Characteristics and Cytological Analysis of Several Novel Allopolyploids and Aneuploids between Brassica oleracea and Raphanus sativus
Abstract
1. Introduction
2. Results
2.1. Determination of Genome Configuration of These Offspring Plants of RRRC
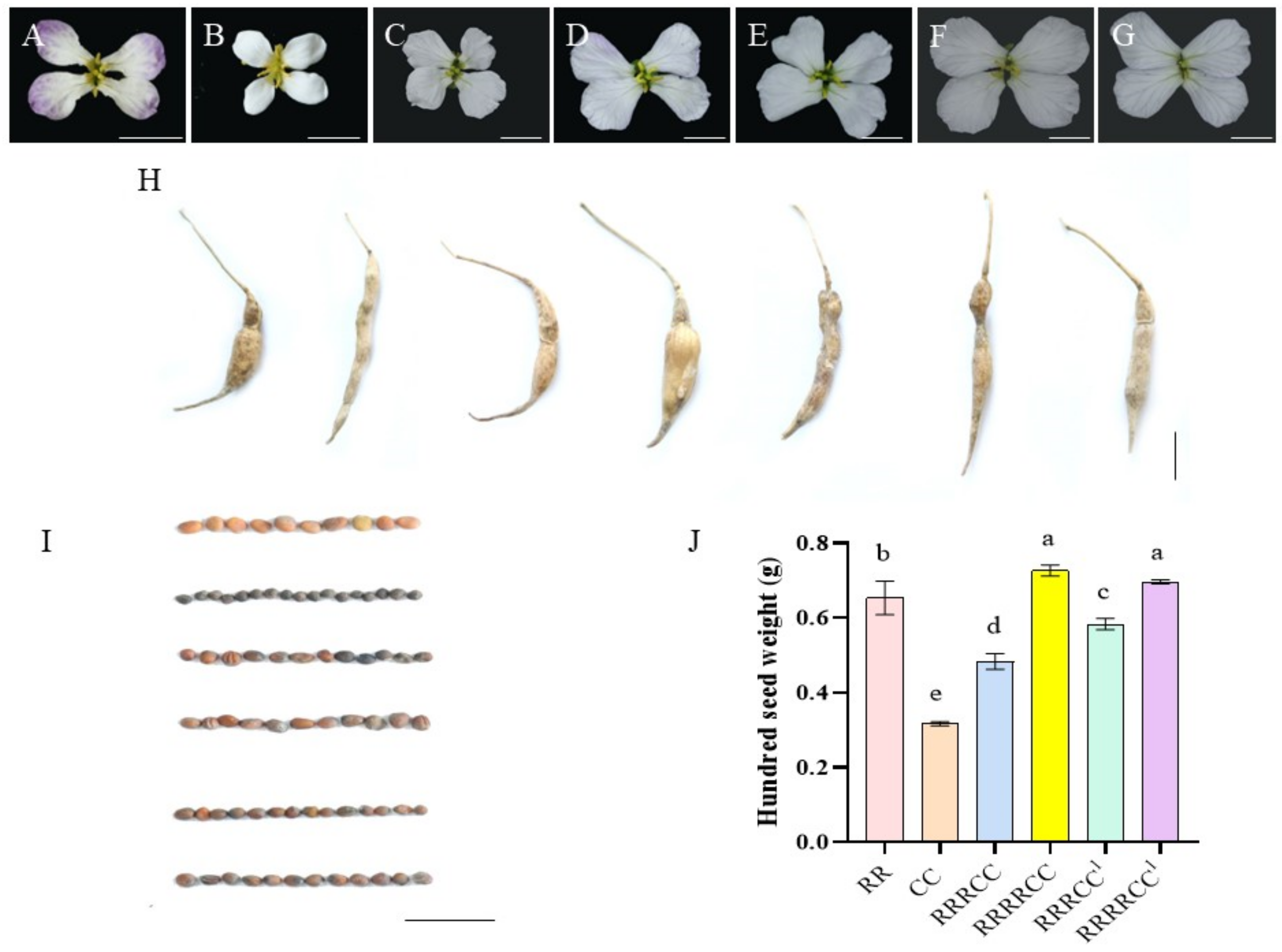
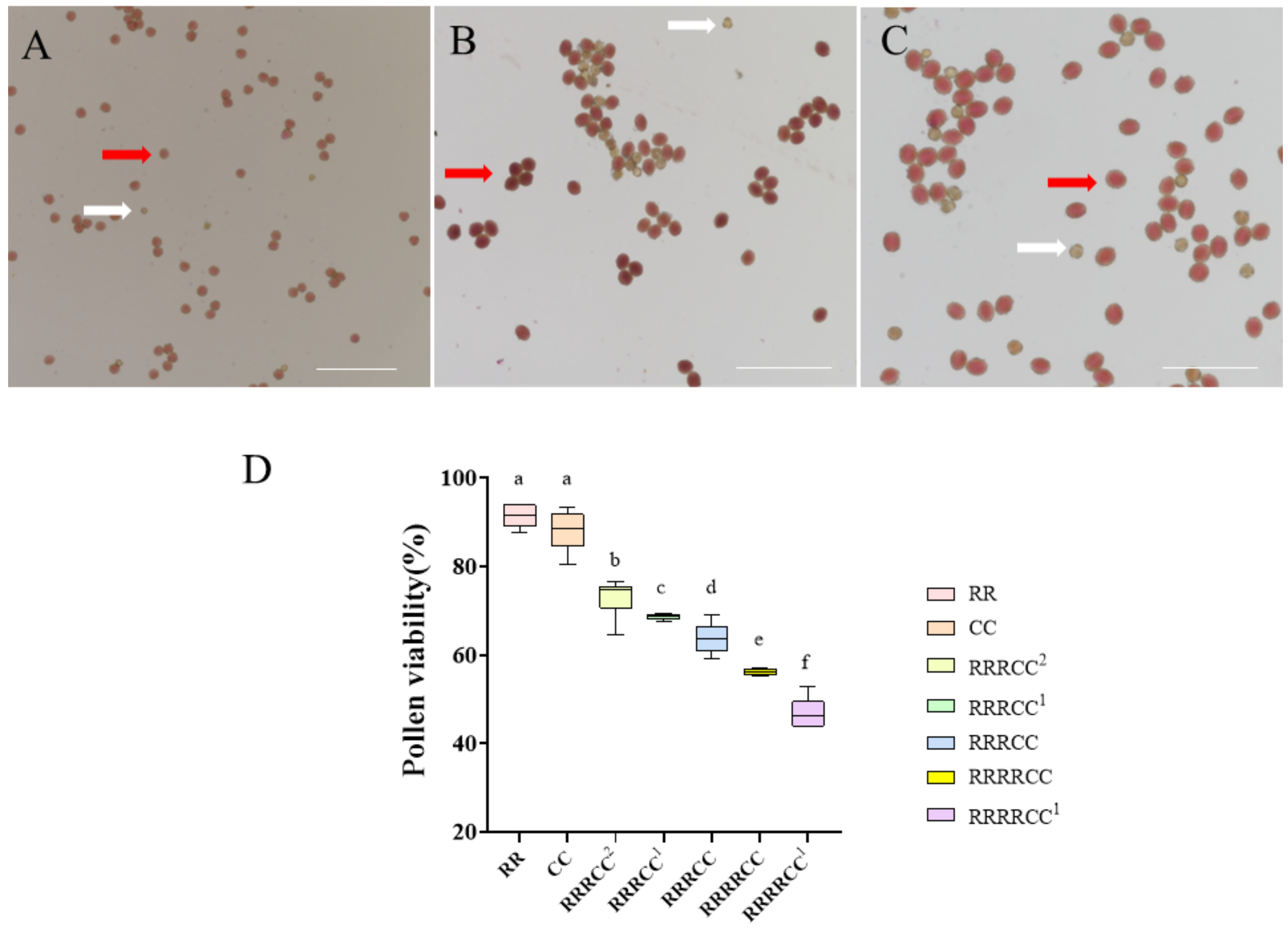
2.2. Morphological Characteristics and Pollen Fertility of Offspring Plants
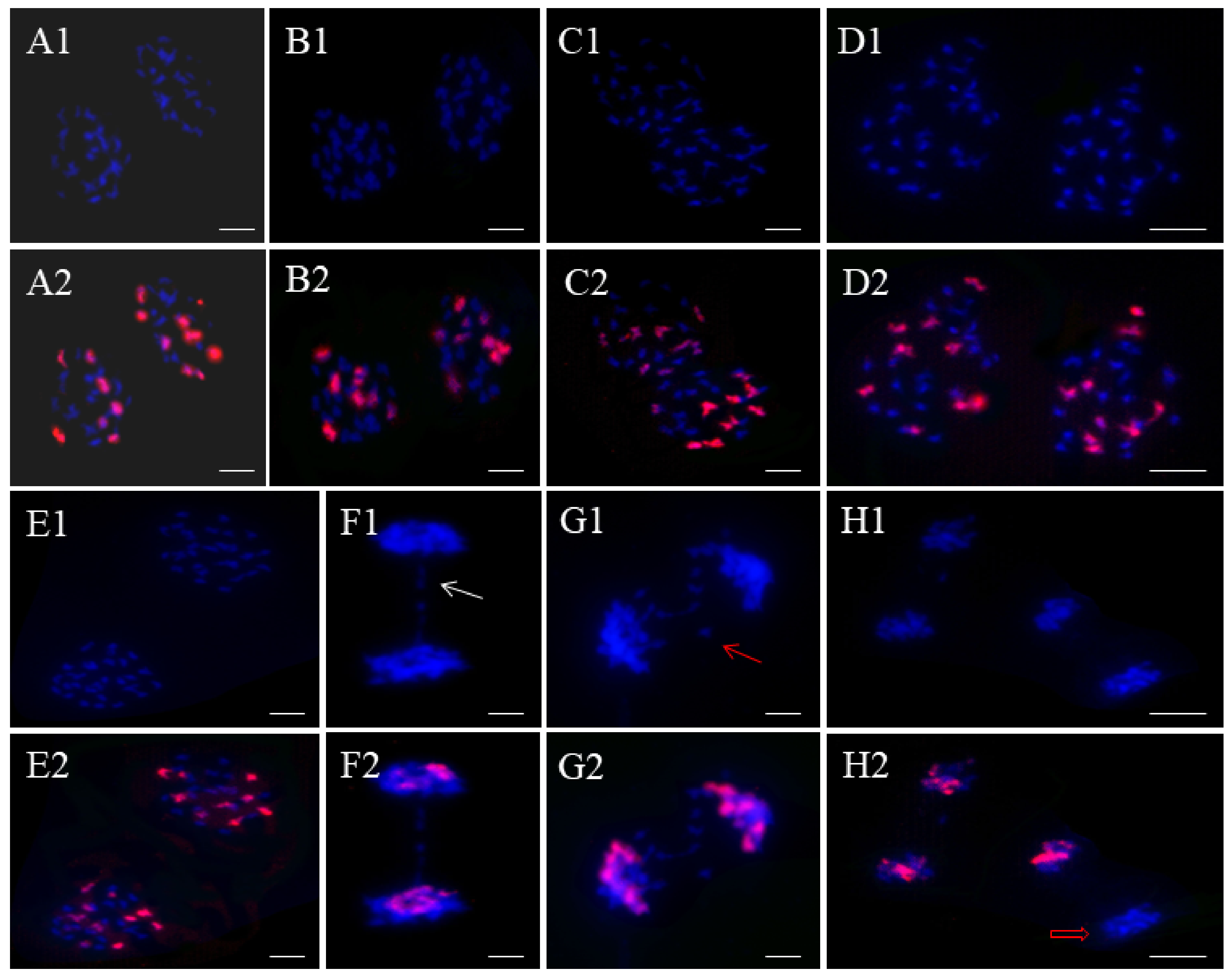
2.3. Chromosome Pairing Patterns of Offspring Plants
2.4. Chromosome Segregations in These Progenies
3. Discussion
4. Materials and Methods
4.1. Chromosome Plant Materials and Cultivation Conditions
4.2. Analysis of Cytology and Pollen Viability
4.3. Probe Preparation and Fluorescence In Situ Hybridization
4.4. Phenotype Evaluation of These Offspring Plants
4.5. Data Statistics Processing and Graphing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, Z.; Li, M.; Wu, X.; Wang, J. The characteristics of mRNA m6A methylomes in allopolyploid Brassica napus and its diploid progenitors. Hortic. Res. 2023, 10, uhac230. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Du, M.j.; Ji, R.z.; Cheng, F.y. Cytological origination of the first found allotriploid tree peony, Paeonia× lemoinei ‘Oukan’(AAB), reveled by molecular karyotype comparation. Sci. Hortic. 2024, 324, 112563. [Google Scholar] [CrossRef]
- Liu, J.; Yao, Y.; Xin, M.; Peng, H.; Ni, Z.; Sun, Q. Shaping polyploid wheat for success: Origins, domestication, and the genetic improvement of agronomic traits. J. Integr. Plant Biol. 2022, 64, 536–563. [Google Scholar] [CrossRef] [PubMed]
- Park, H.R.; Park, J.E.; Kim, J.H.; Shin, H.; Yu, S.H.; Son, S.; Yi, G.; Lee, S.-S.; Kim, H.H.; Huh, J.H. Meiotic chromosome stability and suppression of crossover between non-homologous chromosomes in x Brassicoraphanus, an intergeneric allotetraploid derived from a cross between Brassica rapa and Raphanus sativus. Front. Plant Sci. 2020, 11, 851. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Tu, Y.; Zeng, P.; Ge, X.; Li, Z. Extraction of the constituent subgenomes of the natural allopolyploid rapeseed (Brassica napus L.). Genetics 2016, 204, 1015–1027. [Google Scholar] [CrossRef] [PubMed]
- Bora, L.; Vijayakumar, R.M.; Ganga, M.; Meenakshi Ganesan, N.; Sarkar, M.; Kundu, M. Induction of polyploids in acid lime (Citrus aurantifolia) through colchicine treatment of in vitro derived shoot tip explants. Plant Breed. 2023, 142, 118–127. [Google Scholar] [CrossRef]
- Barker, M.S.; Arrigo, N.; Baniaga, A.E.; Li, Z.; Levin, D.A. On the relative abundance of autopolyploids and allopolyploids. New Phytol. 2016, 210, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Morton, J.A.; Pellicer, J.; Leitch, I.J.; Leitch, A.R. Genome downsizing after polyploidy: Mechanisms, rates and selection pressures. Plant J. 2021, 107, 1003–1015. [Google Scholar] [CrossRef] [PubMed]
- Ihien Katche, E.; Mason, S. Resynthesized rapeseed (Brassica napus): Breeding and genomics. Crit. Rev. Plant Sci. 2023, 42, 65–92. [Google Scholar] [CrossRef]
- Soltis, P.S.; Soltis, D.E. The role of hybridization in plant speciation. Annu. Rev. Plant Biol. 2009, 60, 561–588. [Google Scholar] [CrossRef]
- Mason, A.S.; Batley, J. Creating new interspecific hybrid and polyploid crops. Trends Biotechnol. 2015, 33, 436–441. [Google Scholar] [CrossRef]
- Charlesworth, B.; Charlesworth, D. Elements of Evolutionary Genetics; Springer: Greenwoord Village, CO, USA, 2010; Volume 42. [Google Scholar]
- Ma’arup, R.; Trethowan, R.M.; Ahmed, N.U.; Bramley, H.; Sharp, P.J. Emmer wheat (Triticum dicoccon Schrank) improves water use efficiency and yield of hexaploid bread wheat. Plant Sci. 2020, 295, 110212. [Google Scholar] [CrossRef] [PubMed]
- Kantar, M.B.; Betts, K.; Michno, J.M.; Luby, J.J.; Morrell, P.L.; Hulke, B.S.; Stupar, R.M.; Wyse, D.L. Evaluating an interspecific Helianthus annuus× Helianthus tuberosus population for use in a perennial sunflower breeding program. Field Crops Res. 2014, 155, 254–264. [Google Scholar] [CrossRef]
- Tanhuanpää, P.; Erkkilä, M.; Kalendar, R.; Schulman, A.H.; Manninen, O. Assessment of genetic diversity in Nordic timothy (Phleum pratense L.). Hereditas 2016, 153, 5. [Google Scholar] [CrossRef][Green Version]
- Mei, J.; Liu, J.; Yue, F.; Chen, Y.; Ming, J.; Xiong, Z.; Yu, F.; Li, J.; Qian, W. Broadening the genetic base of Brassica juncea by introducing genomic components from B. rapa and B. nigra via digenomic allohexaploid bridging. Crop J. 2022, 10, 672–679. [Google Scholar] [CrossRef]
- Li, Z.; Li, M.; Wang, J. Asymmetric subgenomic chromatin architecture impacts on gene expression in resynthesized and natural allopolyploid Brassica napus. Commun. Biol. 2022, 5, 762. [Google Scholar] [CrossRef]
- Marsolier-Kergoat, M.C.; Khan, M.M.; Schott, J.; Zhu, X.; Llorente, B. Mechanistic view and genetic control of DNA recombination during meiosis. Mol. Cell 2018, 70, 9–20. [Google Scholar] [CrossRef]
- Zickler, D.; Kleckner, N. Meiosis: Dances between homologs. Annu. Rev. Genet. 2023, 57, 1–63. [Google Scholar] [CrossRef]
- Friedt, W.; Tu, J.; Fu, T. Academic and economic importance of Brassica napus rapeseed. In The Brassica napus Genome; Springer: Cham, Switzerland, 2018; pp. 1–20. [Google Scholar]
- Jabeen, N. Agricultural, economic and societal importance of Brassicaceae plants. In The Plant Family Brassicaceae: Biology and Physiological Responses to Environmental Stresses; Springer: Singapore, 2020; pp. 45–128. [Google Scholar]
- Nagaharu, U.J.J.J.B.; Nagaharu, N. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 1935, 7, 389–452. [Google Scholar]
- Li, X.; Wang, Y.; Cai, C.; Ji, J.; Han, F.; Zhang, L.; Chen, S.; Zhang, L.; Yang, Y.; Tang, Q. Large-scale gene expression alterations introduced by structural variation drive morphotype diversification in Brassica oleracea. Nat. Genet. 2024, 56, 517–529. [Google Scholar] [CrossRef]
- Zhou, R.; Qin, X.; Hou, J.; Liu, Y. Research progress on Brassicaceae plants: A bibliometrics analysis. Front. Plant Sci. 2024, 15, 1285050. [Google Scholar] [CrossRef] [PubMed]
- Arefin, M.N.; Bhuiyan, M.K.A.; Rubayet, M.T. Integrated use of fungicide, plant extract and bio-agent for management of Alternaria blight disease of Radish (Raphanus sativus L.) and quality seed production. Res. Agric. Vet. Sci. 2019, 3, 10–21. [Google Scholar]
- Jaafar, N.A.; Ahmed, A.S.; Al-Sandooq, D.L. Detection of active compounds in radish Raphanus Sativus L. and their various biological effects. Plant Arch 2020, 20, 1647–1650. [Google Scholar]
- Huang, T.; Lu, J.; Zhang, W.; Wang, W.; Wan, Y.; Pei, Y.; Mao, F.; Wang, L.; Li, J. Evaluation of genetic variation of morphological and clubroot-resistance traits of radish and metabonomic analysis of clubroot-resistant cultivar. Sci. Hortic. 2023, 321, 112272. [Google Scholar] [CrossRef]
- Gamba, M.; Asllanaj, E.; Raguindin, P.F.; Glisic, M.; Franco, O.H.; Minder, B.; Bussler, W.; Metzger, B.; Kern, H.; Muka, T. Nutritional and phytochemical characterization of radish (Raphanus sativus): A systematic review. Trends Food Sci. Technol. 2021, 113, 205–218. [Google Scholar] [CrossRef]
- Intawongse, M.; Dean, J.R. Uptake of heavy metals by vegetable plants grown on contaminated soil and their bioavailability in the human gastrointestinal tract. Trends Food Sci. Technol. 2006, 23, 36–48. [Google Scholar] [CrossRef]
- Qi, X.H.; Zhang, M.F. Molecular phylogenetic studies on members of the Brassica and Raphanus genera based on the nuclear ribosomal internal transcribed spacer, the chloroplast trn L intron, trn LF, and cpSSRs. J. Hortic. Sci. Biotechnol. 2012, 87, 149–156. [Google Scholar] [CrossRef]
- Budahn, H.; Peterka, H.; Mousa, M.A.A.; Ding, Y.; Zhang, S.; Li, J. Molecular mapping in oil radish (Raphanus sativus L.) and QTL analysis of resistance against beet cyst nematode (Heterodera schachtii). Theor. Appl. Genet. 2009, 118, 775–782. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Z.; Nwafor, C.C.; Hou, Z.; Gong, J.; Zhu, B.; Jiang, Y.; Zhou, Y.; Wu, J.; Piao, Z.; Tong, Y. Cytological and morphological analysis of hybrids between Brassicoraphanus, and Brassica napus for introgression of clubroot resistant trait into Brassica napus L. PLoS ONE 2017, 12, e0177470. [Google Scholar] [CrossRef]
- Lou, L.; Lou, Q.; Li, Z.; Xu, Y.; Liu, Z.; Su, X. Production and characterization of intergeneric hybrids between turnip (Brassica rapa L. em. Metzg. subsp. rapa) and radish (Raphanus sativus L.). Sci. Hortic. 2017, 220, 57–65. [Google Scholar] [CrossRef]
- Feng, Q.; Yu, J.; Yang, X.; Lv, X.; Lu, Y.; Yuan, J.; Du, X.; Zhu, B.; Li, Z. Development and characterization of an allooctaploid (AABBCCRR) incorporating Brassica and radish genomes via two rounds of interspecific hybridizations. Sci. Hortic. 2022, 293, 110730. [Google Scholar] [CrossRef]
- Yu, J.; Lei, S.; Fang, S.; Tai, N.; Yu, W.; Yang, Z.; Gu, L.; Wang, H.; Du, X.; Zhu, B. Identification, Characterization, and Cytological Analysis of Several Unexpected Hybrids Derived from Reciprocal Crosses between Raphanobrassica and Its Diploid Parents. Plants 2023, 12, 1875. [Google Scholar] [CrossRef] [PubMed]
- Husband, B.C.; Baldwin, S.J.; Suda, J. The Incidence of Polyploidy in Natural Plant Populations: Major Patterns and Evolutionary Processes; Springer: Vienna, Austria, 2013; pp. 255–276. [Google Scholar]
- Doyle, J.J.; Flagel, L.E.; Paterson, A.H.; Rapp, R.A.; Soltis, D.E.; Soltis, P.S.; Wendel, J.F. Evolutionary genetics of genome merger and doubling in plants. Annu. Rev. Genet. 2008, 42, 443–461. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.; Gaeta, R.T.; Pires, J.C. Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc. Natl. Acad. Sci. USA 2011, 108, 7908–7913. [Google Scholar] [CrossRef] [PubMed]
- Siegel, J.J.; Amon, A. New insights into the troubles of aneuploidy. Annu. Rev. Genet. 2012, 28, 189–214. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Tan, C.; Cui, C.; Ge, X.; Li, Z. Distinct subgenome stabilities in synthesized Brassica allohexaploids. Theor. Appl. Genet. 2016, 129, 1257–1271. [Google Scholar] [CrossRef] [PubMed]
- Katche, E.; Gaebelein, R.; Idris, Z.; Vasquez-Teuber, P.; Lo, Y.T.; Nugent, D.; Batley, J.; Mason, A.S. Stable, fertile lines produced by hybridization between allotetraploids Brassica juncea (AABB) and Brassica carinata (BBCC) have merged the A and C genomes. New Phytol. 2021, 230, 1242–1257. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Wang, T.; Yue, F.; Harun, A.; Zhu, B.; Qian, W.; Ge, X.; Li, Z. Production and cytology of Brassica autoallohexaploids with two and four copies of two subgenomes. Theor. Appl. Genet. 2022, 135, 2641–2653. [Google Scholar] [CrossRef] [PubMed]
- Vallejo-Marín, M.; Buggs, R.J.; Cooley, A.M.; Puzey, J.R. Speciation by genome duplication: Repeated origins and genomic composition of the recently formed allopolyploid species Mimulus peregrinus. Evolution 2015, 69, 1487–1500. [Google Scholar] [CrossRef]
- Zhang, L.; He, J.; He, H.; Wu, J.; Li, M. Genome-wide unbalanced expression bias and expression level dominance toward Brassica oleracea in artificially synthesized intergeneric hybrids of Raphanobrassica. Hortic. Res. 2021, 8, 246. [Google Scholar] [CrossRef]
- Cui, C.; Ge, X.; Gautam, M.; Kang, L.; Li, Z. Cytoplasmic and Genomic Effects on Meiotic Pairing in Brassica Hybrids and Allotetraploids from Pair Crosses of Three Cultivated Diploids. Genetics 2012, 191, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.B.; Jong, J.H.D.; Zabel, P. Preparation of tomato meiotic pachytene and mitotic metaphase chromosomes suitable for fluorescence in situ hybridization (FISH). Chromosome Res. 1996, 4, 24–28. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Cui, C.; Xiang, Y.; Ge, X.; Li, Z. Development of Brassica oleracea-nigra monosomic alien addition lines: Genotypic, cytological and morphological analyses. Theor. Appl. Genet. 2017, 130, 2491–2504. [Google Scholar] [CrossRef] [PubMed]
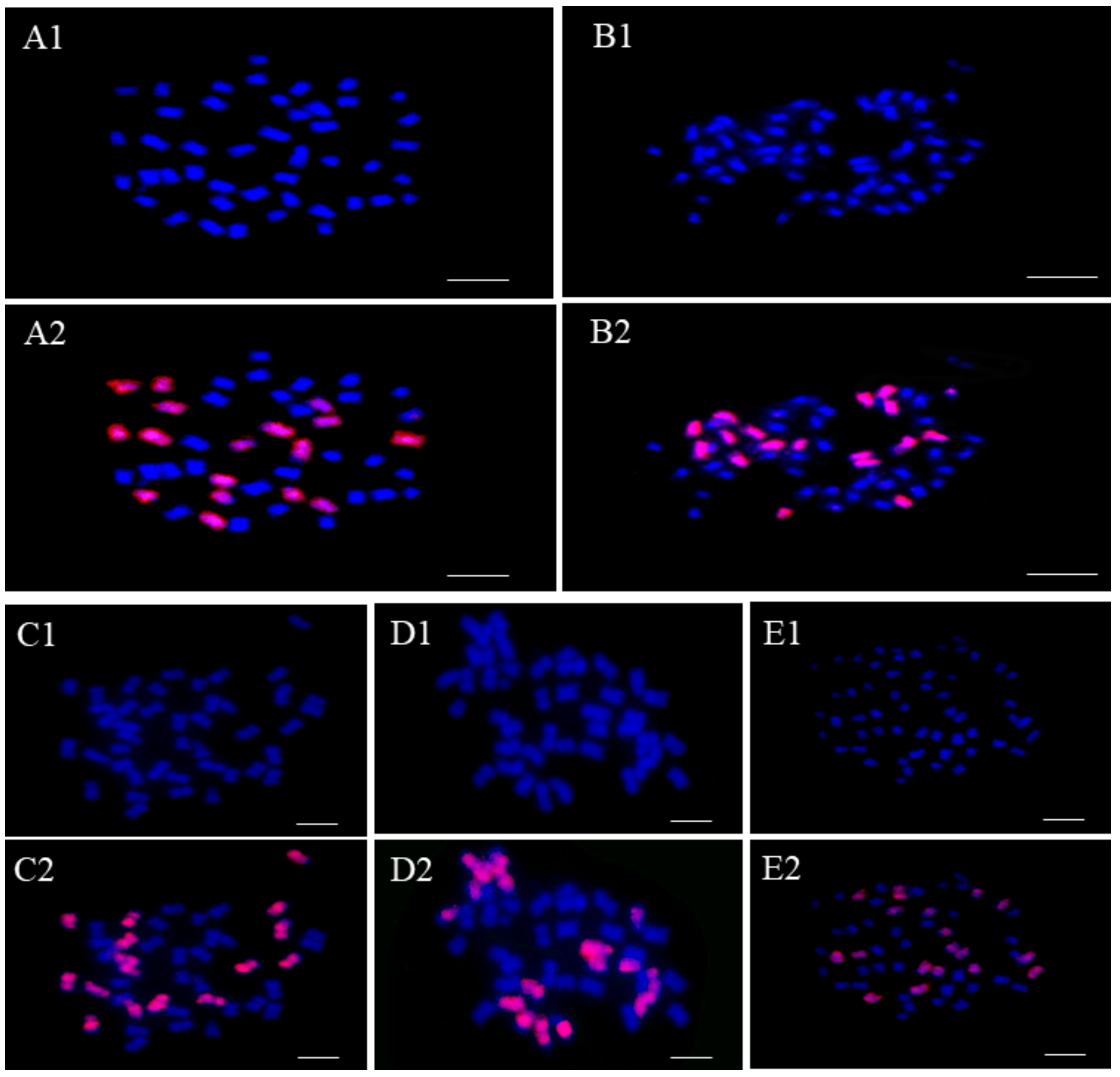


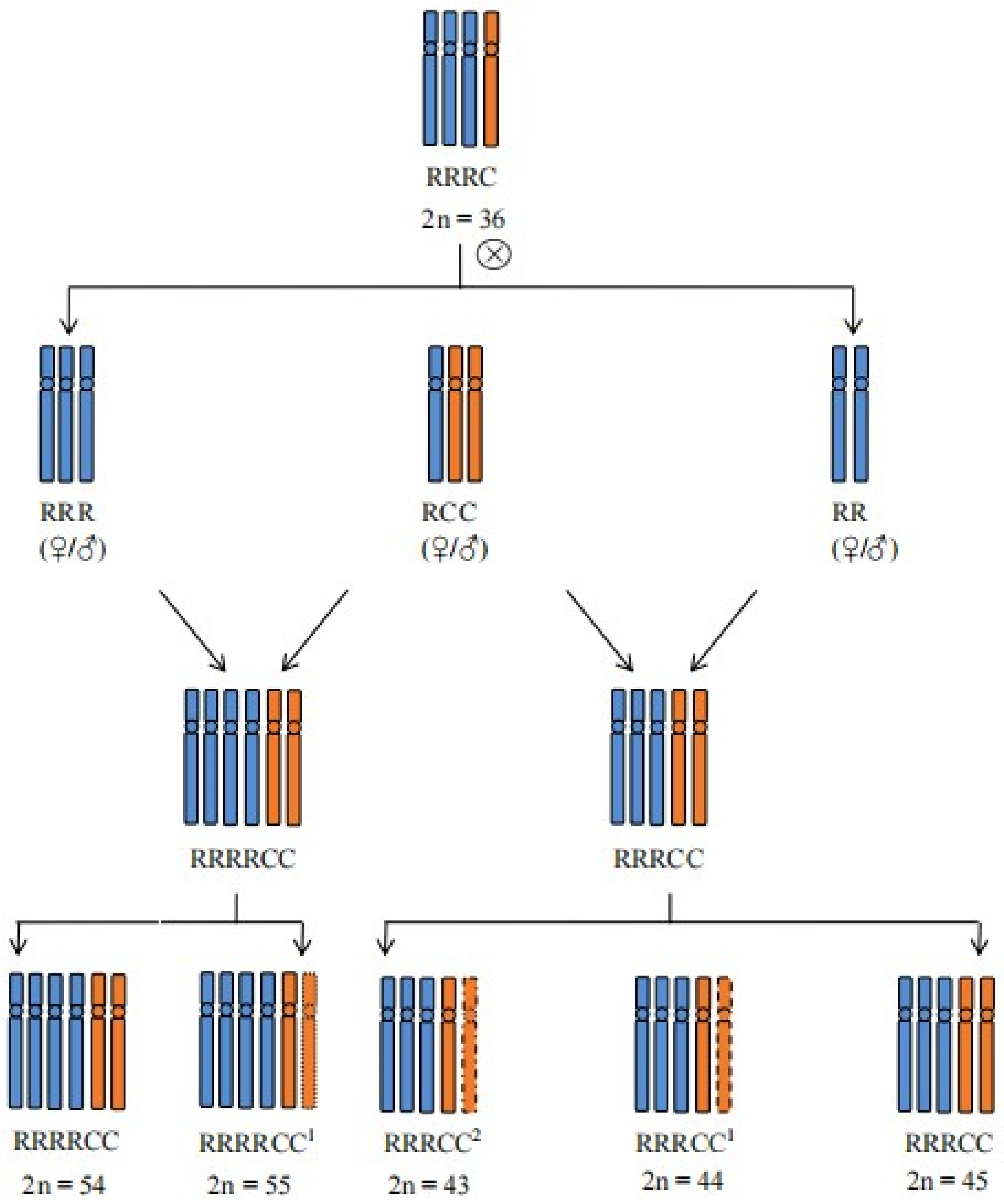
| Plant Types | Total Chromosome Number | Genome Configuration | Number of Plants |
|---|---|---|---|
| RRRCC2 | 43 | 27R + 16C | 4 |
| RRRCC1 | 44 | 27R + 17C | 5 |
| RRRCC | 45 | 27R + 18C | 5 |
| RRRRCC | 54 | 36R + 18C | 1 |
| RRRRCC1 | 55 | 36R + 19C | 1 |
| Total | - | - | 16 |
| Plant Types | Total PMCs | Ratio | Allosyndesis Types | Autosyndesis Types |
|---|---|---|---|---|
| RRRCC2 (c) | 55 | 69.09% | R-C; R-R-C; C-C-R; R-C-R; C-R-C; R-C-C-R; | C-C; C-C-C; R-R; R-R-R; |
| RRRCC1 (d) | 60 | 61.67% | R-C; R-R-C; C-C-R; R-C-R; C-R-C; R-C-C-R; C-C-C-R; C-C-R-R-R-C; | C-C; C-C-C; R-R; R-R-R; R-R-R-R; |
| RRRCC (e) | 63 | 58.73% | R-C; R-R-C; C-C-R; R-C-R; C-R-C; R-C-C-R; C-C-C-R; R-R-C-C-C-C; | C-C; C-C-C; C-C-C-C; R-R; R-R-R; R-R-R-R; |
| RRRRCC (b) | 66 | 74.24% | R-C; R-R-C; C-C-R; R-C-R; C-R-C; R-C-C-R; C-C-C-C-R; R-R-R-R-R-C; | C-C; C-C-C; C-C-C-C; R-R; R-R-R; R-R-R-R; |
| RRRRCC1 (a) | 60 | 78.33% | R-C; R-R-C; C-C-R; R-C-R; C-R-C; R-C-C-R; C-C-C-R; C-C-C-R-R; | C-C; C-C-C; R-R; R-R-R; R-R-R-R; |
| Study | Materials | Hybrid Types | Results |
|---|---|---|---|
| “Genome-wide unbalanced expression bias and expression level dominance toward Brassica oleracea in artificially synthesized intergeneric hybrids of Raphanobrassica” by Zhang et al. [44] | Raphanus sativus Brassica oleracea var. alboglabra Raphanobrassica | Raphanobrassica (2n = 36) | Transcriptome shock was observed in five tissues of Raphanobrassica. Genome-wide unbalanced biased expression and expression level dominance were also discovered, and both of them were toward B. oleracea in Raphanobrassica, which is consistent with the observed phenotypes. |
| “Identification, Characterization, and Cytological Analysis of Several Unexpected Hybrids Derived from Reciprocal Crosses between Raphanobrassica and Its Diploid Parents” by Yu et al. [35] | Raphanus sativus var. Loutouqing Brassica oleracea var. Chijielan Raphanobrassica | Several hybrids including RRC (2n = 27), CCR (2n = 27), RRRC (2n = 36), CCCR (2n = 36), and RRC’ (2n = 26) | Two expected hybrids (RRC and CCR) and three unexpected hybrids (RRRC, CCCR, and RRC’) were reported for the first time, and the study demonstrated that the unexpected hybrids exhibited significantly more intergenomic chromosome pairings than the expected hybrids. |
| Characteristics and Cytological Analysis of Several Novel Allopolyploids and Aneuploids between Brassica oleracea and Raphanus sativus by the authors of the present study | Raphanus sativus var. Loutouqing Brassica oleracea var. Chijielan Two hybrids RRRC and CCCR derived from Yu’s study | Two allopolyploids including RRRCC (2n = 45) and RRRRCC (2n = 54). Three anuepolyploids including RRRCC2 (2n = 43), RRRCC1 (2n = 44), and RRRRCC1 (2n = 55) | Two main genome configurations in these offspring plants, RRRCC (2n = 43, 44, 45) and RRRRCC (2n = 54, 55), were identified for the first time. It was discovered that the frequency of R and C genome pairing varied among different progenies at diakinesis and anaphase II. This observation elucidated the formation of these offspring plants, highlighting the presence of an unreduced single C genome for the first time. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, M.; Fang, S.; Wei, B.; Hu, Q.; Cai, M.; Zeng, T.; Gu, L.; Wang, H.; Du, X.; Zhu, B.; et al. Characteristics and Cytological Analysis of Several Novel Allopolyploids and Aneuploids between Brassica oleracea and Raphanus sativus. Int. J. Mol. Sci. 2024, 25, 8368. https://doi.org/10.3390/ijms25158368
Hu M, Fang S, Wei B, Hu Q, Cai M, Zeng T, Gu L, Wang H, Du X, Zhu B, et al. Characteristics and Cytological Analysis of Several Novel Allopolyploids and Aneuploids between Brassica oleracea and Raphanus sativus. International Journal of Molecular Sciences. 2024; 25(15):8368. https://doi.org/10.3390/ijms25158368
Chicago/Turabian StyleHu, Mingyang, Shiting Fang, Bo Wei, Qi Hu, Mengxian Cai, Tuo Zeng, Lei Gu, Hongcheng Wang, Xuye Du, Bin Zhu, and et al. 2024. "Characteristics and Cytological Analysis of Several Novel Allopolyploids and Aneuploids between Brassica oleracea and Raphanus sativus" International Journal of Molecular Sciences 25, no. 15: 8368. https://doi.org/10.3390/ijms25158368
APA StyleHu, M., Fang, S., Wei, B., Hu, Q., Cai, M., Zeng, T., Gu, L., Wang, H., Du, X., Zhu, B., & Ou, J. (2024). Characteristics and Cytological Analysis of Several Novel Allopolyploids and Aneuploids between Brassica oleracea and Raphanus sativus. International Journal of Molecular Sciences, 25(15), 8368. https://doi.org/10.3390/ijms25158368







