SARS-CoV-2 Rapidly Infects Peripheral Sensory and Autonomic Neurons, Contributing to Central Nervous System Neuroinvasion before Viremia
Abstract
:1. Introduction
2. Results
2.1. Clinical Data
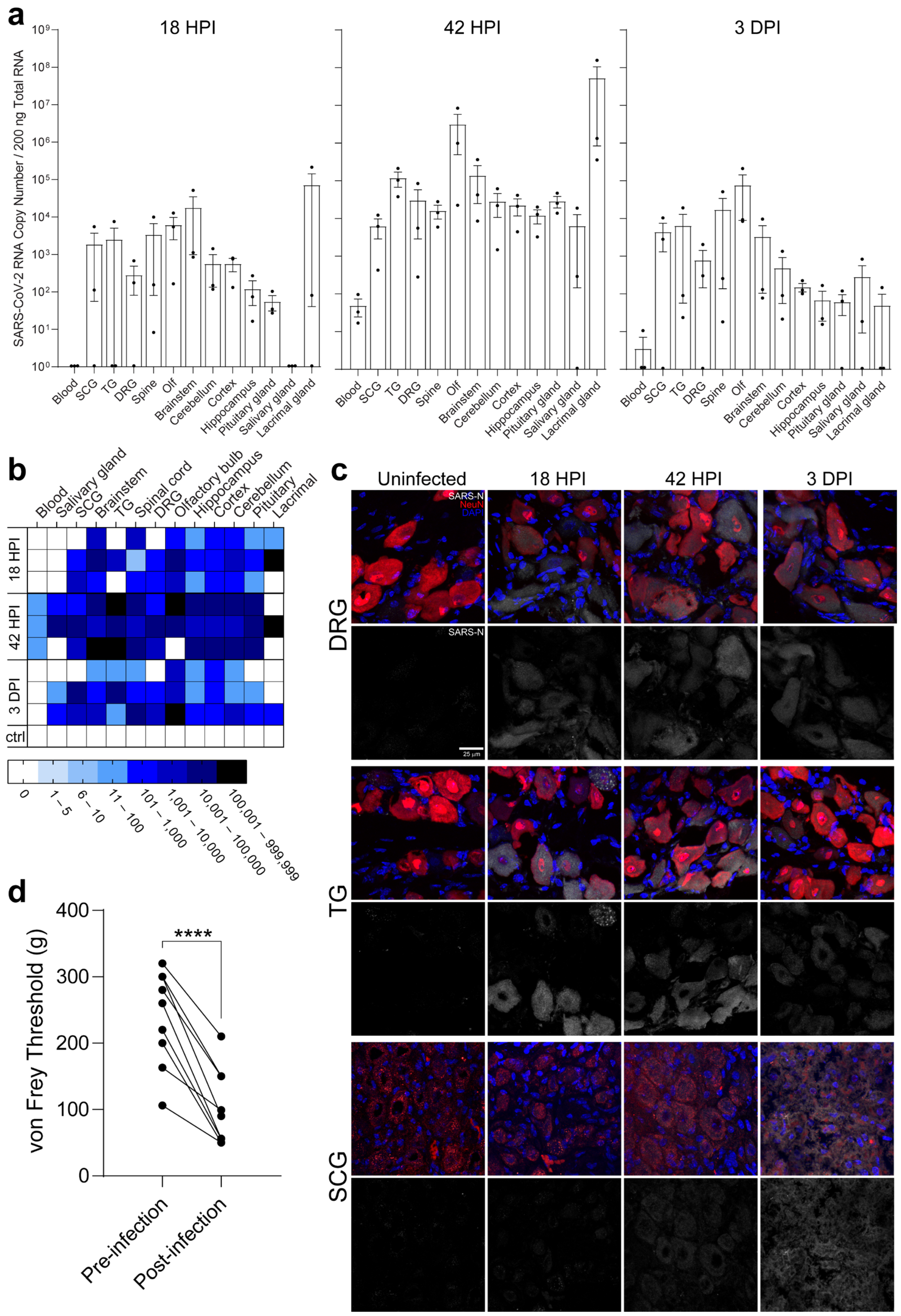
2.2. PNS Sensory Trigeminal Ganglia (TG) and Sympathetic Superior Cervical Ganglia (SCG) Neurons Innervating the Oronasopharynx Are Susceptible to Infection
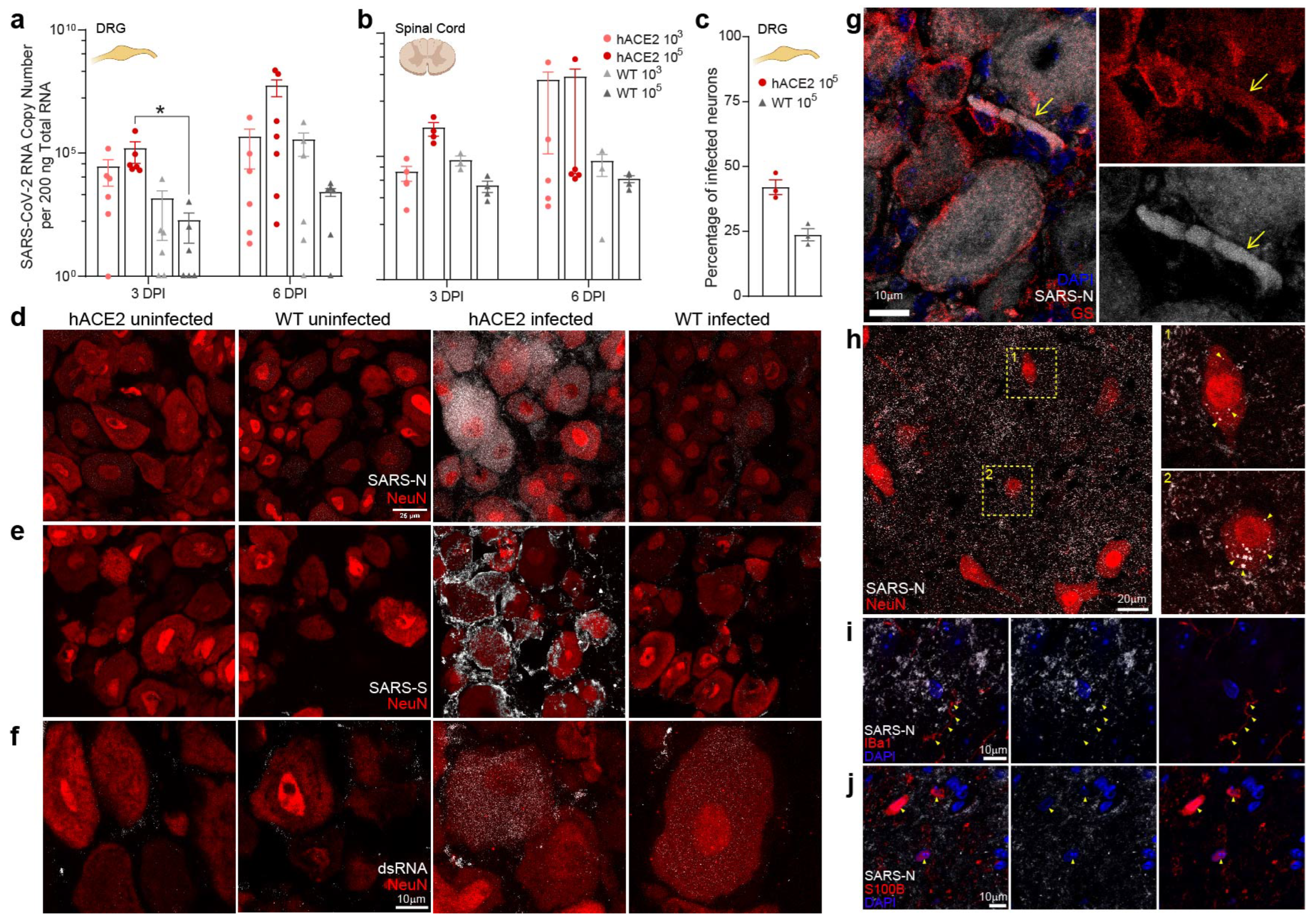
2.3. PNS Sensory Lumbosacral Dorsal Root Ganglia (LS-DRG) and CNS Lumbosacral Spinal Cord (LS-SC) Neurons Are Susceptible to Infection
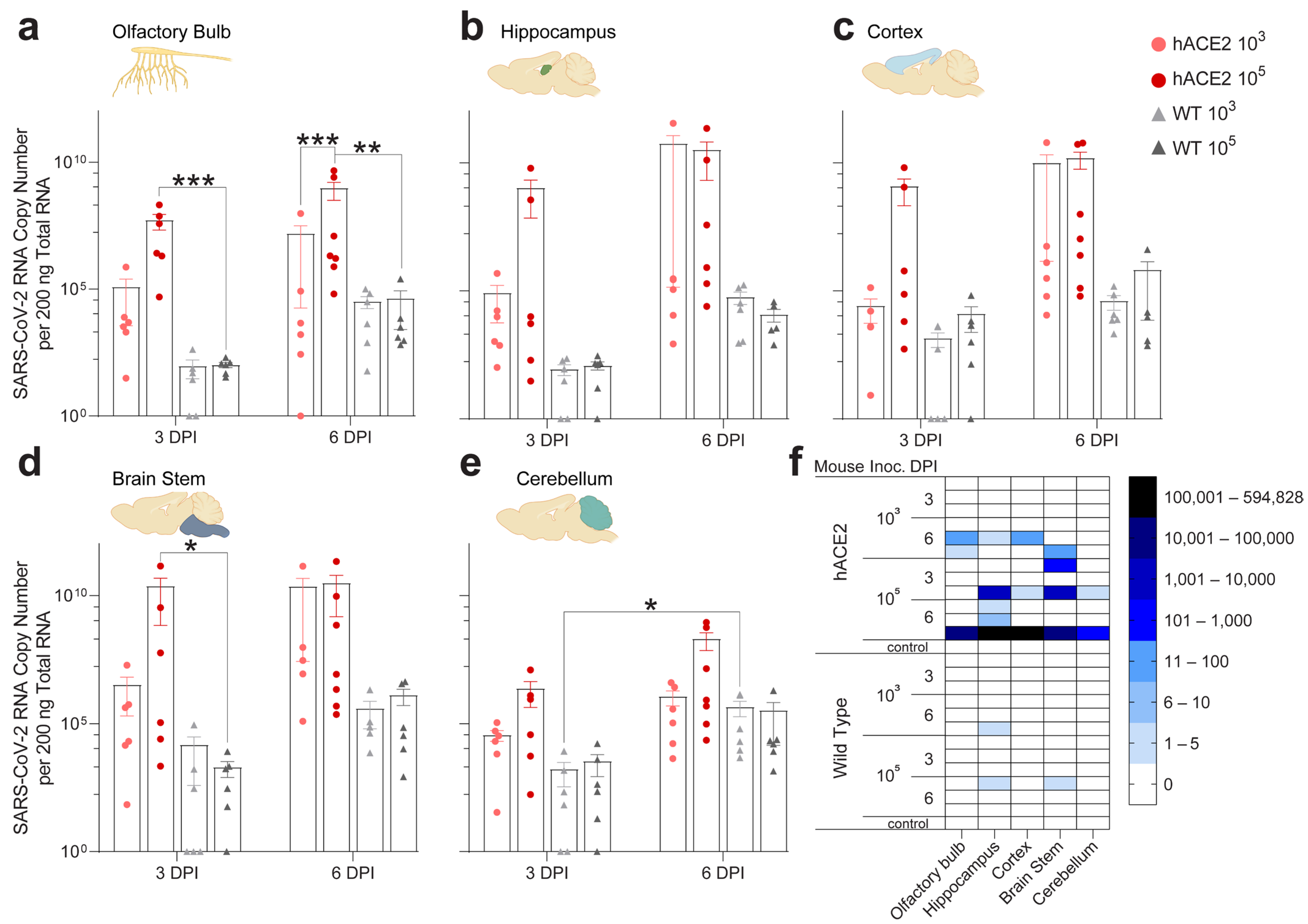
2.4. Individual Brain Regions Support Varying Levels of Viral Invasion and Reproduction
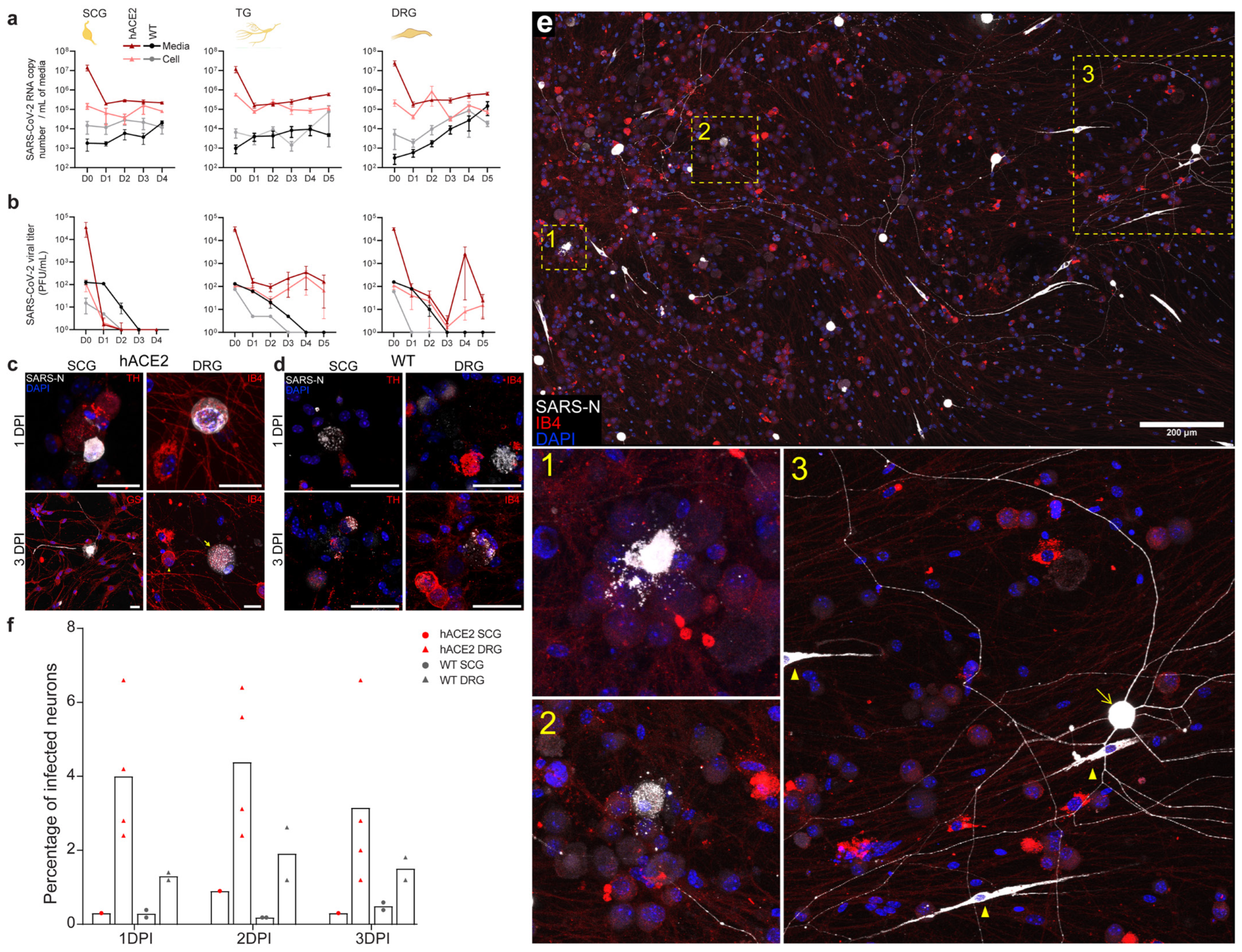
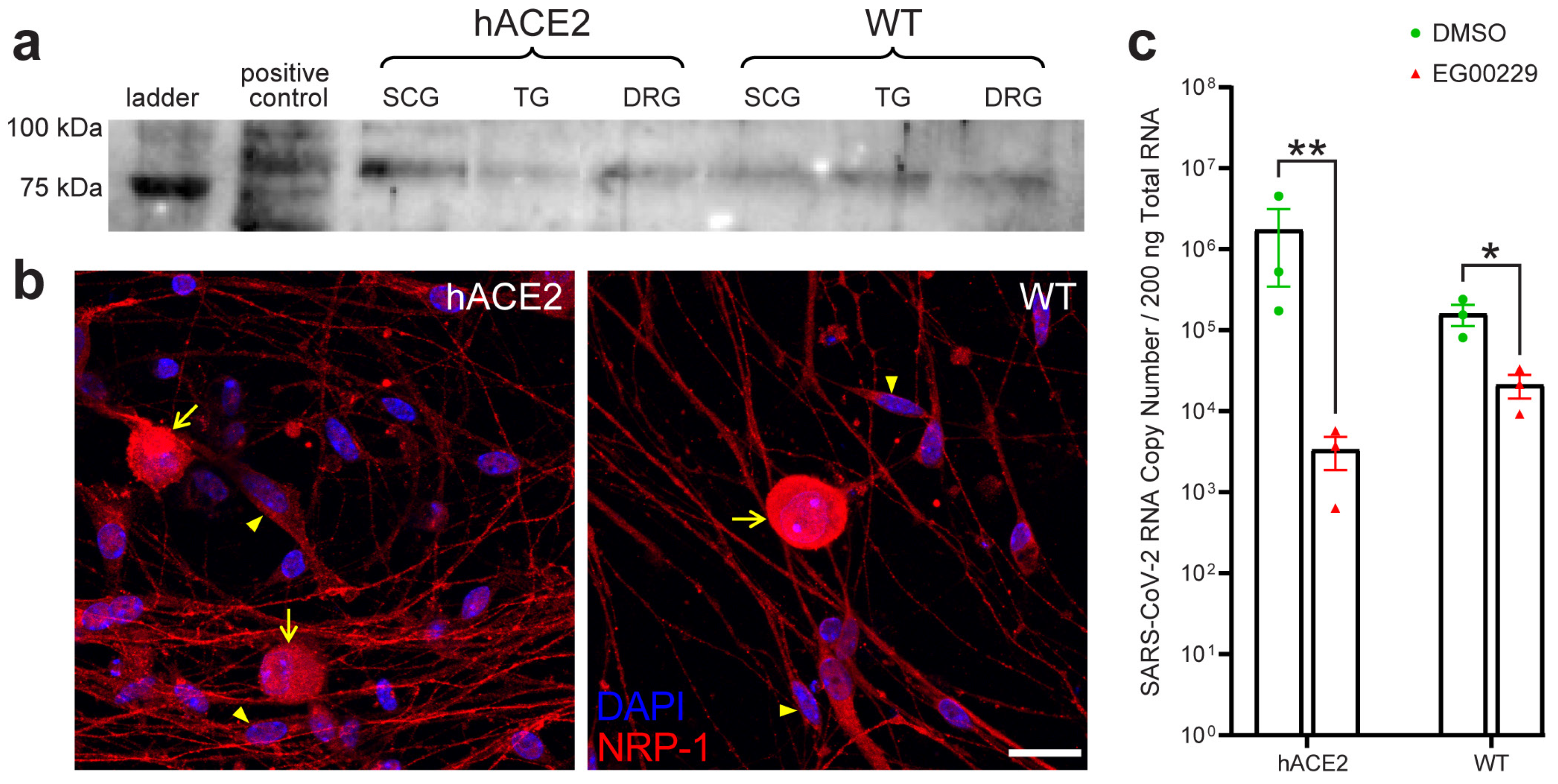
2.5. SARS-CoV-2 Productively Infects Primary Cultured PNS Sensory but Not Autonomic Neurons from Adult Mice
2.6. Neuroinvasion of the PNS and CNS Occurs before Viremia
2.7. Neuronal Entry in the PNS Involves Neuropilin-1 (NRP-1)
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Cell Lines
4.3. Viruses
4.4. Mouse Infections
4.5. Golden Syrian Hamster Infections and Von Frey Assessment for Allodynia
4.6. Primary Neuronal Culture of Peripheral Sensory and Sympathetic Neurons
4.7. Primary Neuronal Culture Infection
4.8. Plaque Assays
4.9. RNA Extraction and SARS-CoV-2 Specific RT-qPCR
4.10. Immunofluorescence
4.11. Antibody Validation
4.12. Confocal Microscopy and Image Analysis
4.13. Quantification of Infection in Autonomic and Sensory Ganglia in Primary Neuronal Culture and Tissues
4.14. Detection of SARS-CoV-2 Neuronal Replication and Release in Primary Neuronal Culture
4.15. Inhibition of SARS-CoV-2 Infection by Neuropilin-1 Blockade in Primary Sensory Neuronal Culture
4.16. Detection of hACE2 and NRP-1 in PNS Ganglia
4.17. Statistics and Reproducibility
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shouman, K.; Vanichkachorn, G.; Cheshire, W.P.; Suarez, M.D.; Shelly, S.; Lamotte, G.J.; Sandroni, P.; Benarroch, E.E.; Berini, S.E.; Cutsforth-Gregory, J.K.; et al. Autonomic dysfunction following COVID-19 infection: An early experience. Clin. Auton. Res. 2021, 31, 385–394. [Google Scholar] [CrossRef]
- Chou, S.H.; Beghi, E.; Helbok, R.; Moro, E.; Sampson, J.; Altamirano, V.; Mainali, S.; Bassetti, C.; Suarez, J.I.; McNett, M.; et al. Global Incidence of Neurological Manifestations Among Patients Hospitalized With COVID-19—A Report for the GCS-NeuroCOVID Consortium and the ENERGY Consortium. JAMA Netw. Open 2021, 4, e2112131. [Google Scholar] [CrossRef]
- Mao, L.; Jin, H.; Wang, M.; Hu, Y.; Chen, S.; He, Q.; Chang, J.; Hong, C.; Zhou, Y.; Wang, D.; et al. Neurologic Manifestations of Hospitalized Patients With Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol. 2020, 77, 683–690. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.C.; Kern, F.; Losada, P.M.; Agam, M.R.; Maat, C.A.; Schmartz, G.P.; Fehlmann, T.; Stein, J.A.; Schaum, N.; Lee, D.P.; et al. Dysregulation of brain and choroid plexus cell types in severe COVID-19. Nature 2021, 595, 565–571. [Google Scholar] [CrossRef]
- Paniz-Mondolfi, A.; Bryce, C.; Grimes, Z.; Gordon, R.E.; Reidy, J.; Lednicky, J.; Sordillo, E.M.; Fowkes, M. Central nervous system involvement by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). J. Med. Virol. 2020, 92, 699–702. [Google Scholar] [CrossRef] [PubMed]
- Puelles, V.G.; Lutgehetmann, M.; Lindenmeyer, M.T.; Sperhake, J.P.; Wong, M.N.; Allweiss, L.; Chilla, S.; Heinemann, A.; Wanner, N.; Liu, S.; et al. Multiorgan and Renal Tropism of SARS-CoV-2. N. Engl. J. Med. 2020, 383, 590–592. [Google Scholar] [CrossRef] [PubMed]
- Virhammar, J.; Kumlien, E.; Fallmar, D.; Frithiof, R.; Jackmann, S.; Skold, M.K.; Kadir, M.; Frick, J.; Lindeberg, J.; Olivero-Reinius, H.; et al. Acute necrotizing encephalopathy with SARS-CoV-2 RNA confirmed in cerebrospinal fluid. Neurology 2020, 95, 445–449. [Google Scholar] [CrossRef]
- Xiang, P.; Xu, X.; Lu, X.; Gao, L.; Wang, H.; Li, Z.; Xiong, H.; Li, R.; Xiong, Y.; Pu, L.; et al. Case Report: Identification of SARS-CoV-2 in Cerebrospinal Fluid by Ultrahigh-Depth Sequencing in a Patient With Coronavirus Disease 2019 and Neurological Dysfunction. Front Med. 2021, 8, 629828. [Google Scholar] [CrossRef] [PubMed]
- Bonavia, A.; Arbour, N.; Yong, V.W.; Talbot, P.J. Infection of primary cultures of human neural cells by human coronaviruses 229E and OC43. J. Virol. 1997, 71, 800–806. [Google Scholar] [CrossRef]
- Arbour, N.; Talbot, P.J. Persistent infection of neural cell lines by human coronaviruses. Adv. Exp. Med. Biol. 1998, 440, 575–581. [Google Scholar] [CrossRef]
- Arbour, N.; Day, R.; Newcombe, J.; Talbot, P.J. Neuroinvasion by human respiratory coronaviruses. J. Virol. 2000, 74, 8913–8921. [Google Scholar] [CrossRef]
- Dube, M.; Le Coupanec, A.; Wong, A.H.M.; Rini, J.M.; Desforges, M.; Talbot, P.J. Axonal Transport Enables Neuron-to-Neuron Propagation of Human Coronavirus OC43. J. Virol. 2018, 92, e00404-18. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Yang, Y.; Gao, J. Infectivity of human coronavirus in the brain. EBioMedicine 2020, 56, 102799. [Google Scholar] [CrossRef] [PubMed]
- Kumari, P.; Rothan, H.A.; Natekar, J.P.; Stone, S.; Pathak, H.; Strate, P.G.; Arora, K.; Brinton, M.A.; Kumar, M. Neuroinvasion and Encephalitis Following Intranasal Inoculation of SARS-CoV-2 in K18-hACE2 Mice. Viruses 2021, 13, 132. [Google Scholar] [CrossRef] [PubMed]
- Golden, J.W.; Cline, C.R.; Zeng, X.; Garrison, A.R.; Carey, B.D.; Mucker, E.M.; White, L.E.; Shamblin, J.D.; Brocato, R.L.; Liu, J.; et al. Human angiotensin-converting enzyme 2 transgenic mice infected with SARS-CoV-2 develop severe and fatal respiratory disease. JCI Insight 2020, 5, e142032. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wong, L.R.; Li, K.; Verma, A.K.; Ortiz, M.E.; Wohlford-Lenane, C.; Leidinger, M.R.; Knudson, C.M.; Meyerholz, D.K.; McCray, P.B., Jr.; et al. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature 2021, 589, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Misra, S.; Kolappa, K.; Prasad, M.; Radhakrishnan, D.; Thakur, K.T.; Solomon, T.; Michael, B.D.; Winkler, A.S.; Beghi, E.; Guekht, A.; et al. Frequency of Neurologic Manifestations in COVID-19: A Systematic Review and Meta-analysis. Neurology 2021, 97, e2269–e2281. [Google Scholar] [CrossRef] [PubMed]
- Pinzon, R.T.; Wijaya, V.O.; Jody, A.A.; Nunsio, P.N.; Buana, R.B. Persistent neurological manifestations in long COVID-19 syndrome: A systematic review and meta-analysis. J. Infect. Public Health 2022, 15, 856–869. [Google Scholar] [CrossRef]
- Reme, B.A.; Gjesvik, J.; Magnusson, K. Predictors of the post-COVID condition following mild SARS-CoV-2 infection. Nat. Commun. 2023, 14, 5839. [Google Scholar] [CrossRef]
- Chandrashekar, A.; Liu, J.; Yu, J.; McMahan, K.; Tostanoski, L.H.; Jacob-Dolan, C.; Mercado, N.B.; Anioke, T.; Chang, A.; Gardner, S.; et al. Prior infection with SARS-CoV-2 WA1/2020 partially protects rhesus macaques against reinfection with B.1.1.7 and B.1.351 variants. Sci. Transl. Med. 2021, 13, eabj2641. [Google Scholar] [CrossRef]
- Siddiqui, S.; Alhamdi, H.W.S.; Alghamdi, H.A. Recent Chronology of COVID-19 Pandemic. Front. Public Health 2022, 10, 778037. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. COVID-19 Weekly Epidemiological Update-29 December 2020; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Aleem, A.; Akbar Samad, A.B.; Vaqar, S. Emerging Variants of SARS-CoV-2 and Novel Therapeutics Against Coronavirus (COVID-19). In StatPearls; StatPearls Publishing: St. Petersburg, FL, USA, 2023. [Google Scholar]
- Oladunni, F.S.; Park, J.G.; Pino, P.A.; Gonzalez, O.; Akhter, A.; Allue-Guardia, A.; Olmo-Fontanez, A.; Gautam, S.; Garcia-Vilanova, A.; Ye, C.; et al. Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat. Commun. 2020, 11, 6122. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.A.; Xie, X.; Bailey, A.L.; Kalveram, B.; Lokugamage, K.G.; Muruato, A.; Zou, J.; Zhang, X.; Juelich, T.; Smith, J.K.; et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature 2021, 591, 293–299. [Google Scholar] [CrossRef]
- Winkler, E.S.; Bailey, A.L.; Kafai, N.M.; Nair, S.; McCune, B.T.; Yu, J.; Fox, J.M.; Chen, R.E.; Earnest, J.T.; Keeler, S.P.; et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020, 21, 1327–1335. [Google Scholar] [CrossRef]
- Rathnasinghe, R.; Strohmeier, S.; Amanat, F.; Gillespie, V.L.; Krammer, F.; Garcia-Sastre, A.; Coughlan, L.; Schotsaert, M.; Uccellini, M.B. Comparison of transgenic and adenovirus hACE2 mouse models for SARS-CoV-2 infection. Emerg. Microbes Infect. 2020, 9, 2433–2445. [Google Scholar] [CrossRef]
- Yinda, C.K.; Port, J.R.; Bushmaker, T.; Offei Owusu, I.; Purushotham, J.N.; Avanzato, V.A.; Fischer, R.J.; Schulz, J.E.; Holbrook, M.G.; Hebner, M.J.; et al. K18-hACE2 mice develop respiratory disease resembling severe COVID-19. PLoS Pathog. 2021, 17, e1009195. [Google Scholar] [CrossRef] [PubMed]
- Frere, J.J.; Serafini, R.A.; Pryce, K.D.; Zazhytska, M.; Oishi, K.; Golynker, I.; Panis, M.; Zimering, J.; Horiuchi, S.; Hoagland, D.A.; et al. SARS-CoV-2 infection in hamsters and humans results in lasting and unique systemic perturbations after recovery. Sci. Transl. Med. 2022, 14, eabq3059. [Google Scholar] [CrossRef] [PubMed]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.E.; Williamson, M.K.; Anton-Plagaro, C.; Shoemark, D.K.; Simon-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865. [Google Scholar] [CrossRef] [PubMed]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef]
- Moutal, A.; Martin, L.F.; Boinon, L.; Gomez, K.; Ran, D.; Zhou, Y.; Stratton, H.J.; Cai, S.; Luo, S.; Gonzalez, K.B.; et al. SARS-CoV-2 spike protein co-opts VEGF-A/neuropilin-1 receptor signaling to induce analgesia. Pain 2021, 162, 243–252. [Google Scholar] [CrossRef]
- Pellegrini, L.; Albecka, A.; Mallery, D.L.; Kellner, M.J.; Paul, D.; Carter, A.P.; James, L.C.; Lancaster, M.A. SARS-CoV-2 Infects the Brain Choroid Plexus and Disrupts the Blood-CSF Barrier in Human Brain Organoids. Cell Stem Cell 2020, 27, 951–961.e5. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F.; Pather, S.R.; Huang, W.K.; Zhang, F.; Wong, S.Z.H.; Zhou, H.; Cubitt, B.; Fan, W.; Chen, C.Z.; Xu, M.; et al. Human Pluripotent Stem Cell-Derived Neural Cells and Brain Organoids Reveal SARS-CoV-2 Neurotropism Predominates in Choroid Plexus Epithelium. Cell Stem Cell 2020, 27, 937–950.e9. [Google Scholar] [CrossRef] [PubMed]
- Rhea, E.M.; Logsdon, A.F.; Hansen, K.M.; Williams, L.M.; Reed, M.J.; Baumann, K.K.; Holden, S.J.; Raber, J.; Banks, W.A.; Erickson, M.A. The S1 protein of SARS-CoV-2 crosses the blood-brain barrier in mice. Nat. Neurosci. 2021, 24, 368–378. [Google Scholar] [CrossRef] [PubMed]
- Buzhdygan, T.P.; DeOre, B.J.; Baldwin-Leclair, A.; Bullock, T.A.; McGary, H.M.; Khan, J.A.; Razmpour, R.; Hale, J.F.; Galie, P.A.; Potula, R.; et al. The SARS-CoV-2 spike protein alters barrier function in 2D static and 3D microfluidic in-vitro models of the human blood-brain barrier. Neurobiol. Dis. 2020, 146, 105131. [Google Scholar] [CrossRef]
- Meinhardt, J.; Radke, J.; Dittmayer, C.; Franz, J.; Thomas, C.; Mothes, R.; Laue, M.; Schneider, J.; Brunink, S.; Greuel, S.; et al. Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19. Nat. Neurosci. 2021, 24, 168–175. [Google Scholar] [CrossRef] [PubMed]
- von Weyhern, C.H.; Kaufmann, I.; Neff, F.; Kremer, M. Early evidence of pronounced brain involvement in fatal COVID-19 outcomes. Lancet 2020, 395, e109. [Google Scholar] [CrossRef]
- Tutuncu, E.E.; Ozgur, D.; Karamese, M. Saliva samples for detection of SARS-CoV-2 in mildly symptomatic and asymptomatic patients. J. Med. Virol. 2021, 93, 2932–2937. [Google Scholar] [CrossRef] [PubMed]
- Ota, K.; Yanagihara, K.; Sasaki, D.; Kaku, N.; Uno, N.; Sakamoto, K.; Kosai, K.; Miyazaki, T.; Hasegawa, H.; Fujita, A.; et al. Detection of SARS-CoV-2 using qRT-PCR in saliva obtained from asymptomatic or mild COVID-19 patients, comparative analysis with matched nasopharyngeal samples. PLoS ONE 2021, 16, e0252964. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.; Perez, P.; Kato, T.; Mikami, Y.; Okuda, K.; Gilmore, R.C.; Conde, C.D.; Gasmi, B.; Stein, S.; Beach, M.; et al. SARS-CoV-2 infection of the oral cavity and saliva. Nat. Med. 2021, 27, 892–903. [Google Scholar] [CrossRef]
- Song, J.; Li, Y.; Huang, X.; Chen, Z.; Li, Y.; Liu, C.; Chen, Z.; Duan, X. Systematic analysis of ACE2 and TMPRSS2 expression in salivary glands reveals underlying transmission mechanism caused by SARS-CoV-2. J. Med. Virol. 2020, 92, 2556–2566. [Google Scholar] [CrossRef]
- Stein, S.R.; Ramelli, S.C.; Grazioli, A.; Chung, J.Y.; Singh, M.; Yinda, C.K.; Winkler, C.W.; Sun, J.; Dickey, J.M.; Ylaya, K.; et al. SARS-CoV-2 infection and persistence in the human body and brain at autopsy. Nature 2022, 612, 758–763. [Google Scholar] [CrossRef] [PubMed]
- Shiers, S.; Ray, P.R.; Wangzhou, A.; Sankaranarayanan, I.; Tatsui, C.E.; Rhines, L.D.; Li, Y.; Uhelski, M.L.; Dougherty, P.M.; Price, T.J. ACE2 and SCARF expression in human dorsal root ganglion nociceptors: Implications for SARS-CoV-2 virus neurological effects. Pain 2020, 161, 2494–2501. [Google Scholar] [CrossRef] [PubMed]
- Garg, R.K.; Paliwal, V.K.; Gupta, A. Spinal cord involvement in COVID-19: A review. J. Spinal Cord. Med. 2021, 46, 390–404. [Google Scholar] [CrossRef]
- Bulfamante, G.; Chiumello, D.; Canevini, M.P.; Priori, A.; Mazzanti, M.; Centanni, S.; Felisati, G. First ultrastructural autoptic findings of SARS -Cov-2 in olfactory pathways and brainstem. Minerva Anestesiol. 2020, 86, 678–679. [Google Scholar] [CrossRef] [PubMed]
- Song, E.; Zhang, C.; Israelow, B.; Lu-Culligan, A.; Prado, A.V.; Skriabine, S.; Lu, P.; Weizman, O.E.; Liu, F.; Dai, Y.; et al. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J. Exp. Med. 2021, 218, e20202135. [Google Scholar] [CrossRef] [PubMed]
- Molina-Gil, J.; Gonzalez-Fernandez, L.; Garcia-Cabo, C. Trigeminal neuralgia as the sole neurological manifestation of COVID-19: A case report. Headache 2021, 61, 560–562. [Google Scholar] [CrossRef] [PubMed]
- Caggia, E.; Bongiorno, J.; Ventura, M.; Lingenti, V.; Maci, V. Trigeminal neuralgia post COVID-19: Preliminary report and clinical consideration. J. Neurol. Sci. 2021, 429, 119882. [Google Scholar] [CrossRef]
- Naor, M.S.; Mathew, P.G.; Sharon, R. Transient Horner syndrome associated with COVID-19: A case report. eNeurologicalSci 2021, 25, 100349. [Google Scholar] [CrossRef] [PubMed]
- Illes, R.; Chochol, J.; Dzubera, A.; Chocholova, A.; Zemkova, E. COVID-19 Worsens Chronic Lumbosacral Radicular Pain-Case Series Report. Int. J. Environ. Res. Public Health 2022, 19, 6410. [Google Scholar] [CrossRef]
- Mondal, R.; Deb, S.; Shome, G.; Ganguly, U.; Lahiri, D.; Benito-Leon, J. COVID-19 and emerging spinal cord complications: A systematic review. Mult. Scler. Relat. Disord. 2021, 51, 102917. [Google Scholar] [CrossRef]
- Liu, L.D.; Duricka, D.L. Stellate ganglion block reduces symptoms of Long COVID: A case series. J. Neuroimmunol. 2022, 362, 577784. [Google Scholar] [CrossRef] [PubMed]
- Talalwah, W.; Dorazi, S. Trigeminal Neuralgia in COVID-19: A meta-analysis. Int. J. Odontostomatol. 2023, 17, 70–76. [Google Scholar] [CrossRef]
- Harcourt, J.; Tamin, A.; Lu, X.; Kamili, S.; Sakthivel, S.K.; Murray, J.; Queen, K.; Tao, Y.; Paden, C.R.; Zhang, J.; et al. Severe Acute Respiratory Syndrome Coronavirus 2 from Patient with Coronavirus Disease, United States. Emerg. Infect. Dis. 2020, 26, 1266–1273. [Google Scholar] [CrossRef] [PubMed]
- Osterrieder, N.; Bertzbach, L.D.; Dietert, K.; Abdelgawad, A.; Vladimirova, D.; Kunec, D.; Hoffmann, D.; Beer, M.; Gruber, A.D.; Trimpert, J. Age-Dependent Progression of SARS-CoV-2 Infection in Syrian Hamsters. Viruses 2020, 12, 779. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Zhu, H.; Zhou, M.; Ma, J.; Chen, R.; Chen, Y.; Chen, L.; Wu, K.; Cai, M.; Hong, J.; et al. Gender associates with both susceptibility to infection and pathogenesis of SARS-CoV-2 in Syrian hamster. Signal Transduct. Target. Ther. 2021, 6, 136. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, S.; Ruiz-Bedoya, C.A.; Zhou, R.; Creisher, P.S.; Villano, J.S.; Littlefield, K.; Ruelas Castillo, J.; Marinho, P.; Jedlicka, A.E.; Ordonez, A.A.; et al. Sex Differences in Lung Imaging and SARS-CoV-2 Antibody Responses in a COVID-19 Golden Syrian Hamster Model. mBio 2021, 12, e0097421. [Google Scholar] [CrossRef] [PubMed]
- Sia, S.F.; Yan, L.M.; Chin, A.W.H.; Fung, K.; Choy, K.T.; Wong, A.Y.L.; Kaewpreedee, P.; Perera, R.; Poon, L.L.M.; Nicholls, J.M.; et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 2020, 583, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Francis, M.E.; Goncin, U.; Kroeker, A.; Swan, C.; Ralph, R.; Lu, Y.; Etzioni, A.L.; Falzarano, D.; Gerdts, V.; Machtaler, S.; et al. SARS-CoV-2 infection in the Syrian hamster model causes inflammation as well as type I interferon dysregulation in both respiratory and non-respiratory tissues including the heart and kidney. PLoS Pathog. 2021, 17, e1009705. [Google Scholar] [CrossRef] [PubMed]
- Bednash, J.S.; Kagan, V.E.; Englert, J.A.; Farkas, D.; Tyurina, Y.Y.; Tyurin, V.A.; Samovich, S.N.; Farkas, L.; Elhance, A.; Johns, F.; et al. Syrian hamsters as a model of lung injury with SARS-CoV-2 infection: Pathologic, physiologic, and detailed molecular profiling. Transl. Res. 2022, 240, 1–16. [Google Scholar] [CrossRef]
- Hawks, S.A.; Prussin, A.J., 2nd; Kuchinsky, S.C.; Pan, J.; Marr, L.C.; Duggal, N.K. Infectious SARS-CoV-2 Is Emitted in Aerosol Particles. mBio 2021, 12, e0252721. [Google Scholar] [CrossRef]
- Bertke, A.S.; Swanson, S.M.; Chen, J.; Imai, Y.; Kinchington, P.R.; Margolis, T.P. A5-positive primary sensory neurons are nonpermissive for productive infection with herpes simplex virus 1 in vitro. J. Virol. 2011, 85, 6669–6677. [Google Scholar] [CrossRef] [PubMed]
- Powell-Doherty, R.D.; Abbott, A.R.N.; Nelson, L.A.; Bertke, A.S. Amyloid-beta and p-Tau Anti-Threat Response to Herpes Simplex Virus 1 Infection in Primary Adult Murine Hippocampal Neurons. J. Virol. 2020, 94, e01874-19. [Google Scholar] [CrossRef] [PubMed]
- Bertke, A.S.; Apakupakul, K.; Ma, A.; Imai, Y.; Gussow, A.M.; Wang, K.; Cohen, J.I.; Bloom, D.C.; Margolis, T.P. LAT region factors mediating differential neuronal tropism of HSV-1 and HSV-2 do not act in trans. PLoS ONE 2012, 7, e53281. [Google Scholar] [CrossRef] [PubMed]
- Bertke, A.S.; Ma, A.; Margolis, M.S.; Margolis, T.P. Different mechanisms regulate productive herpes simplex virus 1 (HSV-1) and HSV-2 infections in adult trigeminal neurons. J. Virol. 2013, 87, 6512–6516. [Google Scholar] [CrossRef] [PubMed]
- Delwig, A.; Chaney, S.Y.; Bertke, A.S.; Verweij, J.; Quirce, S.; Larsen, D.D.; Yang, C.; Buhr, E.; Van Gelder, R.; Gallar, J.; et al. Melanopsin expression in the cornea. Vis. Neurosci. 2018, 35, E004. [Google Scholar] [CrossRef] [PubMed]
- Goswami, P.; Ives, A.M.; Abbott, A.R.N.; Bertke, A.S. Stress Hormones Epinephrine and Corticosterone Selectively Reactivate HSV-1 and HSV-2 in Sympathetic and Sensory Neurons. Viruses 2022, 14, 1115. [Google Scholar] [CrossRef]
- Ives, A.M.; Bertke, A.S. Stress Hormones Epinephrine and Corticosterone Selectively Modulate Herpes Simplex Virus 1 (HSV-1) and HSV-2 Productive Infections in Adult Sympathetic, but Not Sensory, Neurons. J. Virol. 2017, 91, 1115. [Google Scholar] [CrossRef]
- Swartwout, B.K.; Zlotnick, M.G.; Saver, A.E.; McKenna, C.M.; Bertke, A.S. Zika Virus Persistently and Productively Infects Primary Adult Sensory Neurons In Vitro. Pathogens 2017, 6, 49. [Google Scholar] [CrossRef]
- Yanez, A.A.; Harrell, T.; Sriranganathan, H.J.; Ives, A.M.; Bertke, A.S. Neurotrophic Factors NGF, GDNF and NTN Selectively Modulate HSV1 and HSV2 Lytic Infection and Reactivation in Primary Adult Sensory and Autonomic Neurons. Pathogens 2017, 6, 5. [Google Scholar] [CrossRef]
- Jia, M.; Taylor, T.M.; Senger, S.M.; Ovissipour, R.; Bertke, A.S. SARS-CoV-2 Remains Infectious on Refrigerated Deli Food, Meats, and Fresh Produce for up to 21 Days. Foods 2022, 11, 286. [Google Scholar] [CrossRef]
- Joyce, J.D.; Patel, A.K.; Murphy, B.; Carr, D.J.J.; Gershburg, E.; Bertke, A.S. Assessment of Two Novel Live-Attenuated Vaccine Candidates for Herpes Simplex Virus 2 (HSV-2) in Guinea Pigs. Vaccines 2021, 9, 258. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, A.K.; Ewing, D.; Liang, Z.; Jani, V.; Cheng, Y.; Sun, P.; Raviprakash, K.; Wu, S.J.; Petrovsky, N.; Defang, G.; et al. Immunogenicity of Adjuvanted Psoralen-Inactivated SARS-CoV-2 Vaccines and SARS-CoV-2 Spike Protein DNA Vaccines in BALB/c Mice. Pathogens 2021, 10, 626. [Google Scholar] [CrossRef]
- Mu, J.; Fang, Y.; Yang, Q.; Shu, T.; Wang, A.; Huang, M.; Jin, L.; Deng, F.; Qiu, Y.; Zhou, X. SARS-CoV-2 N protein antagonizes type I interferon signaling by suppressing phosphorylation and nuclear translocation of STAT1 and STAT2. Cell Discov. 2020, 6, 65. [Google Scholar] [CrossRef] [PubMed]
- Syed, A.M.; Taha, T.Y.; Tabata, T.; Chen, I.P.; Ciling, A.; Khalid, M.M.; Sreekumar, B.; Chen, P.Y.; Hayashi, J.M.; Soczek, K.M.; et al. Rapid assessment of SARS-CoV-2-evolved variants using virus-like particles. Science 2021, 374, 1626–1632. [Google Scholar] [CrossRef]
- Bourassa, L.; Perchetti, G.A.; Phung, Q.; Lin, M.J.; Mills, M.G.; Roychoudhury, P.; Harmon, K.G.; Reed, J.C.; Greninger, A.L. A SARS-CoV-2 Nucleocapsid Variant that Affects Antigen Test Performance. J. Clin. Virol. 2021, 141, 104900. [Google Scholar] [CrossRef]
- Boson, B.; Legros, V.; Zhou, B.; Siret, E.; Mathieu, C.; Cosset, F.L.; Lavillette, D.; Denolly, S. The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles. J. Biol. Chem. 2021, 296, 100111. [Google Scholar] [CrossRef]
- Ke, Z.; Oton, J.; Qu, K.; Cortese, M.; Zila, V.; McKeane, L.; Nakane, T.; Zivanov, J.; Neufeldt, C.J.; Cerikan, B.; et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 2020, 588, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.; Zhang, S.; Wang, Q.; Anang, S.; Wang, J.; Ding, H.; Kappes, J.C.; Sodroski, J. Spike glycoprotein and host cell determinants of SARS-CoV-2 entry and cytopathic effects. J. Virol. 2021, 95, e02304-20. [Google Scholar] [CrossRef] [PubMed]
- Smither, S.J.; Kempster, S.; Priestnall, S.L.; Suarez-Bonnet, A.; Stapleton, H.; Laws, T.R.; Ferguson, D.; Almond, N.; Lever, M.S.; Williamson, E.D. Early Isolates of SARS-CoV-2 Result in Different Pathogenesis in the Transduced Mouse Model of COVID-19. Viruses 2022, 14, 1769. [Google Scholar] [CrossRef]
- Dent, M.A.; Segura-Anaya, E.; Alva-Medina, J.; Aranda-Anzaldo, A. NeuN/Fox-3 is an intrinsic component of the neuronal nuclear matrix. FEBS Lett. 2010, 584, 2767–2771. [Google Scholar] [CrossRef]
- Shi, L.; Yang, A.C.; Meng, D.W.; Li, S.W.; Liu, H.G.; Li, J.J.; Wang, X.; Zhang, X.; Zhang, J.G. Pathological alterations and stress responses near DBS electrodes after MRI scans at 7.0T, 3.0T and 1.5T: An in vivo comparative study. PLoS ONE 2014, 9, e101624. [Google Scholar] [CrossRef]
- Sriram, D.; Chintala, R.; Parthasaradhi, B.V.V.; Nayak, S.C.; Mariappan, I.; Radha, V. Expression of a novel brain specific isoform of C3G is regulated during development. Sci. Rep. 2020, 10, 18838. [Google Scholar] [CrossRef]
- Wang, G.; Yang, H.; Yan, S.; Wang, C.E.; Liu, X.; Zhao, B.; Ouyang, Z.; Yin, P.; Liu, Z.; Zhao, Y.; et al. Cytoplasmic mislocalization of RNA splicing factors and aberrant neuronal gene splicing in TDP-43 transgenic pig brain. Mol. Neurodegener. 2015, 10, 42. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, M.L.; Bielenberg, D.R.; Gechtman, Z.; Miao, H.Q.; Takashima, S.; Soker, S.; Klagsbrun, M. Identification of a natural soluble neuropilin-1 that binds vascular endothelial growth factor: In vivo expression and antitumor activity. Proc. Natl. Acad. Sci. USA 2000, 97, 2573–2578. [Google Scholar] [CrossRef] [PubMed]
- Mehta, V.; Fields, L.; Evans, I.M.; Yamaji, M.; Pellet-Many, C.; Jones, T.; Mahmoud, M.; Zachary, I. VEGF (Vascular Endothelial Growth Factor) Induces NRP1 (Neuropilin-1) Cleavage via ADAMs (a Disintegrin and Metalloproteinase) 9 and 10 to Generate Novel Carboxy-Terminal NRP1 Fragments That Regulate Angiogenic Signaling. Arter. Thromb. Vasc. Biol. 2018, 38, 1845–1858. [Google Scholar] [CrossRef]
- Shintani, Y.; Takashima, S.; Asano, Y.; Kato, H.; Liao, Y.; Yamazaki, S.; Tsukamoto, O.; Seguchi, O.; Yamamoto, H.; Fukushima, T.; et al. Glycosaminoglycan modification of neuropilin-1 modulates VEGFR2 signaling. EMBO J. 2006, 25, 3045–3055. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Fournier, A.; Nakamura, F.; Wang, L.H.; Murakami, Y.; Kalb, R.G.; Fujisawa, H.; Strittmatter, S.M. Plexin-neuropilin-1 complexes form functional semaphorin-3A receptors. Cell 1999, 99, 59–69. [Google Scholar] [CrossRef]
- Birla, H.; Rai, S.N.; Singh, S.S.; Zahra, W.; Rawat, A.; Tiwari, N.; Singh, R.K.; Pathak, A.; Singh, S.P. Tinospora cordifolia Suppresses Neuroinflammation in Parkinsonian Mouse Model. Neuromolecular Med. 2019, 21, 42–53. [Google Scholar] [CrossRef]
- Goswami, P.; Gupta, S.; Biswas, J.; Sharma, S.; Singh, S. Endoplasmic Reticulum Stress Instigates the Rotenone Induced Oxidative Apoptotic Neuronal Death: A Study in Rat Brain. Mol. Neurobiol. 2016, 53, 5384–5400. [Google Scholar] [CrossRef]
- Petersen, A.; Puschban, Z.; Lotharius, J.; NicNiocaill, B.; Wiekop, P.; O’Connor, W.T.; Brundin, P. Evidence for dysfunction of the nigrostriatal pathway in the R6/1 line of transgenic Huntington’s disease mice. Neurobiol. Dis. 2002, 11, 134–146. [Google Scholar] [CrossRef]
- Stephenson, D.; Ramirez, A.; Long, J.; Barrezueta, N.; Hajos-Korcsok, E.; Matherne, C.; Gallagher, D.; Ryan, A.; Ochoa, R.; Menniti, F.; et al. Quantification of MPTP-induced dopaminergic neurodegeneration in the mouse substantia nigra by laser capture microdissection. J. Neurosci. Methods 2007, 159, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Frieg, B.; Gorg, B.; Homeyer, N.; Keitel, V.; Haussinger, D.; Gohlke, H. Molecular Mechanisms of Glutamine Synthetase Mutations that Lead to Clinically Relevant Pathologies. PLoS Comput. Biol. 2016, 12, e1004693. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Moon, C.; Ahn, M.; Joo, H.G.; Jin, J.K.; Shin, T. Immunohistochemical localization of galectin-3 in the pig retina during postnatal development. Mol. Vis. 2009, 15, 1971–1976. [Google Scholar] [PubMed]
- Toussaint, J.P.; St-Arnaud, M.; Charest, C. Nitrogen transfer and assimilation between the arbuscular mycorrhizal fungus Glomus intraradices Schenck & Smith and Ri T-DNA roots of Daucus carota L. in an in vitro compartmented system. Can. J. Microbiol. 2004, 50, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Vernizzi, L.; Paiardi, C.; Licata, G.; Vitali, T.; Santarelli, S.; Raneli, M.; Manelli, V.; Rizzetto, M.; Gioria, M.; Pasini, M.E.; et al. Glutamine Synthetase 1 Increases Autophagy Lysosomal Degradation of Mutant Huntingtin Aggregates in Neurons, Ameliorating Motility in a Drosophila Model for Huntington’s Disease. Cells 2020, 9, 196. [Google Scholar] [CrossRef]
- Andriessen, E.; Binet, F.; Fournier, F.; Hata, M.; Dejda, A.; Mawambo, G.; Crespo-Garcia, S.; Pilon, F.; Buscarlet, M.; Beauchemin, K.; et al. Myeloid-resident neuropilin-1 promotes choroidal neovascularization while mitigating inflammation. EMBO Mol. Med. 2021, 13, e11754. [Google Scholar] [CrossRef]
- Exner, E.C.; Geurts, A.M.; Hoffmann, B.R.; Casati, M.; Stodola, T.; Dsouza, N.R.; Zimmermann, M.; Lombard, J.H.; Greene, A.S. Interaction between Mas1 and AT1RA contributes to enhancement of skeletal muscle angiogenesis by angiotensin-(1-7) in Dahl salt-sensitive rats. PLoS ONE 2020, 15, e0232067. [Google Scholar] [CrossRef]
- Parsons Aubone, A.M.; Bisiau, C.M.; McCue, P.M.; Bouma, G.J. Presence of Clock genes in equine full-term placenta. J. Anim. Sci. 2020, 98, skaa094. [Google Scholar] [CrossRef]
- Ruehle, M.A.; Eastburn, E.A.; LaBelle, S.A.; Krishnan, L.; Weiss, J.A.; Boerckel, J.D.; Wood, L.B.; Guldberg, R.E.; Willett, N.J. Extracellular matrix compression temporally regulates microvascular angiogenesis. Sci. Adv. 2020, 6, eabb6351. [Google Scholar] [CrossRef]
- Strobel, H.A.; LaBelle, S.A.; Krishnan, L.; Dale, J.; Rauff, A.; Poulson, A.M.t.; Bader, N.; Beare, J.E.; Aliaj, K.; Weiss, J.A.; et al. Stromal Cells Promote Neovascular Invasion Across Tissue Interfaces. Front. Physiol. 2020, 11, 1026. [Google Scholar] [CrossRef]
- Filipek, A.; Jastrzebska, B.; Nowotny, M.; Kuznicki, J. CacyBP/SIP, a calcyclin and Siah-1-interacting protein, binds EF-hand proteins of the S100 family. J. Biol. Chem. 2002, 277, 28848–28852. [Google Scholar] [CrossRef] [PubMed]
- Ponomarenko, A.; Tyrtyshnaia, A.; Ivashkevich, D.; Ermolenko, E.; Dyuizen, I.; Manzhulo, I. Synaptamide Modulates Astroglial Activity in Mild Traumatic Brain Injury. Mar. Drugs 2022, 20, 538. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhu, J.; Xu, H.; Yi, Q.; Yan, L.; Ye, L.; Zhang, X.; Xie, M.; Tan, B. Time-Dependent Internalization of S100B by Mesenchymal Stem Cells via the Pathways of Clathrin- and Lipid Raft-Mediated Endocytosis. Front. Cell Dev. Biol. 2021, 9, 674995. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joyce, J.D.; Moore, G.A.; Goswami, P.; Harrell, T.L.; Taylor, T.M.; Hawks, S.A.; Green, J.C.; Jia, M.; Irwin, M.D.; Leslie, E.; et al. SARS-CoV-2 Rapidly Infects Peripheral Sensory and Autonomic Neurons, Contributing to Central Nervous System Neuroinvasion before Viremia. Int. J. Mol. Sci. 2024, 25, 8245. https://doi.org/10.3390/ijms25158245
Joyce JD, Moore GA, Goswami P, Harrell TL, Taylor TM, Hawks SA, Green JC, Jia M, Irwin MD, Leslie E, et al. SARS-CoV-2 Rapidly Infects Peripheral Sensory and Autonomic Neurons, Contributing to Central Nervous System Neuroinvasion before Viremia. International Journal of Molecular Sciences. 2024; 25(15):8245. https://doi.org/10.3390/ijms25158245
Chicago/Turabian StyleJoyce, Jonathan D., Greyson A. Moore, Poorna Goswami, Telvin L. Harrell, Tina M. Taylor, Seth A. Hawks, Jillian C. Green, Mo Jia, Matthew D. Irwin, Emma Leslie, and et al. 2024. "SARS-CoV-2 Rapidly Infects Peripheral Sensory and Autonomic Neurons, Contributing to Central Nervous System Neuroinvasion before Viremia" International Journal of Molecular Sciences 25, no. 15: 8245. https://doi.org/10.3390/ijms25158245






