Quantitative Proteomic Analysis Reveals Key Proteins Involved in Testicular Development of Yaks
Abstract
:1. Introduction
2. Results
2.1. Testicular Development and Histology
2.2. Comparison of Protein Profiles
2.3. GO Analysis of DEPs
2.4. KEGG Analysis of DEPs
2.5. Validation of TMT Data for Selected Proteins by PRM
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Animals and Sample Preparation
4.3. Testicular Histomorphology
4.4. TMT Proteomic Analysis
4.5. Protein Functional Annotation and Enrichment Analysis
4.6. Protein Validation by 4D-PRM
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Cai, X.; Mipam, T.D.; Zhao, F.F.; Sun, L. SNPs detected in the yak gene and their association with growth traits. Animal 2015, 9, 1097–1103. [Google Scholar] [CrossRef] [PubMed]
- Ruan, C.M.; Wang, J.; Yang, Y.X.; Hu, J.J.; Ma, Y.J.; Zhang, Y.; Zhao, X.X. Proteomic analysis of Tianzhu White Yak (Bos grunniens) testis at different sexual developmental stages. Anim. Sci. J. 2019, 90, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Yan, P.; Xiang, L.; Guo, X.; Bao, P.J.; Jin, S.; Wu, X.Y. The low expression of Dmrt7 is associated with spermatogenic arrest in cattle-yak. Mol. Biol. Rep. 2014, 41, 7255–7263. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.W.; Gong, J.S.; Wang, X.Y.; Wu, X.H.; Li, Y.L.; Ma, Y.J.; Zhang, Y.; Zhao, X.X. Molecular cloning, bioinformatics analysis and expression of insulin-like growth factor 2 from Tianzhu White yak. Int. J. Mol. Sci. 2014, 15, 504–524. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Wei, D.Q.; Wang, K.; Wu, D.; Zhang, J.J.; Che, L.Q.; Xu, S.Y.; Fang, Z.F.; Feng, B.; Li, J.; et al. Proteomic analysis reveals key proteins involved in arginine promotion of testicular development in boars. Theriogenology 2020, 154, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Hassine, A.B.; Basly, M.; Kraoua, L.; Arfaoui, R.; Rachdi, R. Testis development in the absence of SRY: Chromosomal rearrangements at SOX9 and SOX3. Hum. Reprod. 2015, 30, 127. [Google Scholar]
- Bhattacharya, I.; Dey, S. Emerging concepts on Leydig cell development in fetal and adult testis. Front. Endocrinol. 2023, 13, 1086276. [Google Scholar] [CrossRef] [PubMed]
- Johnson, P.F.; McKnight, S.L. Eukaryotic transcriptional regulatory proteins. Annu. Rev. Biochem. 1989, 58, 799–839. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.S.; Du, J.; Han, J.B.; Bao, X.B.; Song, X.R.; Lu, Z.C. Proteomics reveals the preliminary physiological states of the spotted seal (Phoca largha) pups. Sci. Rep. 2020, 10, 18727. [Google Scholar] [CrossRef]
- Ma, C.; Wang, W.W.; Wang, Y.D.; Sun, Y.; Kang, L.; Zhang, Q.; Jiang, Y.L. TMT-labeled quantitative proteomic analyses on the to identify the proteins underlying intramuscular fat content in pigs. J. Proteom. 2020, 213, 103630. [Google Scholar] [CrossRef]
- Xue, Y.P.; Xu, P.F.; Xu, S.J.; Xue, K.; Xu, L.L.; Chen, J.; Xu, J.; Shi, X.Y.; Li, Q.; Gu, L. Peptidomic analysis of endometrial tissue from patients with ovarian endometriosis. Cell Physiol. Biochem. 2018, 47, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Pei, S.W.; Luo, J.; Weng, X.X.; Xu, Y.L.; Bai, J.J.; Li, F.D.; Li, W.H.; Yue, X.P. iTRAQ-based proteomic analysis provides novel insight into the postnatal testicular development of Hu sheep. J. Proteom. 2023, 286, 104956. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, M.A.; Judycka, S.; Zarski, D.; Malinowska, A.; Swiderska, B.; Palinska-Zarska, K.; Zejewski, M.B.; Ciereszko, A. Proteomic analysis of pikeperch seminal plasma provides novel insight into the testicular development of domesticated fish stocks. Animal 2021, 15, 100279. [Google Scholar] [CrossRef] [PubMed]
- Coen, S.; Keogh, K.; Lonergan, P.; Fair, S.; Kenny, D.A. Early life nutrition affects the molecular ontogeny of testicular development in the young bull calf. Sci. Rep. 2023, 13, 6748. [Google Scholar] [CrossRef] [PubMed]
- Beyret, E.; Lin, H.F. Pinpointing the expression of piRNAs and function of the PIWI protein subfamily during spermatogenesis in the mouse. Dev. Biol. 2011, 355, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Durlej, M.; Knapczyk-Stwora, K.; Slomczynska, M. Prenatal and neonatal flutamide administration increases proliferation and reduces apoptosis in large antral follicles of adult pigs. Anim. Reprod. Sci. 2012, 132, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Li, T.T.; Wang, H.H.; Luo, R.R.; An, X.J.; Li, Q.; Su, M.C.; Shi, H.B.; Chen, H.L.; Zhang, Y.; Ma, Y.J. Proteome informatics in Tibetan sheep (Ovis aries) testes suggest the crucial proteins related to development and functionality. Front. Vet. Sci. 2022, 9, 923789. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Mipam, T.D.; Zhao, F.; Liu, W.; Zhao, W.; Wu, S.; Xu, C.; Yu, S.; Cai, X. Comparative testis proteome of cattleyak from different developmental stages. Animal 2017, 11, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Pontelo, T.P.; Miranda, J.R.; Felix, M.A.R.; Pereira, B.A.; Silva, W.E.; Avelar, G.F.; Mariano, F.C.M.Q.; Guimaraes, G.C.; Zangeronimo, M.G. Histological characteristics of the gonads of pig fetuses and their relationship with fetal anatomical measurements. Res. Vet. Sci. 2018, 117, 28–36. [Google Scholar] [CrossRef]
- Kalwar, Q.; Chu, M.; Ahmad, A.A.; Ding, X.Z.; Wu, X.Y.; Bao, P.J.; Yan, P. Morphometric evaluation of spermatogenic cells and seminiferous tubules and exploration of luteinizing hormone beta polypeptide in testis of Datong yak. Animals 2020, 10, 66. [Google Scholar] [CrossRef]
- Faucette, A.N.; Maher, V.A.; Gutierrez, M.A.; Jucker, J.M.; Yates, D.C.; Welsh, T.H.; Amstalden, M.; Newton, G.R.; Nuti, L.C.; Forrest, D.W.; et al. Temporal changes in histomorphology and gene expression in goat testes during postnatal development. J. Anim. Sci. 2014, 92, 4440–4448. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Kuriwaki, R.; Hagino, S.; Shimazaki, M.; Sambuu, R.; Hirata, M.; Tanihara, F.; Takagi, M.; Taniguchi, M.; Otoi, T. Abnormal functions of Leydig cells in crossbred cattle-yak showing infertility. Reprod. Domest. Anim. 2020, 55, 209–216. [Google Scholar] [CrossRef]
- Endo, S.; Yoshitake, H.; Tsukamoto, H.; Matsuura, H.; Kato, K.; Sakuraba, M.; Takamori, K.; Fujiwara, H.; Takeda, S.; Araki, Y. TEX101, a glycoprotein essential for sperm fertility, is required for stable expression of Ly6k on testicular germ cells. Sci. Rep. 2016, 6, 23616. [Google Scholar] [CrossRef]
- Fujihara, Y.; Okabe, M.; Ikawa, M. GPI-anchored protein complex, LY6K/TEX101, is required for sperm migration into the oviduct and male fertility in mice. Biol. Reprod. 2014, 90, 60. [Google Scholar] [CrossRef]
- Fujihara, Y.; Tokuhiro, K.; Muro, Y.; Kondoh, G.; Araki, Y.; Ikawa, M.; Okabe, M. Expression of TEX101, regulated by ACE, is essential for the production of fertile mouse spermatozoa. Proc. Natl. Acad. Sci. USA 2013, 110, 8111–8116. [Google Scholar] [CrossRef]
- Li, M.Y.; Chen, Y.X.; Ou, J.P.; Huang, J.J.; Zhang, X.Y. PDCL2 is essential for spermiogenesis and male fertility in mice. Cell Death Discov. 2022, 8, 419. [Google Scholar] [CrossRef]
- Aisha, J.; Yenugu, S. Characterization of SPINK2, SPACA7 and PDCL2: Effect of immunization on fecundity, sperm function and testicular transcriptome. Reprod. Biol. 2022, 23, 100711. [Google Scholar] [CrossRef] [PubMed]
- Fujihara, Y.; Kobayashi, K.; Abbasi, F.; Endo, T.; Yu, Z.F.; Ikawa, M.; Matzuk, M.M. PDCL2 is essential for sperm acrosome formation and male fertility in mice. Andrology 2023, 11, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Schilit, S.L.P.; Menon, S.; Friedrich, C.; Kammin, T.; Wilch, E.; Hanscom, C.; Jiang, S.Z.; Kliesch, S.; Talkowski, M.E.; Tüttelmann, F.; et al. Translocation- mediated dysregulation and frameshift variants cause human male infertility. Am. J. Hum. Genet. 2020, 106, 41–57. [Google Scholar] [CrossRef]
- Sun, J.; Lin, Y.; Wu, J. Long non-coding RNA expression profiling of mouse testis during postnatal development. PLoS ONE 2013, 8, e75750. [Google Scholar] [CrossRef]
- Aarabi, M.; Modarressi, M.H.; Soltanghoraee, H.; Behjati, R.; Amirjannati, N.; Akhondi, M.M. Testicular expression of synaptonemal complex protein 3 (SYCP3) messenger ribonucleic acid in 110 patients with nonobstructive azoospermia. Fertil. Steril. 2006, 86, 325–331. [Google Scholar] [CrossRef] [PubMed]
- He, Z.P.; Feng, L.X.; Zhang, X.D.; Geng, Y.X.; Parodi, D.A.; Suarez-Quian, C.; Dym, M. Expression of Col1a1, Col1a2 and procollagen I in germ cells of immature and adult mouse testis. Reproduction 2005, 130, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Urriola-Muñoz, P.; Lizama, C.; Lagos-Cabré, R.; Reyes, J.G.; Moreno, R.D. Differential expression and localization of ADAM10 and ADAM17 during rat spermatogenesis suggest a role in germ cell differentiation. Biol. Res. 2014, 47, 31. [Google Scholar] [CrossRef] [PubMed]
- Tabara, M.; Shiraishi, K.; Takii, R.; Fujimoto, M.; Nakai, A.; Matsuyama, H. Testicular localization of activating transcription factor 1 and its potential function during spermatogenesis. Biol. Reprod. 2021, 105, 976–986. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.D.; He, Z.; Yu, C.X.; Guan, Q.B. Role of p38 MAPK signalling in testis development and male fertility. Oxidative Med. Cell Longev. 2022, 2022, 6891897. [Google Scholar] [CrossRef] [PubMed]
- Ni, F.D.; Hao, S.L.; Yang, W.X. Multiple signaling pathways in Sertoli cells: Recent findings in spermatogenesis. Cell Death Dis. 2019, 10, 541. [Google Scholar] [CrossRef] [PubMed]
- Chennathukuzhi, V.; Morales, C.R.; El-Alfy, M.; Hecht, N.B. The kinesin KIF17b and RNA-binding protein TB-RBP transport specific cAMP-responsive element modulator-regulated mRNAs in male germ cells. Proc. Natl. Acad. Sci. USA 2003, 100, 15566–15571. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.W.; Luo, X.; Liu, Y.L.; Jiang, Q.; Qian, X.M.; Guo, Z.Y. R171H missense mutation of in a patient with spermatogenic failure. Eur. J. Med. Genet. 2011, 54, E455–E457. [Google Scholar] [CrossRef] [PubMed]
- Burnicka-Turek, O.; Shirneshan, K.; Paprotta, I.; Grzmil, P.; Meinhardt, A.; Engel, W.; Adham, I.M. Inactivation of insulin-like factor 6 disrupts the progression of spermatogenesis at late meiotic prophase. Endocrinology 2009, 150, 4348–4357. [Google Scholar] [CrossRef]
- Umehara, T.; Kida, S.; Hasegawa, S.; Fujimoto, H.; Horikoshi, M. Restricted expression of a member of the transcription elongation factor S-II family in testicular germ cells during and after meiosis. J. Biochem. 1997, 121, 598–603. [Google Scholar] [CrossRef]
- Ito, T.; Xu, Q.H.; Takeuchi, H.; Kubo, T.; Natori, S. Spermatocyte-specific expression of the gene for mouse testis-specific transcription elongation factor S-II. Febs. Lett. 1996, 385, 21–24. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Seldin, M.F.; Taketo, M.M.; Kubo, T.; Natori, S. Gene structure and chromosome mapping of mouse transcription elongation factor S-II (Tcea1). Gene 2000, 244, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.Y.; Lv, M.; Luo, B.H.; Zhao, S.Z.; Mo, Z.C.; Xie, Y.J. The role of the PI3K/AKT/mTOR signalling pathway in male reproduction. Curr. Mol. Med. 2021, 21, 539–548. [Google Scholar] [PubMed]
- Dong, W.L.; Tan, F.Q.; Yang, W.X. Wnt signaling in testis development: Unnecessary or essential? Gene 2015, 565, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Gao, F.; Guillou, F.; Taketo, M.M.; Huff, V.; Behringer, R.R. Negatively regulates β-catenin signaling during testis development. Development 2008, 135, 1875–1885. [Google Scholar] [CrossRef] [PubMed]
- Correia, B.; Sousa, M.I.; Ramalho-Santos, J. The mTOR pathway in reproduction: From gonadal function to developmental coordination. Reproduction 2020, 159, R173–R188. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Wang, X.; Sun, Y.; Lu, X.; Jiang, W.; Chen, Z.; Huang, Y.; Chi, P. TMT-labelled quantitative proteomic analysis to identify the proteins underlying radiation-induced colorectal fibrosis in rats. J. Proteom. 2020, 223, 103801. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Liu, X.; Peng, Y.; Zhan, R.; Xu, H.; Ge, X. Comparative analysis of methicillin-sensitive and resistant Staphylococcus aureus exposed to emodin based on proteomic profiling. Biochem. Biophys. Res. Commun. 2017, 494, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef]
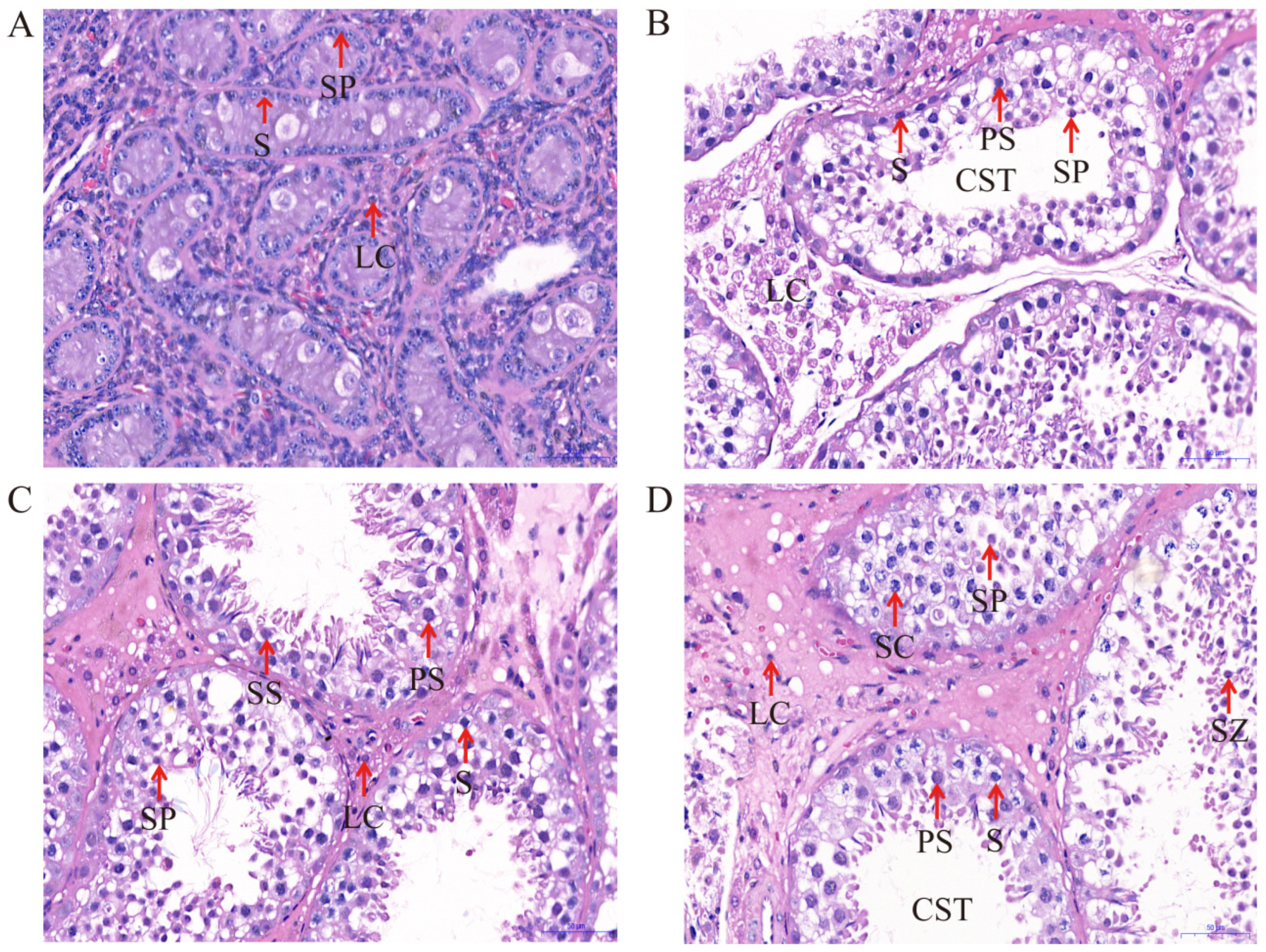
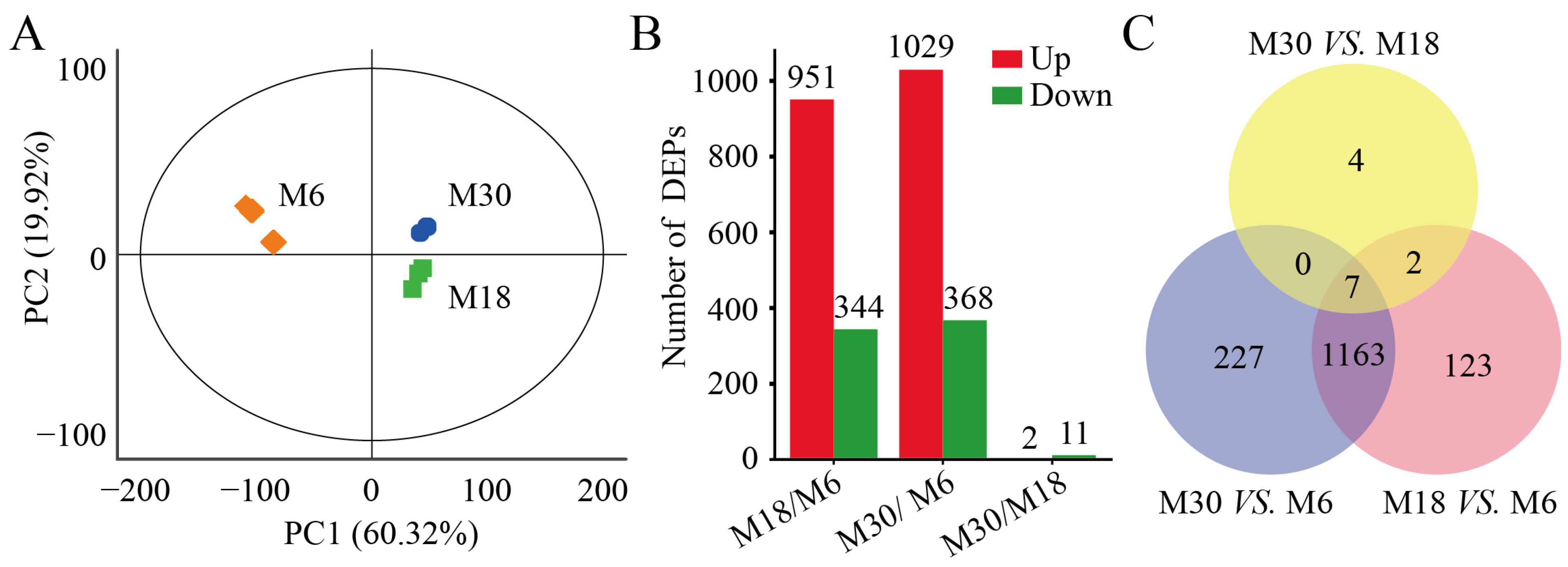
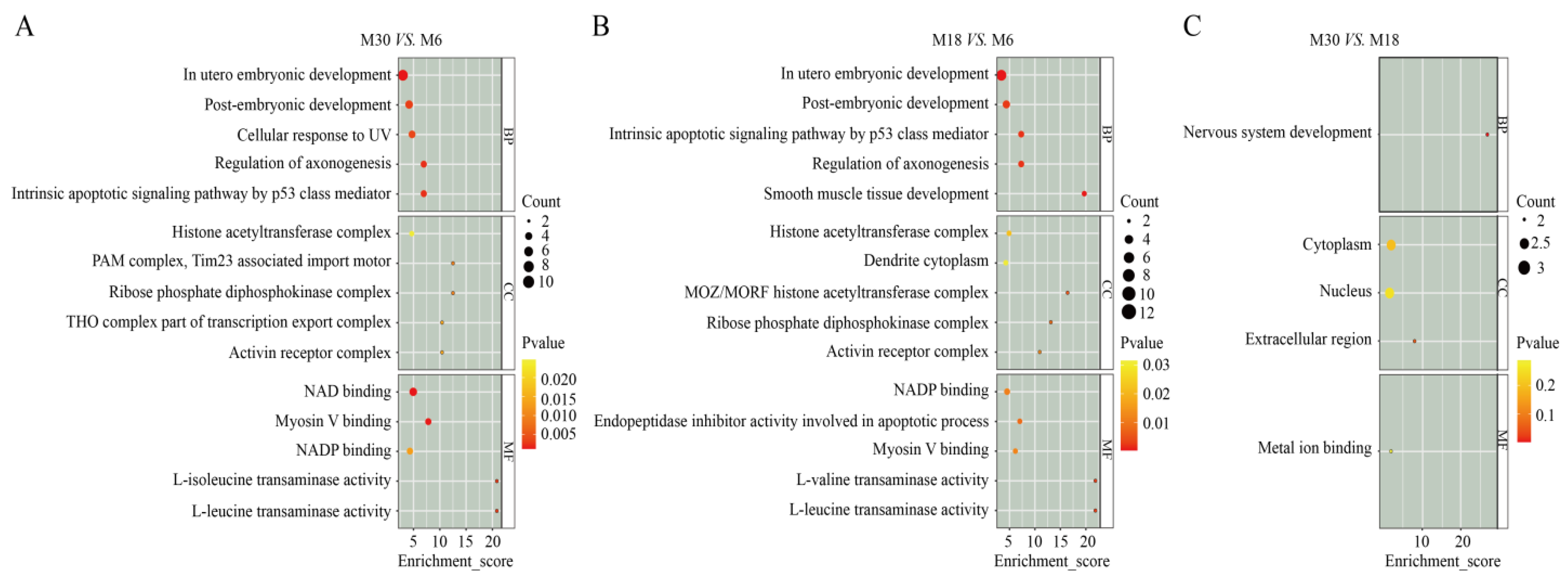
| Types | Months | Rete Tubules | Spermatogonia | Spermatocytes | Sertoli Cells | Leydig Cells |
|---|---|---|---|---|---|---|
| Short diameter | 6 | 43.75 ± 2.343 a | 4.02 ± 0.522 | 6.31 ± 0.891 | 3.17 ± 0.399 a | 4.37 ± 0.578 |
| 18 | 56.30 ± 6.151 b | 3.86 ± 0.300 | 5.40 ± 0.415 | 2.97 ± 0.562 b | 4.47 ± 1.148 | |
| 30 | 140.16 ± 11.678 a | 4.08 ± 0.548 | 5.63 ± 0.715 | 5.01 ± 0.449 c | 4.14 ± 0.791 | |
| 72 | 167.564 ± 8.980 a | 4.57 ± 0.839 | 5.58 ± 1.126 | 4.67 ± 0.729 d | 8.47 ± 0.942 | |
| Long diameter | 6 | 57.76 ± 8.299 a | 4.96 ± 0.405 | 6.46 ± 0.686 | 7.33 ± 0.833 a | 5.40 ± 1.235 |
| 18 | 81.63 ± 6.285 b | 4.78 ± 0.911 | 6.85 ± 0.639 | 5.57 ± 0.562 b | 6.04 ± 1.537 | |
| 30 | 226.62 ± 47.394 a | 5.57 ± 0.716 | 6.59 ± 0.449 | 7.97 ± 0.341 c | 7.31 ± 2.459 | |
| 72 | 209.84 ± 26.630 a | 5.59 ± 0.321 | 7.02 ± 1.357 | 7.07 ± 0.939 c | 6.23 ± 0.754 |
| Map_ID | Map_Name | Test_Seq |
|---|---|---|
| ko04010 | MAPK signaling pathway | KIF17, INSL6, FAM136A, ERK2, JNK3, SHCBP1L, REEP6, ZNF474, MCM5, LACTB2, TCEA2, YWHAQ, PRAK |
| ko04151 | PI3K-Akt signaling pathway | ACRBP, SEPTIN2, PRO768, GPR161, BAG6, FAM71B, ERK2, TMEM5 |
| ko04910 | Insulin signaling pathway | KIF17, RALGPS2, CCT2, GPR161, CCDC167, JNK3, ERK2, IRF2BPL |
| ko04310 | Wnt signaling pathway | GAR1, CFL2, JNK3, LTF, GSTO1, R3HCC1, PEDF, NHLRC3 |
| ko04931 | Insulin resistance | SEPTIN2, CCDC167, TUBG2, JNK3, IRF2BPL, RAB3D |
| ko03010 | Ribosome | MACROH2A1, FAM175B, PDIA5, SNU13, TUSC3, ACE, KIF5A, CCNL2 |
| ko04150 | mTOR signaling pathway | KIF17, GAR1, GPR161, R3HCC1, FEM1A, ERK2 |
| ko04350 | TGF-beta signaling pathway | UBAP2, TBC1D23, GFPT2, ERK2, YWHAQ |
| ko04140 | Autophagy-animal | GPR161, JNK3, TRIM36, RNF146, ERK2 |
| ko03008 | Ribosome biogenesis in eukaryotes | CALD1, STK33, FAM32A, DAZAP1 |
| ko01522 | Endocrine resistance | KIF17, LIN7C, JNK3, ERK2 |
| ko04152 | AMPK signaling pathway | SEPTIN2, IRF2BPL, GPR161, CCDC167 |
| ko00520 | Amino sugar and nucleotide sugar metabolism | DNAJB1, RALGPS2, ESX1 |
| ko04662 | B cell receptor signaling pathway | ERK2, GTSF1, FAM136A |
| ko04371 | Apelin signaling pathway | ERK2, YWHAQ |
| ko00100 | Steroid biosynthesis | FBXW9 |
| Description (NCBI) | Gene Name | Fold Change (TMT) | Fold Change (4D-PRM) |
|---|---|---|---|
| Acrosin binding protein | ACRBP | 4.36 | 1.89 |
| Retinoic acid receptor responder 2 | RARRES2 | 4.54 | 2.04 |
| Synaptotagmin like 2 | SYTL2 | 5.03 | 2.13 |
| LYR motif containing 1 | LYRM1 | 5.26 | 2.07 |
| Protein disulfide isomerase like, testis expressed | PDILT | 3.89 | 1.68 |
| Phosducin like 2 | PDCL2 | 3.79 | 1.37 |
| Tyrosylprotein sulfotransferase 2 | TPST2 | 0.32 | 0.62 |
| ADAM metallopeptidase domain 10 | ADAM10 | 0.50 | 0.44 |
| STAG3 cohesin complex component | STAG3 | 0.45 | 0.29 |
| Storkhead box 1 | STOX1 | 0.24 | 0.27 |
| Fascin actin-bundling protein 1 | FSCN1 | 0.39 | 0.08 |
| Mitogen-activated protein kinase kinase 1 | MAP2K1 | 0.51 | 0.38 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
La, Y.; Ma, X.; Bao, P.; Chu, M.; Yan, P.; Guo, X.; Liang, C. Quantitative Proteomic Analysis Reveals Key Proteins Involved in Testicular Development of Yaks. Int. J. Mol. Sci. 2024, 25, 8433. https://doi.org/10.3390/ijms25158433
La Y, Ma X, Bao P, Chu M, Yan P, Guo X, Liang C. Quantitative Proteomic Analysis Reveals Key Proteins Involved in Testicular Development of Yaks. International Journal of Molecular Sciences. 2024; 25(15):8433. https://doi.org/10.3390/ijms25158433
Chicago/Turabian StyleLa, Yongfu, Xiaoming Ma, Pengjia Bao, Min Chu, Ping Yan, Xian Guo, and Chunnian Liang. 2024. "Quantitative Proteomic Analysis Reveals Key Proteins Involved in Testicular Development of Yaks" International Journal of Molecular Sciences 25, no. 15: 8433. https://doi.org/10.3390/ijms25158433
APA StyleLa, Y., Ma, X., Bao, P., Chu, M., Yan, P., Guo, X., & Liang, C. (2024). Quantitative Proteomic Analysis Reveals Key Proteins Involved in Testicular Development of Yaks. International Journal of Molecular Sciences, 25(15), 8433. https://doi.org/10.3390/ijms25158433






