Absence of Aquaporin-4 (AQP4) Prolongs the Presence of a CD11c+ Microglial Population during Postnatal Corpus Callosum Development
Abstract
:1. Introduction
2. Results
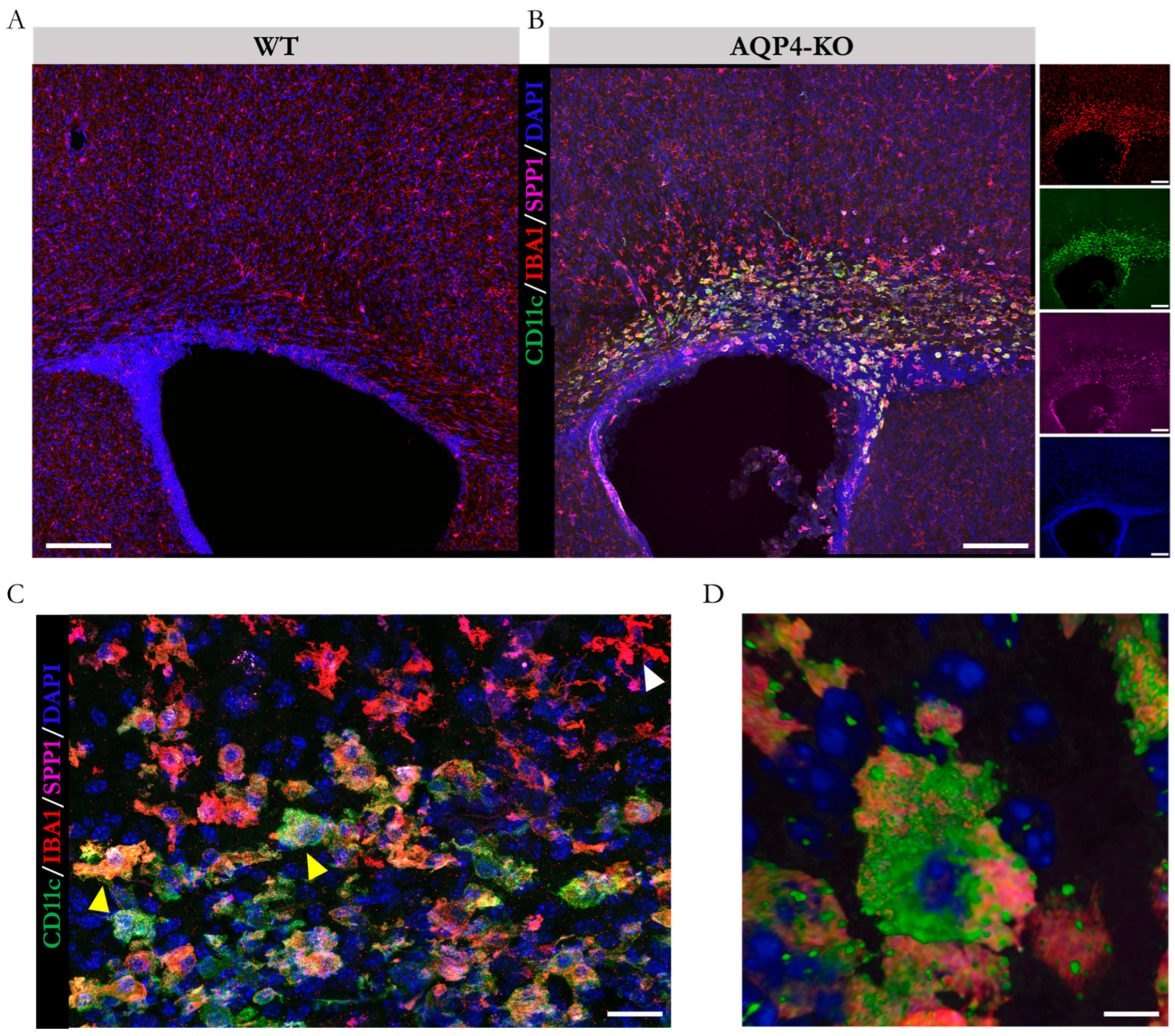
2.1. Comparative Time Course Analysis of the Presence of Postnatal CD11c+ Microglia in the Corpus Callosum of WT and AQP4-KO Animals
2.2. Comparative Transcriptomic Analysis of Corpus Callosum Tissue in WT Versus AQP4-KO Animals
2.3. Differential Expression of Genes Associated with Immune Processes and Identification of Cellular Origin of the Ssp1 Transcript
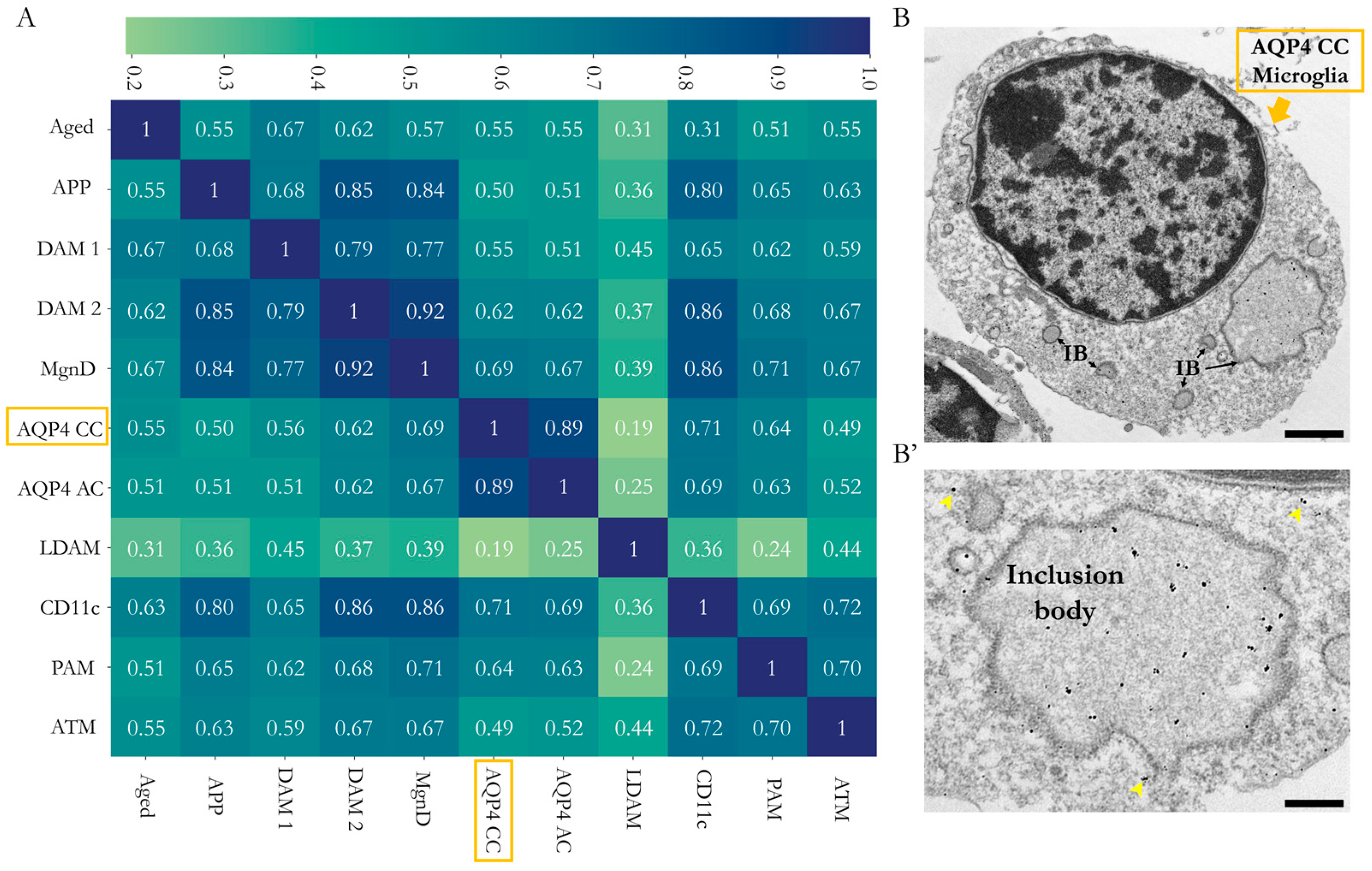
2.4. Comparative Analysis of the CD11c+ Microglia in the Corpus Callosum with Other Microglia
3. Discussion
4. Materials and Methods
4.1. Immunohistochemistry and Imaging Quantification
4.2. In Situ Hybridisation (RNAscope)
4.3. Transmission Electron Microscopy and Immunogold
4.4. Microarray-Based Transcriptomic Analysis
4.5. RNA Extraction and Quantitative Reverse-Transcription Polymerase Chain Reaction Analysis
4.6. Comparative Transcriptomic Analysis of the Microglial Population
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Ginhoux, F.; Lim, S.; Hoeffel, G.; Low, D.; Huber, T. Origin and differentiation of microglia. Front. Cell Neurosci. 2013, 7, 45. [Google Scholar] [CrossRef] [PubMed]
- Gomez Perdiguero, E.; Klapproth, K.; Schulz, C.; Busch, K.; Azzoni, E.; Crozet, L.; Garner, H.; Trouillet, C.; de Bruijn, M.F.; Geissmann, F.; et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 2015, 518, 547–551. [Google Scholar] [CrossRef]
- Benmamar-Badel, A.; Owens, T.; Wlodarczyk, A. Protective Microglial Subset in Development, Aging, and Disease: Lessons From Transcriptomic Studies. Front. Immunol. 2020, 11, 430. [Google Scholar] [CrossRef]
- Lenz, K.M.; Nelson, L.H. Microglia and Beyond: Innate Immune Cells As Regulators of Brain Development and Behavioral Function. Front. Immunol. 2018, 9, 698. [Google Scholar] [CrossRef]
- Sierra, A.; Paolicelli, R.C.; Kettenmann, H. Cien Años de Microglía: Milestones in a Century of Microglial Research. Trends Neurosci. 2019, 42, 778–792. [Google Scholar] [CrossRef] [PubMed]
- Paolicelli, R.C.; Sierra, A.; Stevens, B.; Tremblay, M.E.; Aguzzi, A.; Ajami, B.; Wyss-Coray, T. Microglia states and nomenclature: A field at its crossroads. Neuron 2022, 110, 3458–3483. [Google Scholar] [CrossRef]
- Stratoulias, V.; Venero, J.L.; Tremblay, M.È.; Joseph, B. Microglial subtypes: Diversity within the microglial community. EMBO J. 2019, 38, e101997. [Google Scholar] [CrossRef]
- Li, Q.; Cheng, Z.; Zhou, L.; Darmanis, S.; Neff, N.F.; Okamoto, J.; Gulati, G.; Bennett, M.L.; Sun, L.O.; Clarke, L.E.; et al. Developmental Heterogeneity of Microglia and Brain Myeloid Cells Revealed by Deep Single-Cell RNA Sequencing. Neuron 2019, 101, 207–223.e10. [Google Scholar] [CrossRef]
- Hammond, T.R.; Dufort, C.; Dissing-Olesen, L.; Giera, S.; Young, A.; Wysoker, A.; Walker, A.J.; Gergits, F.; Segel, M.; Nemesh, J.; et al. Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity 2019, 50, 253–271.e6. [Google Scholar] [CrossRef]
- Tan, Y.L.; Yuan, Y.; Tian, L. Microglial regional heterogeneity and its role in the brain. Mol. Psychiatry 2020, 25, 351–367. [Google Scholar] [CrossRef]
- Thion, M.S.; Garel, S. Microglial ontogeny, diversity and neurodevelopmental functions. Curr. Opin. Genet. Dev. 2020, 65, 186–194. [Google Scholar] [CrossRef]
- Amor, S.; McNamara, N.B.; Gerrits, E.; Marzin, M.C.; Kooistra, S.M.; Miron, V.E.; Nutma, E. White matter microglia heterogeneity in the CNS. Acta Neuropathol. 2022, 143, 125–141. [Google Scholar] [CrossRef] [PubMed]
- Wlodarczyk, A.; Holtman, I.R.; Krueger, M.; Yogev, N.; Bruttger, J.; Khorooshi, R.; Benmamar-Badel, A.; de Boer-Bergsma, J.J.; Martin, N.A.; Karram, K.; et al. A novel microglial subset plays a key role in myelinogenesis in developing brain. EMBO J. 2017, 36, 3292–3308. [Google Scholar] [CrossRef] [PubMed]
- Holtman, I.R.; Raj, D.D.; Miller, J.A.; Schaafsma, W.; Yin, Z.; Brouwer, N.; Wes, P.D.; Möller, T.; Orre, M.; Kamphuis, W.; et al. Induction of a common microglia gene expression signature by aging and neurodegenerative conditions: A co-expression meta-analysis. Acta Neuropathol. Commun. 2015, 3, 31. [Google Scholar] [CrossRef] [PubMed]
- Keren-Shaul, H.; Spinrad, A.; Weiner, A.; Matcovitch-Natan, O.; Dvir-Szternfeld, R.; Ulland, T.K.; David, E.; Baruch, K.; Lara-Astaiso, D.; Toth, B.; et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 2017, 169, 1276–1290.e17. [Google Scholar] [CrossRef] [PubMed]
- Krasemann, S.; Madore, C.; Cialic, R.; Baufeld, C.; Calcagno, N.; El Fatimy, R.; Beckers, L.; O’Loughlin, E.; Xu, Y.; Fanek, Z.; et al. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity 2017, 47, 566–581.e9. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Qiu, Y.; Wight, A.E.; Kim, H.J.; Cantor, H. Definition of a mouse microglial subset that regulates neuronal development and proinflammatory responses in the brain. Proc. Natl. Acad. Sci. USA 2022, 119, e2116241119. [Google Scholar] [CrossRef] [PubMed]
- Ghena, N.R.; Anderson, S.R.; Roberts, J.M.; Irvin, E.; Schwakopf, J.; Bosco, A.; Vetter, M.L. CD11c-expressing microglia are transient, driven by interactions with apoptotic cells. bioRxiv 2024, preprints. [Google Scholar]
- Ueno, M.; Fujita, Y.; Tanaka, T.; Nakamura, Y.; Kikuta, J.; Ishii, M.; Yamashita, T. Layer V cortical neurons require microglial support for survival during postnatal development. Nat. Neurosci. 2013, 16, 543–551. [Google Scholar] [CrossRef]
- Mayo, F.; González-Vinceiro, L.; Hiraldo-González, L.; Calle-Castillejo, C.; Morales-Alvarez, S.; Ramírez-Lorca, R.; Echevarría, M. Aquaporin-4 Expression Switches from White to Gray Matter Regions during Postnatal Development of the Central Nervous System. Int. J. Mol. Sci. 2023, 24, 3048. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Echevarría, M.; García-Miranda, P. Special Issue “Aquaporins in Brain Disease”. Int. J. Mol. Sci. 2024, 25, 3513. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Francisco, M.; González-Vinceiro, L.; González, L.H.; Rodríguez-Gómez, F.D.; Castillejo, C.C.; Mayo, M.; Netti, V.; Lorca, R.R.; Echevarría, M. Impact of Aquaporin-4 and CD11c+ microglia in the development of ependymal cells in the aqueduct. Inferences to hydrocephalus. Fluids Barriers CNS 2024, 21, 53. [Google Scholar] [CrossRef]
- Nomaki, K.; Fujikawa, R.; Masuda, T.; Tsuda, M. Spatiotemporal dynamics of the CD11c+ micro-glial population in the mouse brain and spinal cord from developmental to adult stages. Mol. Brain 2024, 17, 24. [Google Scholar] [CrossRef] [PubMed]
- Nemes-Baran, A.D.; White, D.R.; DeSilva, T.M. Fractalkine-Dependent Microglial Pruning of Viable Oligodendrocyte Progenitor Cells Regulates Myelination. Cell Rep. 2020, 32, 108047. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Traiffort, E.; Kassoussi, A.; Zahaf, A.; Laouarem, Y. Astrocytes and Microglia as Major Players of Myelin Production in Normal and Pathological Conditions. Front. Cell Neurosci. 2020, 14, 79. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hagemeyer, N.; Hanft, K.M.; Akriditou, M.A.; Unger, N.; Park, E.S.; Stanley, E.R.; Staszewski, O.; Dimou, L.; Prinz, M. Microglia contribute to normal myelinogenesis and to oligodendrocyte progenitor maintenance during adulthood. Acta Neuropathol. 2017, 134, 441–458. [Google Scholar] [CrossRef]
- Poliani, P.L.; Wang, Y.; Fontana, E.; Robinette, M.L.; Yamanishi, Y.; Gilfillan, S.; Colonna, M. TREM2 sustains microglial expansion during aging and response to demyelination. J. Clin. Investig. 2015, 125, 2161–2170. [Google Scholar] [CrossRef]
- Lampron, A.; Larochelle, A.; Laflamme, N.; Préfontaine, P.; Plante, M.M.; Sánchez, M.G.; Rivest, S. Inefficient clearance of myelin debris by microglia impairs remyelinating processes. J. Exp. Med. 2015, 212, 481–495. [Google Scholar] [CrossRef]
- Sato-Hashimoto, M.; Nozu, T.; Toriba, R.; Horikoshi, A.; Akaike, M.; Kawamoto, K.; Ohnishi, H. Microglial SIRPα regulates the emergence of CD11c+ microglia and demyelination damage in white matter. Elife 2019, 8, e42025. [Google Scholar] [CrossRef]
- Selvaraju, R.; Bernasconi, L.; Losberger, C.; Graber, P.; Kadi, L.; Avellana-Adalid, V.; Picard-Riera, N.; Baron Van Evercooren, A.; Cirillo, R.; Kosco-Vilbois, M.; et al. Osteopontin is upregulated during in vivo demyelination and remyelination and enhances myelin formation in vitro. Mol. Cell Neurosci. 2004, 25, 707–721. [Google Scholar] [CrossRef]
- Thomas, L.; Pasquini, L.A. Galectin-3-Mediated Glial Crosstalk Drives Oligodendrocyte Differentiation and (Re)myelination. Front. Cell Neurosci. 2018, 12, 297. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Trillo-Contreras, J.L.; Toledo-Aral, J.J.; Echevarría, M.; Villadiego, J. AQP1 and AQP4 Contribution to Cerebrospinal Fluid Homeostasis. Cells 2019, 8, 197. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Orre, M.; Kamphuis, W.; Osborn, L.M.; Jansen, A.H.P.; Kooijman, L.; Bossers, K.; Hol, E.M. Isolation of glia from Alzheimer’s mice reveals inflammation anddysfunction. Neurobiol. Aging 2014, 35, 2746–2760. [Google Scholar] [CrossRef] [PubMed]
- Marschallinger, J.; Iram, T.; Zardeneta, M.; Lee, S.E.; Lehallier, B.; Haney, M.S.; Pluvinage, J.V.; Mathur, V.; Hahn, O.; Morgens, D.W.; et al. Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat. Neurosci. 2020, 23, 194–208. [Google Scholar] [CrossRef] [PubMed]
- D’haeseleer, P.; Liang, S.; Somogyi, R. Genetic network inference: From co-expression clustering to reverse engineering. Bioinformatics 2000, 16, 707–726. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.M.; Johnson, K.F. Methods of Microarray Data Analysis 2; Kluwer Academic Publishers: Norwell, MA, USA, 2002. [Google Scholar]
- Speed, T. Statistical analysis of gene expression microarray data. In Interdisciplinary Statistics; Chapman & Hall/CRC: Boca Raton, FL, USA, 2003. [Google Scholar]
- Jiang, D.; Tang, C.; Zhang, A. Cluster analysis for gene expression data: A survey. IEEE Trans. Knowl. Data Eng. 2004, 16, 1370–1386. [Google Scholar] [CrossRef]
- Priness, I.; Maimon, O.; Ben-Gal, I. Evaluation of gene-expression clustering via mutual information distance measure. BMC Bioinform. 2007, 8, 111. [Google Scholar] [CrossRef]
- Mead, A. Review of the Development of Multidimensional Scaling Methods. J. R. Stat. Society. Ser. D (Stat.) 1992, 41, 27–39. [Google Scholar] [CrossRef]
- Green, P.E.; Carmone, F.J.; Smith, S.M. Multidimensional Scaling: Concepts and Applications; Allyn and Bacon: Needham Heights, MA, 1989. [Google Scholar]
- Borg, I.; Groenen, P. Modern Multidimensional Scaling: Theory and Applications; Springer: New York, NY, USA, 1997. [Google Scholar]
- Cox, T.F.; Cox, M.A.A. Multidimensional Scaling, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2000. [Google Scholar]
- Chen, Y.; Meltzer, P.S. Gene expression analysis via multidimensional scaling. Curr. Protoc. Bioinform. 2005, 7, 7–11. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mayo, F.; González-Vinceiro, L.; Hiraldo-González, L.; Calle-Castillejo, C.; Torres-Rubio, I.; Mayo, M.; Ramírez-Lorca, R.; Echevarría, M. Absence of Aquaporin-4 (AQP4) Prolongs the Presence of a CD11c+ Microglial Population during Postnatal Corpus Callosum Development. Int. J. Mol. Sci. 2024, 25, 8332. https://doi.org/10.3390/ijms25158332
Mayo F, González-Vinceiro L, Hiraldo-González L, Calle-Castillejo C, Torres-Rubio I, Mayo M, Ramírez-Lorca R, Echevarría M. Absence of Aquaporin-4 (AQP4) Prolongs the Presence of a CD11c+ Microglial Population during Postnatal Corpus Callosum Development. International Journal of Molecular Sciences. 2024; 25(15):8332. https://doi.org/10.3390/ijms25158332
Chicago/Turabian StyleMayo, Francisco, Lourdes González-Vinceiro, Laura Hiraldo-González, Claudia Calle-Castillejo, Ismael Torres-Rubio, Manuel Mayo, Reposo Ramírez-Lorca, and Miriam Echevarría. 2024. "Absence of Aquaporin-4 (AQP4) Prolongs the Presence of a CD11c+ Microglial Population during Postnatal Corpus Callosum Development" International Journal of Molecular Sciences 25, no. 15: 8332. https://doi.org/10.3390/ijms25158332





