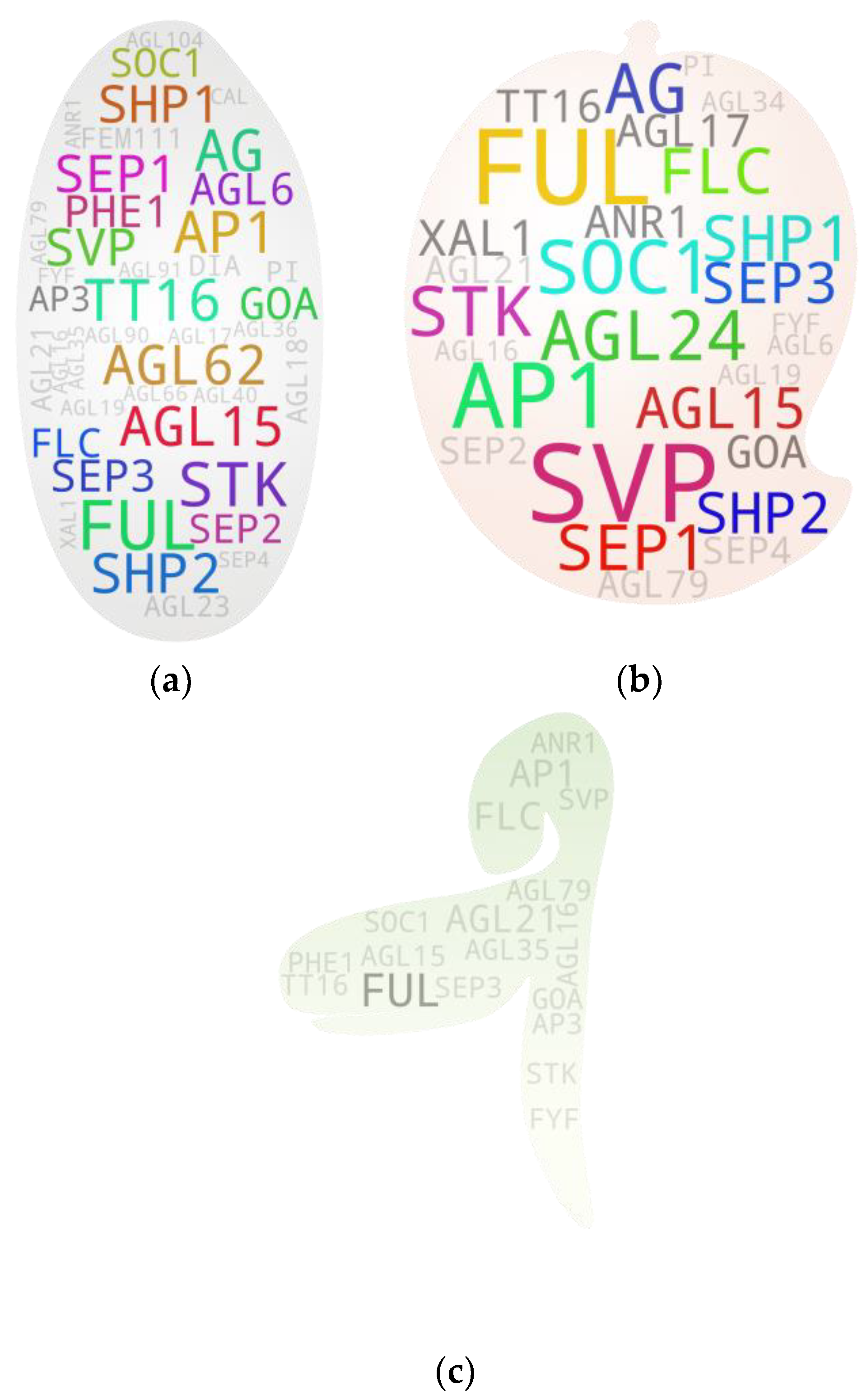
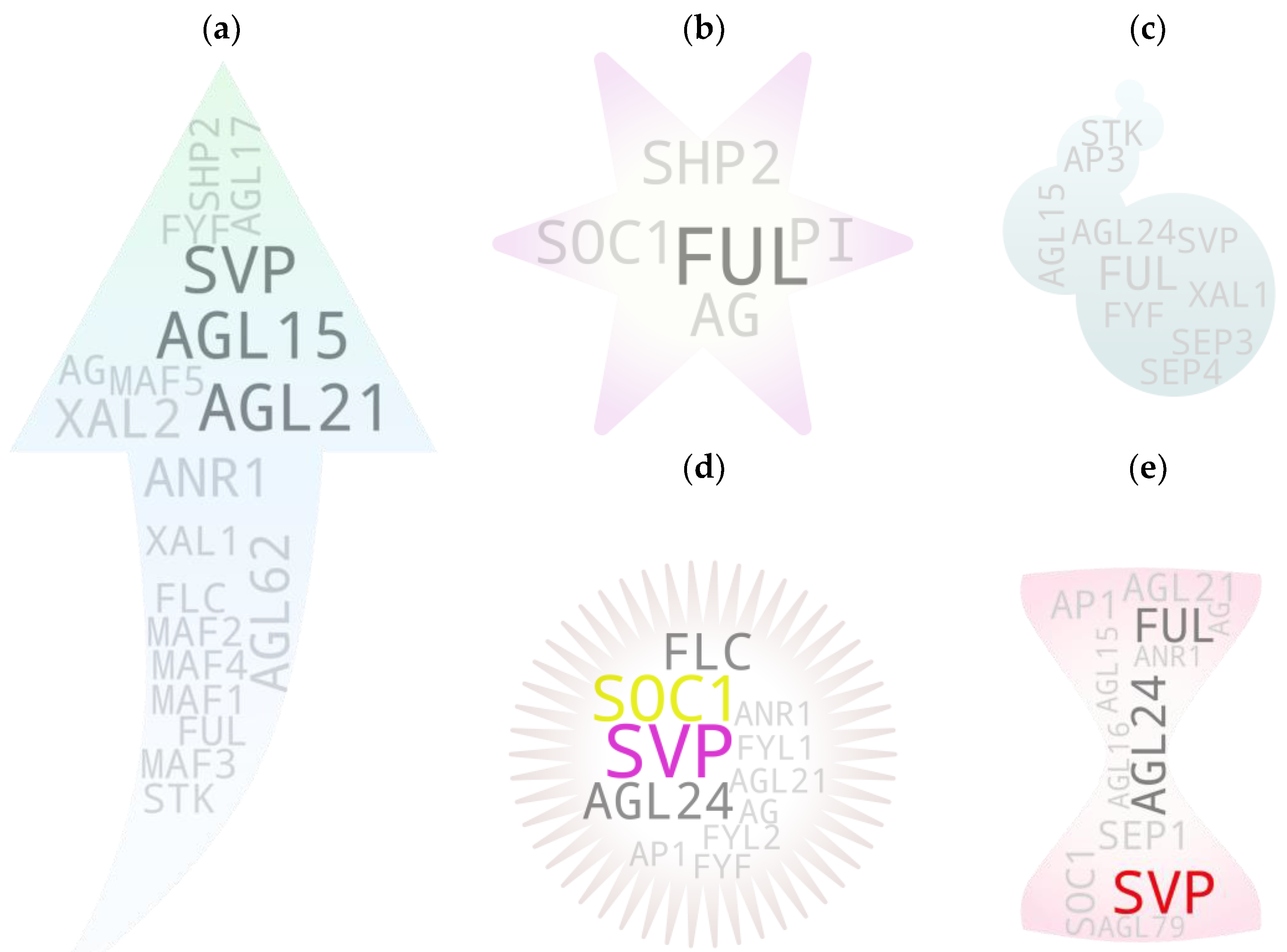
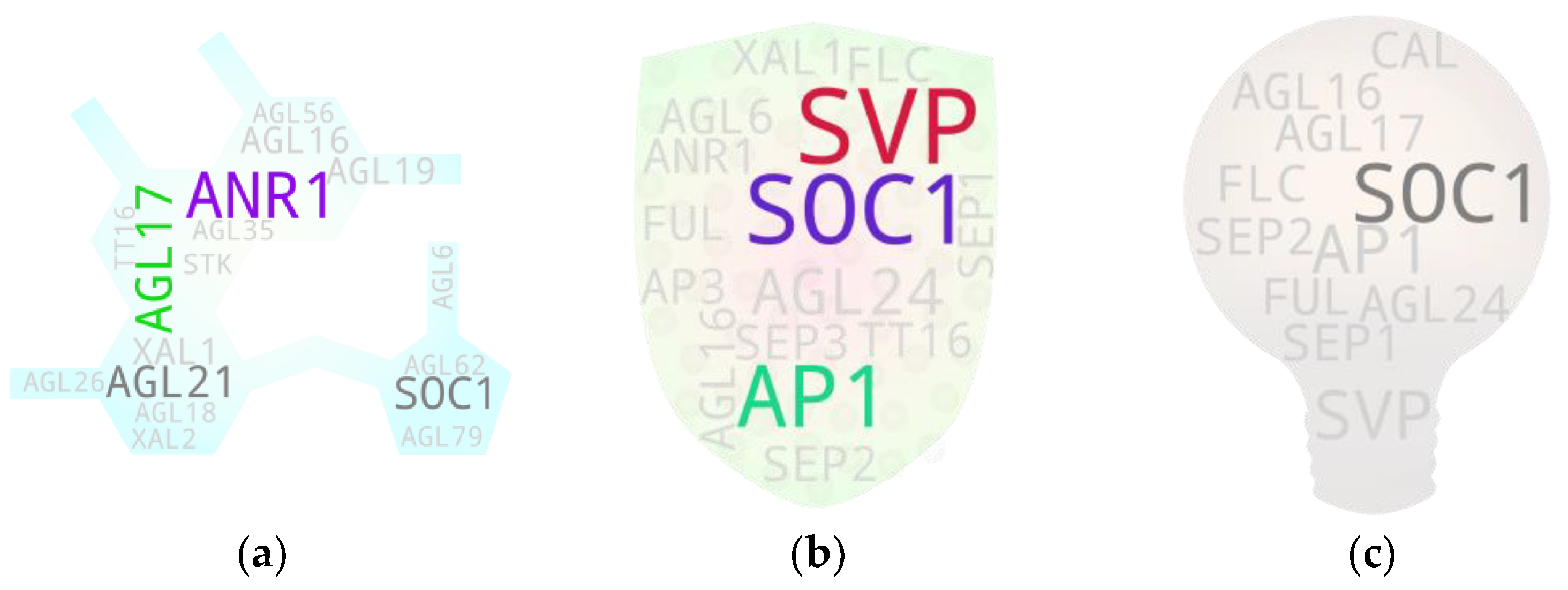
4. Screening Potential Pleiotropics: Sorting Threads from Haystack
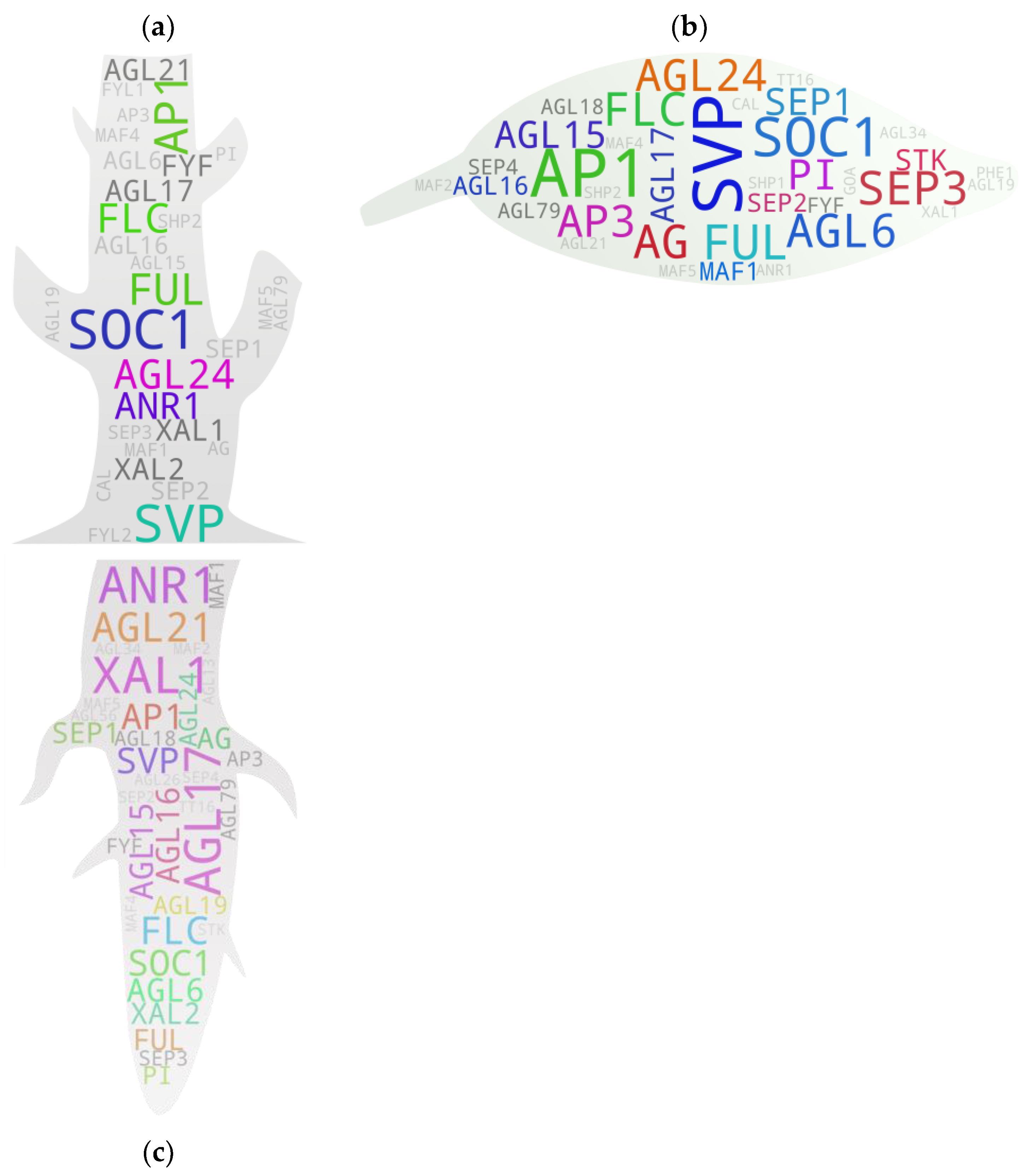
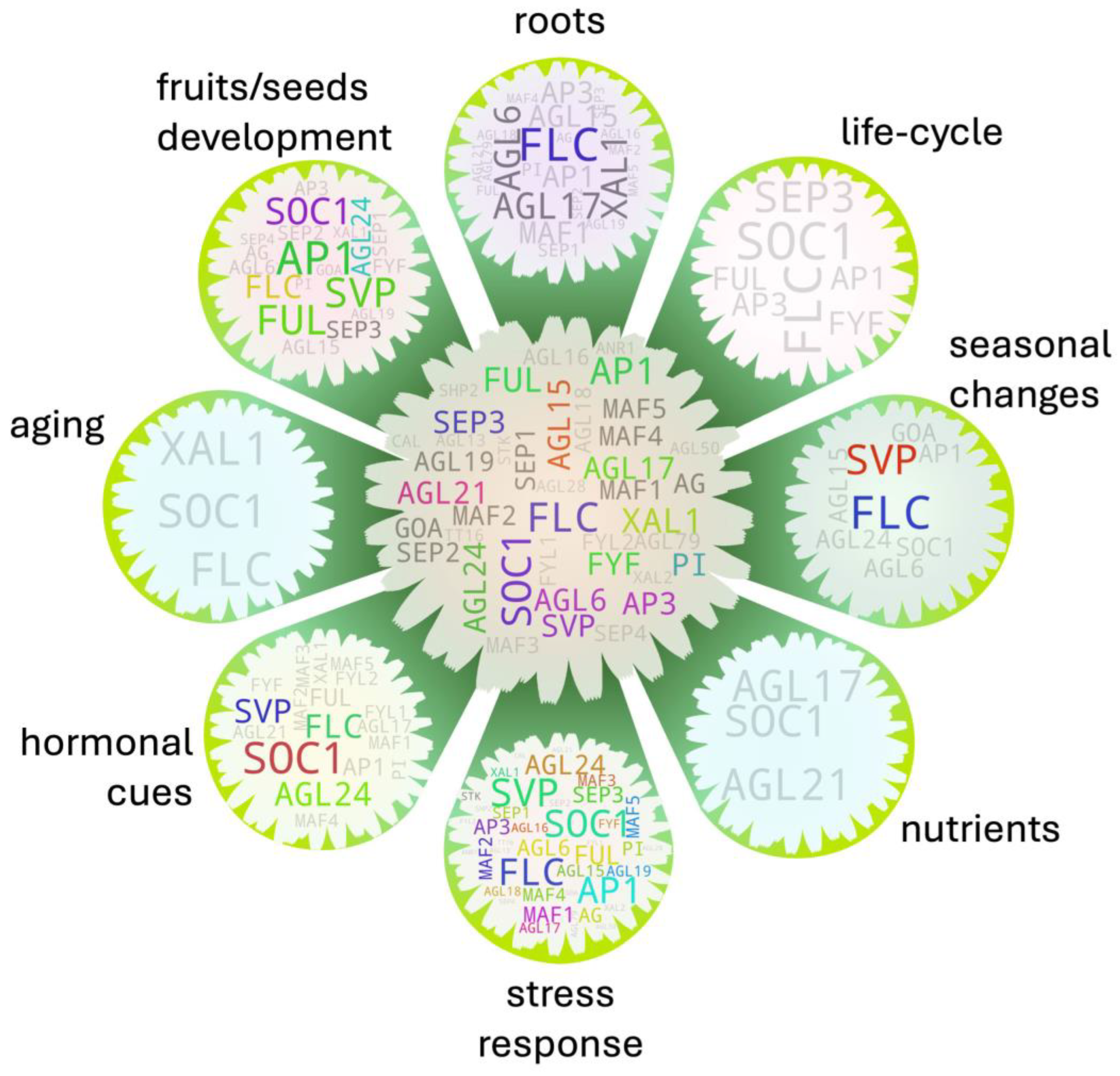
5. Gene-to-Major Tissue Growth Associations
5.1. Shoots
5.2. Leaves
5.3. Roots
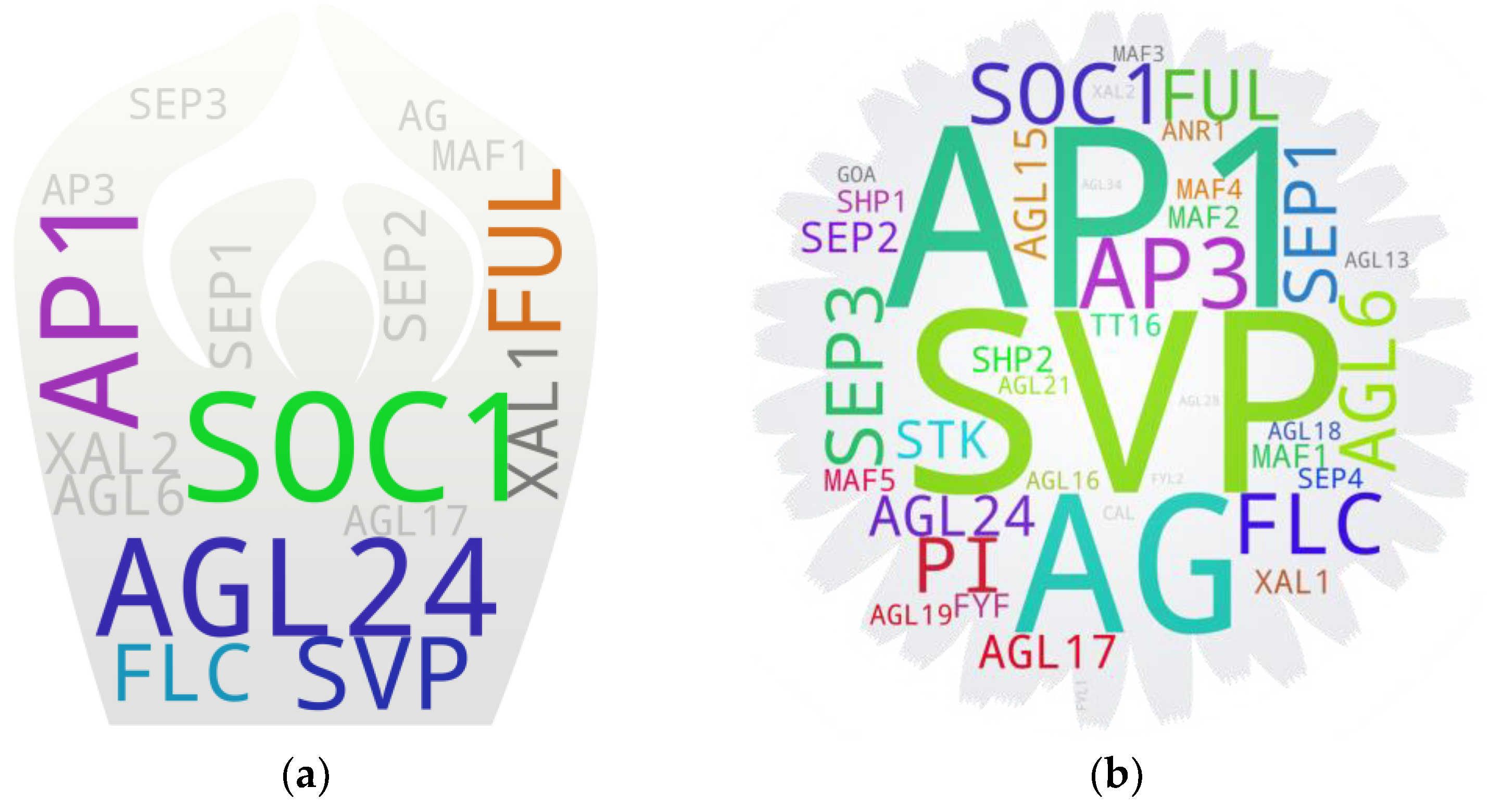
5.4. Apical Meristem
5.5. Flowers
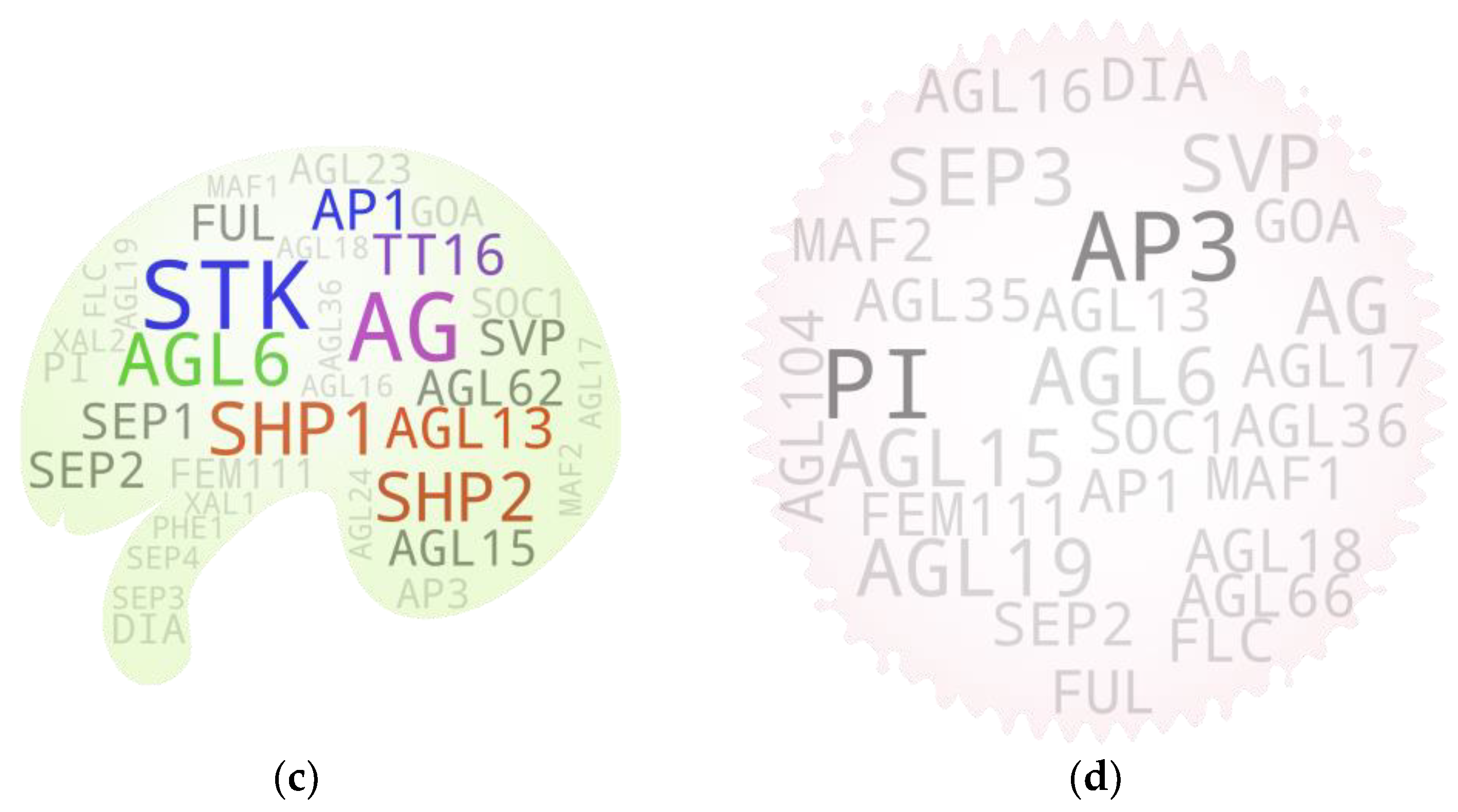
5.6. Ovules
5.7. Pollen
5.8. Seeds
5.9. Fruits
5.10. Seed Germination
6. Gene-to-Factor Associations
6.1. MADS Member–Hormone Association
6.2. MADS Members—Biotic/Abiotic Factor Association
7. Trait-to-Factor Associations Bridged by MADS
8. Optimization Considerations for the Approach
9. Significance and Application of the Approach
10. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schwarz-Sommer, Z.; Huijser, P.; Nacken, W.; Saedler, H.; Sommer, H. Genetic control of flower development by homeotic genes in Antirrhinum majus. Science 1990, 250, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Winter, K.-U.; Meyer, B.; Saedler, H.; Theißen, G. MADS-Box Gene Diversity in Seed Plants 300 Million Years ago. Mol. Biol. Evol. 2000, 17, 1425–1434. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Buylla, E.R.; Corvera-Poiré, A.; Garay-Arroyo, A.; García-Ponce, B.; Jaimes-Miranda, F.; Pérez-Ruiz, R.V. A MADS view of plant development and evolution. In Topics in Animal and Plant Development: From Cell Differentiation to Morphogenesis; Chimal-Monroy, J., Ed.; Transworld Research Network: Kerala, India, 2011; pp. 181–220. [Google Scholar]
- Theissen, G.; Kim, J.T.; Saedler, H. Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J. Mol. Evol. 1996, 43, 484–516. [Google Scholar] [CrossRef] [PubMed]
- Shen, G.; Yang, C.H.; Shen, C.Y.; Huang, K.S. Origination and selection of ABCDE and AGL6 subfamily MADS-box genes in gymnosperms and angiosperms. Biol. Res. 2019, 52, 25. [Google Scholar] [CrossRef]
- Shen, G.; Jia, Y.; Wang, W.L. Evolutionary divergence of motifs in B-class MADS-box proteins of seed plants. J. Biol. Res. 2021, 28, 12. [Google Scholar] [CrossRef]
- Shan, H.; Zahn, L.; Guindon, S.; Wall, P.K.; Kong, H.; Ma, H.; DePamphilis, C.W.; Leebens-Mack, J. Evolution of plant MADS box transcription factors: Evidence for shifts in selection associated with early angiosperm diversification and concerted gene duplications. Mol. Biol. Evol. 2009, 26, 2229–2244. [Google Scholar] [CrossRef]
- Qiu, Y.; Li, Z.; Walther, D.; Kohler, C. Updated phylogeny and protein structure predictions revise the hypothesis on the origin of MADS-box transcription factors in land plants. Mol. Biol. Evol. 2023, 40, msad194. [Google Scholar] [CrossRef]
- Preston, J.C.; Christensen, A.; Malcomber, S.T.; Kellogg, E.A. MADS-box gene expression and implications for developmental origins of the grass spikelet. Am. J. Bot. 2009, 96, 1419–1429. [Google Scholar] [CrossRef]
- Ng, M.; Yanofsky, M.F. Function and evolution of the plant MADS-box gene family. Nat. Rev. Genet. 2001, 2, 186–195. [Google Scholar] [CrossRef]
- Nam, J.; Kim, J.; Lee, S.; An, G.; Ma, H.; Nei, M. Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proc. Natl. Acad. Sci. USA 2004, 101, 1910–1915. [Google Scholar] [CrossRef]
- Smaczniak, C.; Immink, R.G.; Angenent, G.C.; Kaufmann, K. Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development 2012, 139, 3081–3098. [Google Scholar] [CrossRef] [PubMed]
- Mueller, A.; Fillion-Robin, J.-C.; Boidol, R.; Tian, F.; Nechifor, P.; Rampin, R.; Corvellec, M.; Medina, J.; Dai, Y.; Petrushev, B. Amueller/word_cloud-1.9.3. Zenodo 2018. [Google Scholar] [CrossRef]
- Gu, Q.; Ferrandiz, C.; Yanofsky, M.F.; Martienssen, R. The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 1998, 125, 1509–1517. [Google Scholar] [CrossRef] [PubMed]
- Ferrándiz, C.; Liljegren, S.J.; Yanofsky, M.F. Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 2000, 289, 436–438. [Google Scholar] [CrossRef] [PubMed]
- Liljegren, S.J.; Ditta, G.S.; Eshed, Y.; Savidge, B.; Bowman, J.L.; Yanofsky, M.F. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 2000, 404, 766–770. [Google Scholar] [CrossRef] [PubMed]
- Balanzà, V.; Martínez-Fernández, I.; Sato, S.; Yanofsky, M.F.; Ferrándiz, C. Inflorescence meristem fate is dependent on seed development and fruitfull in Arabidopsis thaliana. Front. Plant Sci. 2019, 10, 1622. [Google Scholar] [CrossRef] [PubMed]
- Fuhrer, M.; Gaidora, A.; Venhuizen, P.; Dobrogojski, J.; Beziat, C.; Feraru, M.I.; Kleine-Vehn, J.; Kalyna, M.; Barbez, E. FRUITFULL is a repressor of apical hook opening in Arabidopsis thaliana. Int. J. Mol. Sci. 2020, 21, 6438. [Google Scholar] [CrossRef]
- Elo, A.; Lemmetyinen, J.; Turunen, M.L.; Tikka, L.; Sopanen, T. Three MADS-box genes similar to APETALA1 and FRUITFULL from silver birch (Betula pendula). Physiol. Plant. 2001, 112, 95–103. [Google Scholar] [CrossRef]
- Fujisawa, M.; Shima, Y.; Nakagawa, H.; Kitagawa, M.; Kimbara, J.; Nakano, T.; Kasumi, T.; Ito, Y. Transcriptional regulation of fruit ripening by tomato FRUITFULL homologs and associated MADS box proteins. Plant Cell 2014, 26, 89–101. [Google Scholar] [CrossRef]
- Jiang, X.; Lubini, G.; Hernandes-Lopes, J.; Rijnsburger, K.; Veltkamp, V.; de Maagd, R.A.; Angenent, G.C.; Bemer, M. FRUITFULL-like genes regulate flowering time and inflorescence architecture in tomato. Plant Cell 2022, 34, 1002–1019. [Google Scholar] [CrossRef]
- Kobayashi, K.; Yasuno, N.; Sato, Y.; Yoda, M.; Yamazaki, R.; Kimizu, M.; Yoshida, H.; Nagamura, Y.; Kyozuka, J. Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell 2012, 24, 1848–1859. [Google Scholar] [CrossRef]
- Yin, X.; Liu, X.; Xu, B.; Lu, P.; Dong, T.; Yang, D.; Ye, T.; Feng, Y.Q.; Wu, Y. OsMADS18, a membrane-bound MADS-box transcription factor, modulates plant architecture and the abscisic acid response in rice. J. Exp. Bot. 2019, 70, 3895–3909. [Google Scholar] [CrossRef]
- Yue, Y.; Sun, S.; Li, J.; Yu, H.; Wu, H.; Sun, B.; Li, T.; Han, T.; Jiang, B. GmFULa improves soybean yield by enhancing carbon assimilation without altering flowering time or maturity. Plant Cell Rep. 2021, 40, 1875–1888. [Google Scholar] [CrossRef]
- Liu, G.; Li, C.; Yu, H.; Tao, P.; Yuan, L.; Ye, J.; Chen, W.; Wang, Y.; Ge, P.; Zhang, J.; et al. GREEN STRIPE, encoding methylated TOMATO AGAMOUS-LIKE 1, regulates chloroplast development and Chl synthesis in fruit. New Phytol. 2020, 228, 302–317. [Google Scholar] [CrossRef]
- Ji, D.; Cui, X.; Qin, G.; Chen, T.; Tian, S. SlFERL interacts with S-adenosylmethionine synthetase to regulate fruit ripening. Plant Physiol. 2020, 184, 2168–2181. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.s.; Guo, P.y.; Zhang, J.l.; Xie, Q.l.; Shen, H.; Hu, Z.l.; Chen, G.p. Overexpression of the MADS-box gene SIMBP21 alters leaf morphology and affects reproductive development in tomato. J. Integr. Agric. 2021, 20, 3170–3185. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, Z.; Yao, Q.; Guo, X.; Nguyen, V.; Li, F.; Chen, G. A tomato MADS-box protein, SlCMB1, regulates ethylene biosynthesis and carotenoid accumulation during fruit ripening. Sci. Rep. 2018, 8, 3413. [Google Scholar] [CrossRef] [PubMed]
- Parenicova, L.; de Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B.; et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef]
- Bemer, M.; van Mourik, H.; Muiño, J.M.; Ferrándiz, C.; Kaufmann, K.; Angenent, G.C. FRUITFULL controls SAUR10 expression and regulates Arabidopsis growth and architecture. J. Exp. Bot. 2017, 68, 3391–3403. [Google Scholar] [CrossRef]
- Gregis, V.; Sessa, A.; Colombo, L.; Kater, M.M. AGL24, SHORT VEGETATIVE PHASE, and APETALA1 redundantly control AGAMOUS during early stages of flower development in Arabidopsis. Plant Cell 2006, 18, 1373–1382. [Google Scholar] [CrossRef]
- Gomez-Soto, D.; Ramos-Sanchez, J.M.; Alique, D.; Conde, D.; Triozzi, P.M.; Perales, M.; Allona, I. Overexpression of a SOC1-related gene promotes bud break in ecodormant poplars. Front. Plant Sci. 2021, 12, 670497. [Google Scholar] [CrossRef] [PubMed]
- Ahn, M.-S.; Kim, Y.-S.; Han, J.Y.; Yoon, E.S.; Choi, Y.E. Panax ginseng PgMADS1, an AP1/FUL-like MADS-box gene, is activated by hormones and is involved in inflorescence growth. Plant Cell Tissue Organ Cult. 2015, 122, 161–173. [Google Scholar] [CrossRef]
- Gan, Y.; Bernreiter, A.; Filleur, S.; Abram, B.; Forde, B.G. Overexpressing the ANR1 MADS-box gene in transgenic plants provides new insights into its role in the nitrate regulation of root development. Plant Cell Physiol. 2012, 53, 1003–1016. [Google Scholar] [CrossRef] [PubMed]
- Digel, B.; Tavakol, E.; Verderio, G.; Tondelli, A.; Xu, X.; Cattivelli, L.; Rossini, L.; von Korff, M. Photoperiod-H1 (Ppd-H1) controls leaf size. Plant Physiol. 2016, 172, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Willmann, M.R.; Poethig, R.S. The effect of the floral repressor FLC on the timing and progression of vegetative phase change in Arabidopsis. Development 2011, 138, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, U.; Höhmann, S.; Nettesheim, K.; Wisman, E.; Saedler, H.; Huijser, P. Molecular cloning of SVP: A negative regulator of the floral transition in Arabidopsis. Plant J. 2000, 21, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Ó’Maoiléidigh, D.S.; Wuest, S.E.; Rae, L.; Raganelli, A.; Ryan, P.T.; Kwaśniewska, K.; Das, P.; Lohan, A.J.; Loftus, B.; Graciet, E.; et al. Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. Plant Cell 2013, 25, 2482–2503. [Google Scholar] [CrossRef]
- O’Maoileidigh, D.S.; Stewart, D.; Zheng, B.; Coupland, G.; Wellmer, F. Floral homeotic proteins modulate the genetic program for leaf development to suppress trichome formation in flowers. Development 2018, 145, dev157784. [Google Scholar] [CrossRef]
- Fernandez, D.E.; Wang, C.T.; Zheng, Y.; Adamczyk, B.J.; Singhal, R.; Hall, P.K.; Perry, S.E. The MADS-domain factors AGAMOUS-LIKE15 and AGAMOUS-LIKE18, along with short vegetative phase and AGAMOUS-LIKE24, are necessary to block floral gene expression during the vegetative phase. Plant Physiol. 2014, 165, 1591–1603. [Google Scholar] [CrossRef]
- Yoo, S.K.; Hong, S.M.; Lee, J.S.; Ahn, J.H. A genetic screen for leaf movement mutants identifies a potential role for AGAMOUS-LIKE 6 (AGL6) in circadian-clock control. Mol. Cells 2011, 31, 281–287. [Google Scholar] [CrossRef]
- Huang, N.C.; Tien, H.C.; Yu, T.S. Arabidopsis leaf-expressed AGAMOUS-LIKE 24 mRNA systemically specifies floral meristem differentiation. New Phytol. 2024, 241, 504–515. [Google Scholar] [CrossRef]
- Gan, Y.; Filleur, S.; Rahman, A.; Gotensparre, S.; Forde, B.G. Nutritional regulation of ANR1 and other root-expressed MADS-box genes in Arabidopsis thaliana. Planta 2005, 222, 730–742. [Google Scholar] [CrossRef]
- Alvarez-Buylla, E.R.; Garcia-Ponce, B.; Sanchez, M.P.; Espinosa-Soto, C.; Garcia-Gomez, M.L.; Pineyro-Nelson, A.; Garay-Arroyo, A. MADS-box genes underground becoming mainstream: Plant root developmental mechanisms. New Phytol. 2019, 223, 1143–1158. [Google Scholar] [CrossRef]
- Kim, S.H.; Mizuno, K.; Fujimura, T. Isolation of MADS-box genes from sweet potato (Ipomoea batatas (L.) Lam.) expressed specifically in vegetative tissues. Plant Cell Physiol. 2002, 43, 314–322. [Google Scholar] [CrossRef]
- Nasrollahi, V.; Yuan, Z.C.; Lu, Q.S.M.; McDowell, T.; Kohalmi, S.E.; Hannoufa, A. Deciphering the role of SPL12 and AGL6 from a genetic module that functions in nodulation and root regeneration in Medicago sativa. Plant Mol. Biol. 2022, 110, 511–529. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Hamada, T.; Otani, M.; Shimada, T. Isolation and characterization of MADS box genes possibly related to root development in sweetpotato (Ipomoea batatas L. Lam.). J. Plant Biol. 2005, 48, 387–393. [Google Scholar] [CrossRef]
- Alvarez-Buylla, E.R.; Liljegren, S.J.; Pelaz, S.; Gold, S.E.; Burgeff, C.; Ditta, G.S.; Vergara-Silva, F.; Yanofsky, M.F. MADS-box gene evolution beyond flowers: Expression in pollen, endosperm, guard cells, roots and trichomes. Plant J. 2008, 24, 457–466. [Google Scholar] [CrossRef]
- Adamczyk, B.J.; Lehti Shiu, M.D.; Fernandez, D.E. The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J. 2007, 50, 1007–1019. [Google Scholar] [CrossRef]
- Shah, L.; Sohail, A.; Ahmad, R.; Cheng, S.; Cao, L.; Wu, W. The roles of MADS-box genes from root growth to maturity in Arabidopsis and rice. Agronomy 2022, 12, 582. [Google Scholar] [CrossRef]
- Zhang, X.; Cui, Y.; Yu, M.; Su, B.; Gong, W.; Baluška, F.; Komis, G.; Šamaj, J.; Shan, X.; Lin, J. Phosphorylation-mediated dynamics of nitrate transceptor NRT1.1 regulate auxin flux and nitrate signaling in lateral root growth. Plant Physiol. 2019, 181, 480–498. [Google Scholar] [CrossRef] [PubMed]
- Gan, Y.; Zhou, Z.; An, L.; Bao, S.; Liu, Q.; Srinivasan, M.; Goddard, P. The effects of fluctuations in the nutrient supply on the expression of ANR1 and 11 other MADS box genes in shoots and roots of Arabidopsis thaliana. Botany 2010, 88, 1023–1031. [Google Scholar] [CrossRef]
- Nawy, T.; Lee, J.Y.; Colinas, J.; Wang, J.Y.; Thongrod, S.C.; Malamy, J.E.; Birnbaum, K.; Benfey, P.N. Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell 2005, 17, 1908–1925. [Google Scholar] [CrossRef]
- Burgeff, C.; Liljegren, S.J.; Tapia-Lopez, R.; Yanofsky, M.F.; Alvarez-Buylla, E.R. MADS-box gene expression in lateral primordia, meristems and differentiated tissues of Arabidopsis thaliana roots. Planta 2002, 214, 365–372. [Google Scholar] [CrossRef]
- Han, P.; Garcia-Ponce, B.; Fonseca-Salazar, G.; Alvarez-Buylla, E.R.; Yu, H. AGAMOUS-LIKE 17, a novel flowering promoter, acts in a FT-independent photoperiod pathway. Plant J. 2008, 55, 253–265. [Google Scholar] [CrossRef]
- Zhao, P.X.; Zhang, J.; Chen, S.Y.; Wu, J.; Xia, J.Q.; Sun, L.Q.; Ma, S.S.; Xiang, C.B. AGL16 negatively modulates stress response to balance with growth. bioRxiv 2021. [Google Scholar] [CrossRef]
- Garay-Arroyo, A.; Ortiz-Moreno, E.; de la Paz Sánchez, M.; Murphy, A.S.; García-Ponce, B.; Marsch-Martínez, N.; de Folter, S.; Corvera-Poiré, A.; Jaimes-Miranda, F.; Pacheco-Escobedo, M.A.; et al. The MADS transcription factor XAL2/AGL14 modulates auxin transport during Arabidopsis root development by regulating PIN expression. EMBO J. 2013, 32, 2884–2895. [Google Scholar] [CrossRef]
- Tapia-Lopez, R.; Garcia-Ponce, B.; Dubrovsky, J.G.; Garay-Arroyo, A.; Perez-Ruiz, R.V.; Kim, S.H.; Acevedo, F.; Pelaz, S.; Alvarez-Buylla, E.R. An AGAMOUS-related MADS-box gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis. Plant Physiol. 2008, 146, 1182–1192. [Google Scholar] [CrossRef]
- Johansen, B.; Pedersen, L.B.; Skipper, M.; Frederiksen, S. MADS-box gene evolution-structure and transcription patterns. Mol. Phylogenet. Evol. 2002, 23, 458–480. [Google Scholar] [CrossRef]
- Kinoshita, A.; Vayssieres, A.; Richter, R.; Sang, Q.; Roggen, A.; van Driel, A.D.; Smith, R.S.; Coupland, G. Regulation of shoot meristem shape by photoperiodic signaling and phytohormones during floral induction of Arabidopsis. eLife 2020, 9, e60661. [Google Scholar] [CrossRef]
- Gregis, V.; Sessa, A.; Dorca-Fornell, C.; Kater, M.M. The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. Plant J. 2009, 60, 626–637. [Google Scholar] [CrossRef]
- Martinez-Fernandez, I.; Menezes de Moura, S.; Alves-Ferreira, M.; Ferrandiz, C.; Balanza, V. Identification of players controlling meristem arrest downstream of the FRUITFULL-APETALA2 pathway. Plant Physiol. 2020, 184, 945–959. [Google Scholar] [CrossRef]
- Richter, R.; Kinoshita, A.; Vincent, C.; Martinez-Gallegos, R.; Gao, H.; van Driel, A.D.; Hyun, Y.; Mateos, J.L.; Coupland, G. Floral regulators FLC and SOC1 directly regulate expression of the B3-type transcription factor TARGET OF FLC AND SVP 1 at the Arabidopsis shoot apex via antagonistic chromatin modifications. PLoS Genet. 2019, 15, e1008065. [Google Scholar] [CrossRef] [PubMed]
- Gramzow, L.; Theissen, G. A hitchhiker’s guide to the MADS world of plants. Genome Biol. 2010, 11, 214. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Theissen, G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 2003, 29, 464–489. [Google Scholar] [CrossRef] [PubMed]
- Nam, J.; dePamphilis, C.W.; Ma, H.; Nei, M. Antiquity and evolution of the MADS-box gene family controlling flower development in plants. Mol. Biol. Evol. 2003, 20, 1435–1447. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, P.B.; Liu, X.; Wu, X.; Zhu, S.; Kasahara, R.D. Fertilization in flowering plants: An odyssey of sperm cell delivery. Plant Mol. Biol. 2020, 103, 9–32. [Google Scholar] [CrossRef] [PubMed]
- Dreni, L.; Zhang, D. Flower development: The evolutionary history and functions of the AGL6 subfamily MADS-box genes. J. Exp. Bot. 2016, 67, 1625–1638. [Google Scholar] [CrossRef] [PubMed]
- Simonini, S.; Roig-Villanova, I.; Gregis, V.; Colombo, B.; Colombo, L.; Kater, M.M. BASIC PENTACYSTEINE proteins mediate MADS domain complex binding to the DNA for tissue-specific expression of target genes in Arabidopsis. Plant Cell 2012, 24, 4163–4172. [Google Scholar] [CrossRef]
- Yang, F.; Xu, F.; Wang, X.; Liao, Y.; Chen, Q.; Meng, X. Characterization and functional analysis of a MADS-box transcription factor gene (GbMADS9) from Ginkgo biloba. Sci. Hortic. 2016, 212, 104–114. [Google Scholar] [CrossRef]
- Portereiko, M.F.; Lloyd, A.; Steffen, J.G.; Punwani, J.A.; Otsuga, D.; Drews, G.N. AGL80 Is required for central cell and endosperm development in Arabidopsis. Plant Cell 2006, 18, 1862–1872. [Google Scholar] [CrossRef]
- Steffen, J.G.; Kang, I.H.; Portereiko, M.F.; Lloyd, A.; Drews, G.N. AGL61 interacts with AGL80 and is required for central cell development in Arabidopsis. Plant Physiol. 2008, 148, 259–268. [Google Scholar] [CrossRef]
- Colombo, M.; Masiero, S.; Vanzulli, S.; Lardelli, P.; Kater, M.M.; Colombo, L. AGL23, a type I MADS-box gene that controls female gametophyte and embryo development in Arabidopsis. Plant J. 2008, 54, 1037–1048. [Google Scholar] [CrossRef] [PubMed]
- Immink, R.G.; Posé, D.; Ferrario, S.; Ott, F.; Kaufmann, K.; Valentim, F.L.; de Folter, S.; van der Wal, F.; van Dijk, A.D.; Schmid, M.; et al. Characterization of SOC1’s central role in flowering by the identification of its upstream and downstream regulators. Plant Physiol. 2012, 160, 433–449. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Sakai, H.; Meyerowitz, E.M. Whorl-specific expression of the SUPERMAN gene of Arabidopsis is mediated by cis elements in the transcribed region. Curr. Biol. 2003, 13, 1524–1530. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Smyth, D.R.; Meyerowitz, E.M. Genes directing flower development in Arabidopsis. Plant Cell 1989, 1, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Hsu, W.H.; Yeh, T.J.; Huang, K.Y.; Li, J.Y.; Chen, H.Y.; Yang, C.H. AGAMOUS-LIKE13, a putative ancestor for the E functional genes, specifies male and female gametophyte morphogenesis. Plant J. 2014, 77, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Hao, Z.; Long, X.; Wang, Z.; Zheng, X.; Ye, D.; Peng, Y.; Wu, W.; Hu, X.; Wang, G.; et al. The transcriptome of Cunninghamia lanceolata male/female cone reveal the association between MIKC MADS-box genes and reproductive organs development. BMC Plant Biol. 2020, 20, 508. [Google Scholar] [CrossRef] [PubMed]
- Myat, A.A.; Zhou, Y.; Gao, Y.; Zhao, X.; Liang, C.; Abid, M.A.; Wang, P.; Akram, U.; Abbas, M.; Askari, M.; et al. Overexpression of GhKTI12 enhances seed yield and biomass production in Nicotiana tabacum. Genes 2022, 13, 426. [Google Scholar] [CrossRef]
- Zhang, S.; Yao, J.; Wang, L.; Wu, N.; van Nocker, S.; Li, Z.; Gao, M.; Wang, X. Role of grapevine SEPALLATA-related MADS-box gene VvMADS39 in flower and ovule development. Plant J. 2022, 111, 1565–1579. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Fernández, I.; Fourquin, C.; Lindsay, D.; Berbel, A.; Balanzà, V.; Huang, S.; Dalmais, M.; LeSignor, C.; Bendahmane, A.; Warkentin, T.D.; et al. Analysis of pea mutants reveals the conserved role of FRUITFULL controlling the end of flowering and its potential to boost yield. Proc. Natl. Acad. Sci. USA 2024, 121, e2321975121. [Google Scholar] [CrossRef]
- Shah, S.; Karunarathna, N.L.; Jung, C.; Emrani, N. An APETALA1 ortholog affects plant architecture and seed yield component in oilseed rape (Brassica napus L.). BMC Plant Biol. 2018, 18, 380. [Google Scholar] [CrossRef]
- Samach, A.; Onouchi, H.; Gold, S.E.; Ditta, G.S.; Schwarz-Sommer, Z.; Yanofsky, M.F.; Coupland, G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 2000, 288, 1613–1616. [Google Scholar] [CrossRef]
- Lee, J.; Lee, I. Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 2010, 61, 2247–2254. [Google Scholar] [CrossRef]
- Chen, W.H.; Lin, P.T.; Hsu, W.H.; Hsu, H.F.; Li, Y.C.; Tsao, C.W.; Hsu, M.C.; Mao, W.T.; Yang, C.H. Regulatory network for FOREVER YOUNG FLOWER-like genes in regulating Arabidopsis flower senescence and abscission. Commun. Biol. 2022, 5, 662. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Long, H.; Qiu, F.; Wang, Y.; Zhang, M.; Chao, Y.; Chen, L. MADS-box protein MtSOC1c regulates flowering and seed development in Medicago truncatula. Ind. Crops Prod. 2023, 193, 116125. [Google Scholar] [CrossRef]
- Papaefthimiou, D.; Kapazoglou, A.; Tsaftaris, A.S. Cloning and characterization of SOC1 homologs in barley (Hordeum vulgare) and their expression during seed development and in response to vernalization. Physiol. Plant. 2012, 146, 71–85. [Google Scholar] [CrossRef]
- Kennedy, A.; Geuten, K. The role of FLOWERING LOCUS C relatives in cereals. Front. Plant Sci. 2020, 11, 617340. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Y.; Zhang, J.; Torrance, A.; Watanabe, N.; Adamski, N.M.; Uauy, C. The Triticum ispahanicum elongated glume locus P2 maps to chromosome 6A and is associated with the ectopic expression of SVP-A1. Theor. Appl. Genet. 2022, 135, 2313–2331. [Google Scholar] [CrossRef]
- Fan, C.M.; Wang, X.; Wang, Y.W.; Hu, R.B.; Zhang, X.M.; Chen, J.X.; Fu, Y.F. Genome-wide expression analysis of soybean MADS genes showing potential function in the seed development. PLoS ONE 2013, 8, e62288. [Google Scholar] [CrossRef]
- Chiang, G.C.; Barua, D.; Kramer, E.M.; Amasino, R.M.; Donohue, K. Major flowering time gene, FLOWERING LOCUS C, regulates seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2009, 106, 11661–11666. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Veerappan, V.; Abdelmageed, H.; Kang, M.; Allen, R.D. HSI2/VAL1 silences AGL15 to regulate the developmental transition from seed maturation to vegetative growth in Arabidopsis. Plant Cell 2018, 30, 600–619. [Google Scholar] [CrossRef] [PubMed]
- Bemer, M.; Heijmans, K.; Airoldi, C.; Davies, B.; Angenent, G.C. An atlas of type I MADS box gene expression during female gametophyte and seed development in Arabidopsis. Plant Physiol. 2010, 154, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Ehlers, K.; Bhide, A.S.; Tekleyohans, D.G.; Wittkop, B.; Snowdon, R.J.; Becker, A. The MADS box genes ABS, SHP1, and SHP2 are essential for the coordination of cell divisions in ovule and seed coat development and for endosperm formation in Arabidopsis thaliana. PLoS ONE 2016, 11, e0165075. [Google Scholar] [CrossRef] [PubMed]
- Coen, O.; Fiume, E.; Xu, W.; De Vos, D.; Lu, J.; Pechoux, C.; Lepiniec, L.; Magnani, E. Developmental patterning of the sub-epidermal integument cell layer in Arabidopsis seeds. Development 2017, 144, 1490–1497. [Google Scholar] [CrossRef] [PubMed]
- Heo, S.; Chung, Y.S. Validation of MADS-box genes from apple fruit pedicels during early fruit abscission by transcriptome analysis and real-time PCR. Genes Genom. 2019, 41, 1241–1251. [Google Scholar] [CrossRef]
- Mao, L.; Begum, D.; Chuang, H.W.; Budiman, M.A.; Szymkowiak, E.J.; Irish, E.E.; Wing, R.A. JOINTLESS is a MADS-box gene controlling tomato flower abscission zone development. Nature 2000, 406, 910–913. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Kimbara, J.; Fujisawa, M.; Kitagawa, M.; Ihashi, N.; Maeda, H.; Kasumi, T.; Ito, Y. MACROCALYX and JOINTLESS Interact in the transcriptional regulation of tomato fruit abscission zone development. Plant Physiol. 2011, 158, 439–450. [Google Scholar] [CrossRef] [PubMed]
- Pi, M.; Hu, S.; Cheng, L.; Zhong, R.; Cai, Z.; Liu, Z.; Yao, J.L.; Kang, C. The MADS-box gene FveSEP3 plays essential roles in flower organogenesis and fruit development in woodland strawberry. Hortic. Res. 2021, 8, 247. [Google Scholar] [CrossRef] [PubMed]
- Kofler, J.; Milyaev, A.; Capezzone, F.; Stojnić, S.; Mićić, N.; Flachowsky, H.; Hanke, M.-V.; Wünsche, J.-N. High crop load and low temperature delay the onset of bud initiation in apple. Sci. Rep. 2019, 9, 17986. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, D.E.; Heck, G.R.; Perry, S.E.; Patterson, S.E.; Bleecker, A.B.; Fang, S.C. The embryo MADS domain factor AGL15 acts postembryonically. Inhibition of perianth senescence and abscission via constitutive expression. Plant Cell 2000, 12, 183–198. [Google Scholar] [CrossRef]
- Fang, S.-C.; Fernandez, D.E. Effect of regulated overexpression of the MADS domain factor AGL15 on flower senescence and fruit maturation. Plant Physiol. 2002, 130, 78–89. [Google Scholar] [CrossRef]
- Busi, M.V.; Bustamante, C.; D’Angelo, C.; Hidalgo-Cuevas, M.; Boggio, S.B.; Valle, E.M.; Zabaleta, E. MADS-box genes expressed during tomato seed and fruit development. Plant Mol. Biol. 2003, 52, 801–815. [Google Scholar] [CrossRef]
- Wang, B.; Hu, W.; Fang, Y.; Feng, X.; Fang, J.; Zou, T.; Zheng, S.; Ming, R.; Zhang, J. Comparative analysis of the MADS-box genes revealed their potential functions for flower and fruit development in Longan (Dimocarpus longan). Front. Plant Sci. 2021, 12, 813798. [Google Scholar] [CrossRef]
- Li, C.X.; Lu, X.F.; Xu, J.R.; Liu, Y.Z. Regulation of fruit ripening by MADS-box transcription factors. Sci. Hortic. 2023, 314, 111950. [Google Scholar] [CrossRef]
- Huang, X.; Lu, Z.; Zhai, L.; Li, N.; Yan, H. The Small Auxin-Up RNA SAUR10 is involved in the promotion of seedling growth in rice. Plants 2023, 12, 3880. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.H.; Yu, L.H.; Xiang, C.B. ARABIDOPSIS NITRATE REGULATED 1 acts as a negative modulator of seed germination by activating ABI3 expression. New Phytol. 2020, 225, 835–847. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.H.; Wu, J.; Zhang, Z.S.; Miao, Z.Q.; Zhao, P.X.; Wang, Z.; Xiang, C.B. Arabidopsis MADS-box transcription factor AGL21 acts as environmental surveillance of seed germination by regulating ABI5 expression. Mol. Plant 2017, 10, 834–845. [Google Scholar] [CrossRef] [PubMed]
- Soppe, W.J.J.; Vinegra de la Torre, N.; Albani, M.C. The diverse roles of FLOWERING LOCUS C in annual and perennial Brassicaceae species. Front. Plant Sci. 2021, 12, 627258. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.X.; Zhang, J.; Chen, S.Y.; Wu, J.; Xia, J.Q.; Sun, L.Q.; Ma, S.S.; Xiang, C.B. Arabidopsis MADS-box factor AGL16 is a negative regulator of plant response to salt stress by downregulating salt-responsive genes. New Phytol. 2021, 232, 2418–2439. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.M.; Kim, D.W.; Woo, M.S.; Jeong, H.S.; Son, Y.S.; Akhter, S.; Choi, G.J.; Bahk, J.D. Functional characterization of Arabidopsis HsfA6a as a heat-shock transcription factor under high salinity and dehydration conditions. Plant Cell Environ. 2014, 37, 1202–1222. [Google Scholar] [CrossRef]
- Nesi, N.; Debeaujon, I.; Jond, C.; Stewart, A.J.; Jenkins, G.I.; Caboche, M.; Lepiniec, L. The TRANSPARENT TESTA16 locus encodes the ARABIDOPSIS BSISTER MADS domain protein and is required for proper development and pigmentation of the seed coat. Plant Cell 2002, 14, 2463–2479. [Google Scholar] [CrossRef]
- Dai, Y.; Wang, Y.; Zeng, L.; Jia, R.; He, L.; Huang, X.; Zhao, H.; Liu, D.; Zhao, H.; Hu, S.; et al. Genomic andtranscriptomic insights into the evolution and divergence of MIKC-type MADS-box genes in Carica papaya. Int. J. Mol. Sci. 2023, 24, 14039. [Google Scholar] [CrossRef]
- Ezquer, I.; Mizzotti, C.; Nguema-Ona, E.; Gotté, M.; Beauzamy, L.; Viana, V.E.; Dubrulle, N.; Costa de Oliveira, A.; Caporali, E.; Koroney, A.-S.; et al. The developmental regulator SEEDSTICK controls structural and mechanical properties of the Arabidopsis seed coat. Plant Cell 2016, 28, 2478–2492. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, Z.; Zhao, X.; Liu, L.; Tang, Q.; Fu, J.; Tang, X.; Yang, R.; Lin, J.; Liu, X.; et al. Receptor-Like cytoplasmic kinase stk confers salt tolerance in rice. Rice 2023, 16, 21. [Google Scholar] [CrossRef] [PubMed]
- Bjerkan, K.N.; Alling, R.M.; Myking, I.V.; Brysting, A.K.; Grini, P.E. Genetic and environmental manipulation of Arabidopsis hybridization barriers uncovers antagonistic functions in endosperm cellularization. Front. Plant Sci. 2023, 14, 1229060. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, D.D.; Batista, R.A.; Roszak, P.J.; Hennig, L.; Kohler, C. Auxin production in the endosperm drives seed coat development in Arabidopsis. eLife 2016, 5, e20542. [Google Scholar] [CrossRef]
- Liu, X.; Sun, Z.; Dong, W.; Wang, Z.; Zhang, L. Expansion and functional divergence of the SHORT VEGETATIVE PHASE (SVP) genes in eudicots. Genome Biol. Evol. 2018, 10, 3026–3037. [Google Scholar] [CrossRef]
- Chen, W.; Tamada, Y.; Yamane, H.; Matsushita, M.; Osako, Y.; Gao-Takai, M.; Luo, Z.; Tao, R. H3K4me3 plays a key role in establishing permissive chromatin states during bud dormancy and bud break in apple. Plant J. 2022, 111, 1015–1031. [Google Scholar] [CrossRef]
- Huang, B.; Routaboul, J.M.; Liu, M.; Deng, W.; Maza, E.; Mila, I.; Hu, G.; Zouine, M.; Frasse, P.; Vrebalov, J.T.; et al. Overexpression of the class D MADS-box gene Sl-AGL11 impacts fleshy tissue differentiation and structure in tomato fruits. J. Exp. Bot. 2017, 68, 4869–4884. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.; Wang, L.; Li, J.; Ruan, Y.L. Cell wall invertase is essential for ovule development through sugar signaling rather than provision of carbon nutrients. Plant Physiol. 2020, 183, 1126–1144. [Google Scholar] [CrossRef]
- Wilson, D.C.; Kempthorne, C.J.; Carella, P.; Liscombe, D.K.; Cameron, R.K. Age-related resistance in Arabidopsis thaliana involves the MADS-domain transcription factor SHORT VEGETATIVE PHASE and direct action of salicylic acid on Pseudomonas syringae. Mol. Plant-Microbe Interact. 2017, 30, 919–929. [Google Scholar] [CrossRef]
- Zhou, F.Y.; Yu, Q.; Zhang, Y.; Yao, C.C.; Han, Y.J. StMADS11 Subfamily Gene PfMADS16 from Polypogon fugax Regulates Early Flowering and Seed Development. Front. Plant Sci. 2020, 11, 525. [Google Scholar] [CrossRef] [PubMed]
- Song, G.Q.; Chen, Q. Overexpression of the MADS-box gene K-domain increases the yield potential of blueberry. Plant Sci. 2018, 276, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.X.; Gao, J.; Wei, Y.L.; Ren, R.; Zhang, G.Q.; Lu, C.Q.; Jin, J.P.; Ai, Y.; Wang, Y.Q.; Chen, L.J.; et al. The genome of Cymbidium sinense revealed the evolution of orchid traits. Plant Biotechnol. J. 2021, 19, 2501–2516. [Google Scholar] [CrossRef] [PubMed]
- Jing, Y.; Shi, L.; Li, X.; Zheng, H.; Gao, J.; Wang, M.; He, L.; Zhang, W. OXS2 is required for salt tolerance mainly through associating with salt inducible genes, CA1 and ARAPORT11, in Arabidopsis. Sci. Rep. 2019, 9, 20341. [Google Scholar] [CrossRef] [PubMed]
- Blanvillain, R.; Wei, S.; Wei, P.; Kim, J.H.; Ow, D.W. Stress tolerance to stress escape in plants: Role of the OXS2 zinc-finger transcription factor family. EMBO J. 2011, 30, 3812–3822. [Google Scholar] [CrossRef] [PubMed]
- Castañón-Suárez, C.A.; Arrizubieta, M.; Castelán-Muñoz, N.; Sánchez-Rodríguez, D.B.; Caballero-Cordero, C.; Zluhan-Martínez, E.; Patiño-Olvera, S.C.; Arciniega-González, J.A.; García-Ponce, B.; Sánchez, M.P.; et al. The MADS-box genes SOC1 and AGL24 antagonize XAL2 functions in Arabidopsis thaliana root development. Front. Plant Sci. 2024, 15, 1331269. [Google Scholar] [CrossRef] [PubMed]
- Endo, T.; Shimada, T.; Fujii, H.; Kobayashi, Y.; Araki, T.; Omura, M. Ectopic expression of an FT homolog from citrus confers an early flowering phenotype on trifoliate orange (Poncirus trifoliata L. Raf.). Transgenic Res. 2005, 14, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Schlathölter, I.; Jänsch, M.; Flachowsky, H.; Broggini, G.A.L.; Hanke, M.-V.; Patocchi, A. Generation of advanced fire blight-resistant apple (Malus × domestica) selections of the fifth generation within 7 years of applying the early flowering approach. Planta 2018, 247, 1475–1488. [Google Scholar] [CrossRef]
- Flachowsky, H.; Szankowski, I.; Waidmann, S.; Peil, A.; Tränkner, C.; Hanke, M.-V. The MdTFL1 gene of apple (Malus × domestica Borkh.) reduces vegetative growth and generation time. Tree Physiol. 2012, 32, 1288–1301. [Google Scholar] [CrossRef]
- Flachowsky, H.; Hättasch, C.; Höfer, M.; Peil, A.; Hanke, M.-V. Overexpression of LEAFY in apple leads to a columnar phenotype with shorter internodes. Planta 2010, 231, 251–263. [Google Scholar] [CrossRef]
- Guardiola, J.L. Overview of flower bud induction, flowering and fruit set. In Proceedings of the Citrus Flowering and Fruit Short Course, IFAS, Lake Alfred, FL, USA, 9–10 April 1997; Citrus Research and Education Center, University of Florida: Lake Alfred, FL, USA, 1997; pp. 5–21. [Google Scholar]
- Jones, H.G. Repeat flowering in apple caused by water stress or defoliation. Trees 1987, 1, 135–138. [Google Scholar] [CrossRef]
- Jaudal, M.; Zhang, L.; Che, C.; Li, G.; Tang, Y.; Wen, J.; Mysore, K.S.; Putterill, J. A SOC1-like gene MtSOC1a promotes flowering and primary stem elongation in Medicago. J. Exp. Bot. 2018, 69, 4867–4880. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Wang, D.; Song, G.Q. Expression of a maize SOC1 gene enhances soybean yield potential through modulating plant growth and flowering. Sci. Rep. 2021, 11, 12758. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Bolanos, M.; Martinez, T.; Juarez, S.; Quiroz, S.; Dominguez, A.; Garay-Arroyo, A.; Sanchez, M.P.; Alvarez-Buylla, E.R.; Garcia-Ponce, B. XAANTAL1 reveals an additional level of flowering regulation in the shoot apical meristem in response to light and increased temperature in Arabidopsis. Int. J. Mol. Sci. 2023, 24, 12773. [Google Scholar] [CrossRef] [PubMed]
- Dang, X.; Zhang, Y.; Li, Y.; Chen, S.; Liu, E.; Fang, B.; Liu, Q.; She, D.; Dong, Z.; Fan, Z.; et al. SYL3-k increases style length and yield of F(1) seeds via enhancement of endogenous GA(4) content in Oryza sativa L. pistils. Theor. Appl. Genet. 2022, 135, 321–336. [Google Scholar] [CrossRef]
- ChatGPT, (Mar 14 version) [Large Language Model]; OpenAI: San Francisco, CA, USA, 2023.


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adhikari, P.B.; Kasahara, R.D. An Overview on MADS Box Members in Plants: A Meta-Review. Int. J. Mol. Sci. 2024, 25, 8233. https://doi.org/10.3390/ijms25158233
Adhikari PB, Kasahara RD. An Overview on MADS Box Members in Plants: A Meta-Review. International Journal of Molecular Sciences. 2024; 25(15):8233. https://doi.org/10.3390/ijms25158233
Chicago/Turabian StyleAdhikari, Prakash Babu, and Ryushiro Dora Kasahara. 2024. "An Overview on MADS Box Members in Plants: A Meta-Review" International Journal of Molecular Sciences 25, no. 15: 8233. https://doi.org/10.3390/ijms25158233






