Neutrophil-like Monocytes Increase in Patients with Colon Cancer and Induce Dysfunctional TIGIT+ NK Cells
Abstract
1. Introduction
2. Results
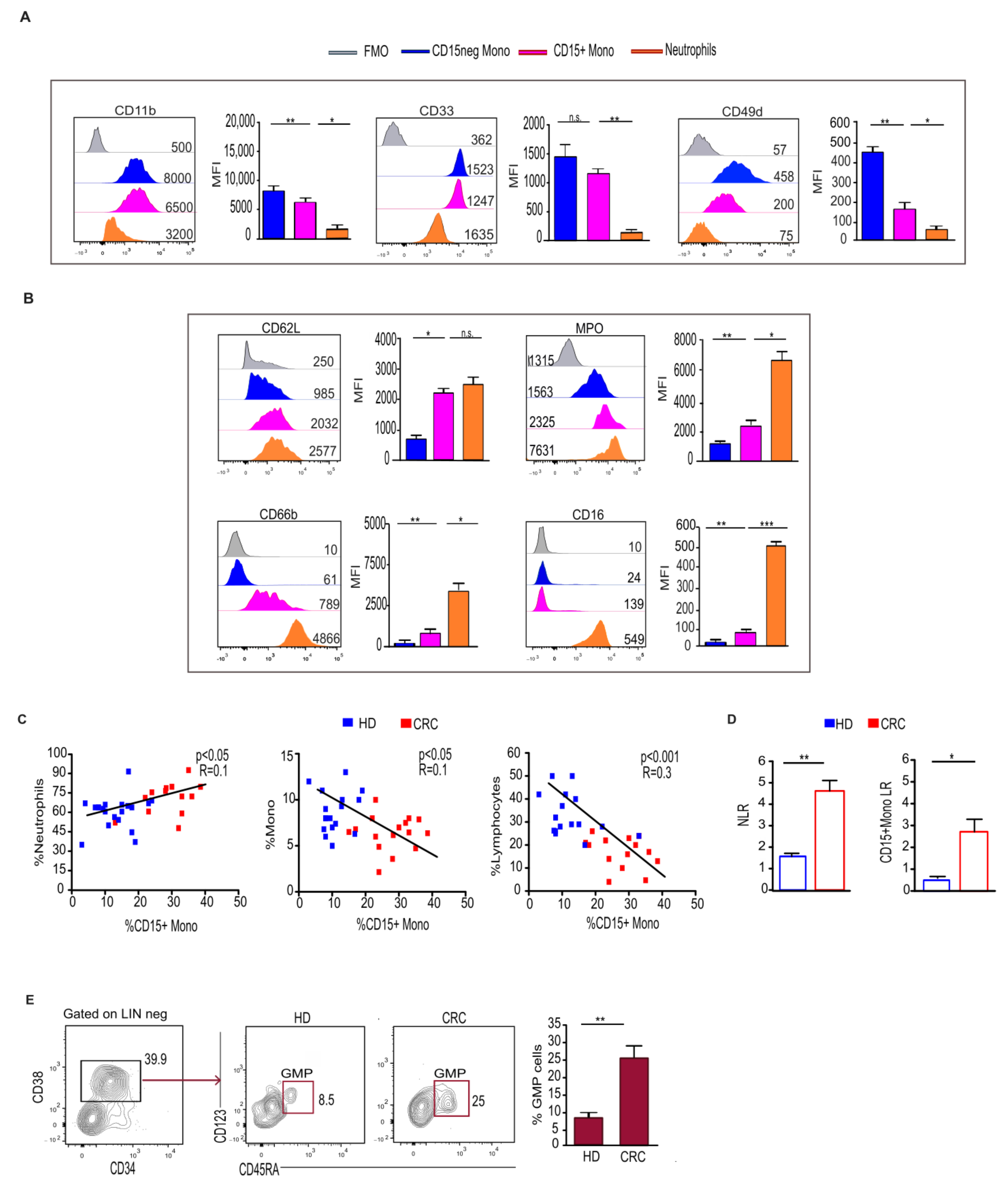
2.1. Monocytes Expressing the Granulocytic Marker CD15 Increase in the Blood and Tumor Tissue of CRC Patients
2.2. CD15+ Monocytes Display Neutrophil-like Features and Are Associated with a Higher Frequency of Circulating Granulocyte–Monocyte Precursors
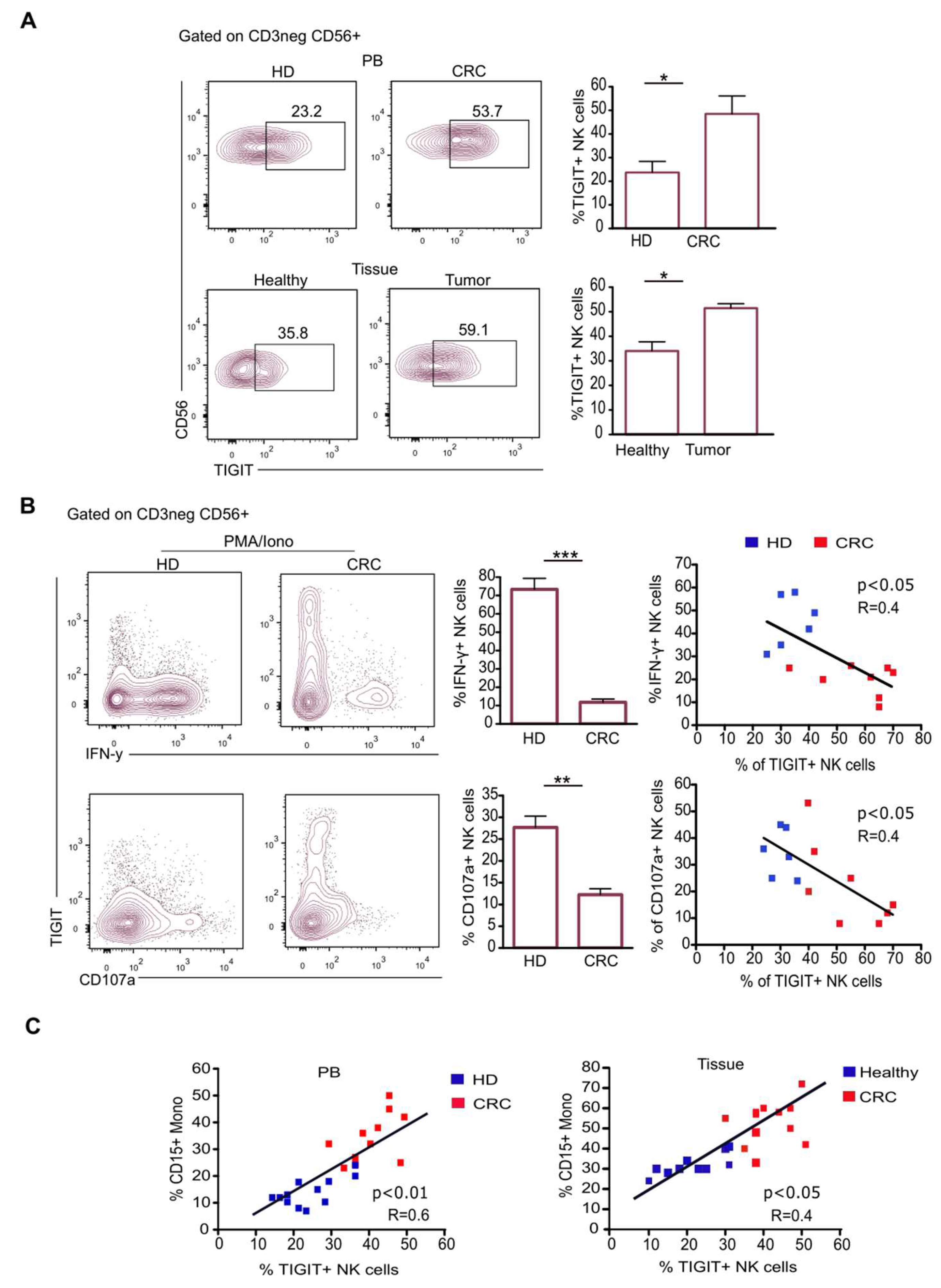
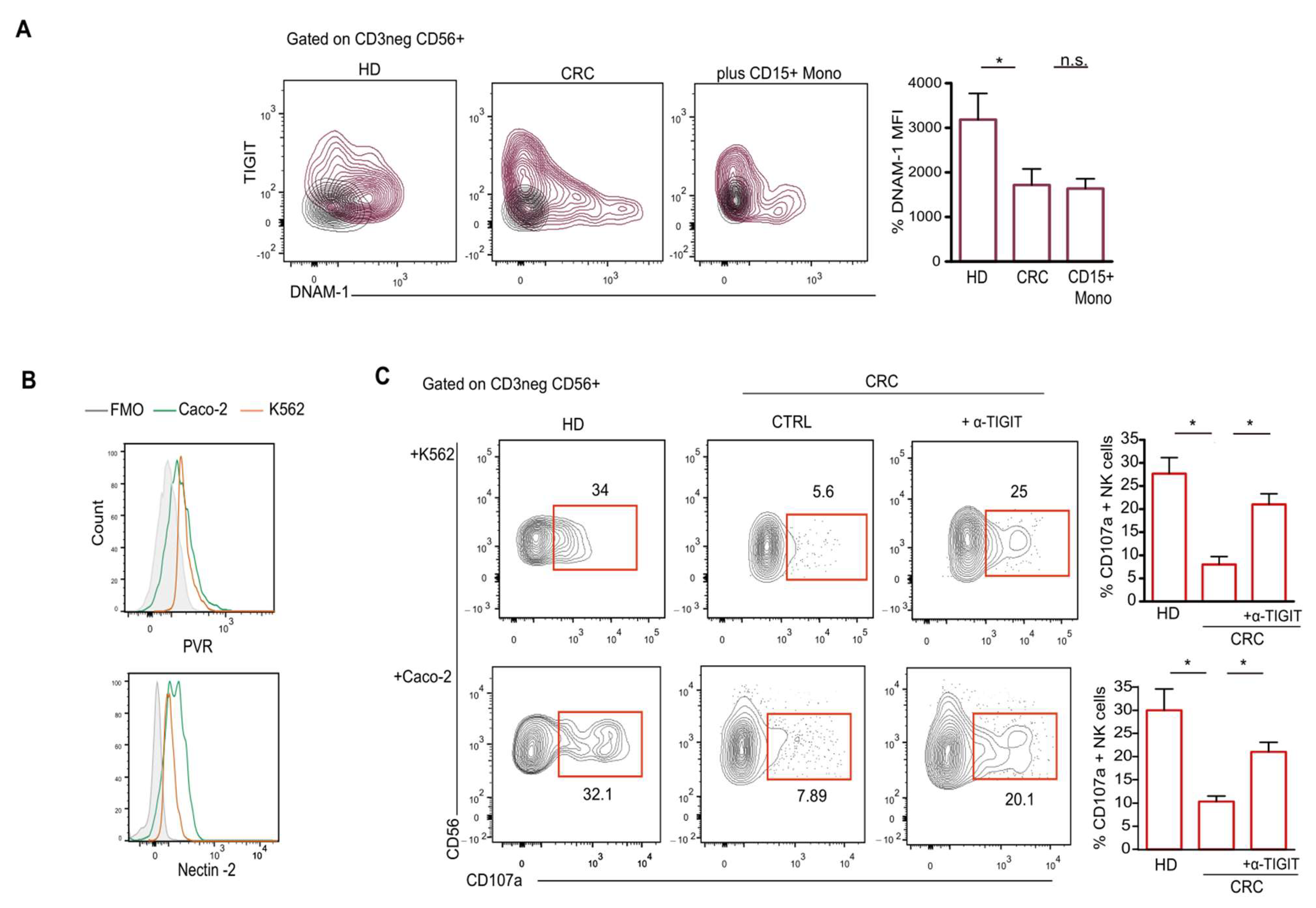
2.3. CD15+ Monocytes Inhibit NK Cell Activity via TIGIT Induction and NKp30 Engagement
2.4. TIGIT Expression on NK Cells Reduces Their Antitumor Ability
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Cell Isolation and Phenotypical Analysis
4.3. Cell Culture Assay
4.4. Imaging Flow Cytometry
4.5. Enzyme-Linked Immunosorbent Assay
4.6. Flow Cytometry
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ghorani, E.; Swanton, C.; Quezada, S.A. Cancer cell-intrinsic mechanisms driving acquired immune tolerance. Immunity 2023, 56, 2270–2295. [Google Scholar] [CrossRef] [PubMed]
- Zou, W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat. Rev. Cancer 2005, 5, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Zou, W. Immune regulation in the tumor microenvironment and its relevance in cancer therapy. Cell. Mol. Immunol. 2022, 19, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Lasser, S.A.; Ozbay Kurt, F.G.; Arkhypov, I.; Utikal, J.; Umansky, V. Myeloid-derived suppressor cells in cancer and cancer therapy. Nat. Rev. Clin. Oncol. 2024, 21, 147–164. [Google Scholar] [CrossRef] [PubMed]
- Corzo, C.A.; Cotter, M.J.; Cheng, P.; Cheng, F.; Kusmartsev, S.; Sotomayor, E.; Padhya, T.; McCaffrey, T.V.; McCaffrey, J.C.; Gabrilovich, D.I. Mechanism regulating reactive oxygen species in tumor-induced myeloid-derived suppressor cells. J. Immunol. 2009, 182, 5693–5701. [Google Scholar] [CrossRef] [PubMed]
- Yaseen, M.M.; Abuharfeil, N.M.; Darmani, H.; Daoud, A. Mechanisms of Immune Suppression by Myeloid-Derived Suppressor Cells: The Role of Interleukin-10 as a Key Immunoregulatory Cytokine. Open Biol. 2020, 10, 200111. [Google Scholar] [CrossRef]
- Wrzesinski, S.H.; Wan, Y.Y.; Flavell, R.A. Transforming growth factor-β and the immune response: Implications for anticancer therapy. Clin. Cancer Res. 2007, 13, 5262–5270. [Google Scholar] [CrossRef] [PubMed]
- Raber, P.; Ochoa, A.C.; Rodriguez, P.C. Metabolism of L-arginine by myeloid-derived suppressor cells in cancer: Mechanisms of T cell suppression and therapeutic perspectives. Immunol. Investig. 2012, 41, 614–634. [Google Scholar] [CrossRef]
- Kumar, V.; Patel, S.; Tcyganov, E.; Gabrilovich, D.I. The Nature of Myeloid-Derived Suppressor Cells in the Tumor Microenvironment. Trends Immunol. 2016, 37, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Bronte, V.; Brandau, S.; Chen, S.H.; Colombo, M.P.; Frey, A.B.; Greten, T.F.; Mandruzzato, S.; Murray, P.J.; Ochoa, A.; Ostrand-Rosenberg, S.; et al. Recommendations for Myeloid-Derived Suppressor Cell Nomenclature and Characterization Standards. Nat. Commun. 2016, 7, 12150. [Google Scholar] [CrossRef]
- Gabrilovich, D.I.; Nagaraj, S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009, 9, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Serafini, P.; Borrello, I.; Bronte, V. Myeloid suppressor cells in cancer: Recruitment, phenotype, properties, and mechanisms of immune suppression. Semin. Cancer Biol. 2006, 16, 53–65. [Google Scholar] [CrossRef]
- Cassetta, L.; Bruderek, K.; Skrzeczynska-Moncznik, J.; Osiecka, O.; Hu, X.; Rundgren, I.M.; Lin, A.; Santegoets, K.; Horzum, U. Differential expansion of circulating human MDSC subsets in patients with cancer, infection and inflammation. J. Immunother. Cancer 2020, 8, e001223. [Google Scholar] [CrossRef]
- Vetsika, E.K.; Koinis, F.; Gioulbasani, M.; Aggouraki, D.; Koutoulaki, A.; Skalidaki, E.; Mavroudis, D.; Georgoulias, V.; Kotsakis, A. A circulating subpopulation of monocytic myeloid-derived suppressor cells as an independent prognostic/predictive factor in untreated non-small lung cancer patients. J. Immunol. Res. 2014, 2014, 659294. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.H.; Benner, B.; Savardekar, H.; Lapurga, G.; Good, L.; Abood, D.; Nagle, E.; Duggan, M.; Stiff, A.; DiVincenzo, M.J.; et al. Effect of Immune Checkpoint Blockade on Myeloid-Derived Suppressor Cell Populations in Patients with Melanoma. Front. Immunol. 2021, 12, 740890. [Google Scholar] [CrossRef] [PubMed]
- Horzum, U.; Yoyen-Ermis, D.; Taskiran, E.Z.; Yilmaz, K.B.; Hamaloglu, E.; Karakoc, D.; Esendagli, G. CD66b+ monocytes represent a proinflammatory myeloid subpopulation in cancer. Cancer Immunol. Immunother. 2021, 70, 75–87. [Google Scholar] [CrossRef]
- Mastio, J.; Condamine, T.; Dominguez, G.; Kossenkov, A.V.; Donthireddy, L.; Veglia, F.; Lin, C.; Wang, F.; Fu, S.; Zhou, J.; et al. Identification of monocyte-like precursors of granulocytes in cancer as a mechanism for accumulation of PMN-MDSCs. J. Exp. Med. 2019, 216, 2150–2169. [Google Scholar] [CrossRef]
- Wu, W.C.; Sun, H.W.; Chen, H.T.; Liang, J.; Yu, X.J.; Wu, C.; Wang, Z.; Zheng, L. Circulating hematopoietic stem and progenitor cells are myeloid-biased in cancer patients. Proc. Natl. Acad. Sci. USA 2014, 111, 4221–4226. [Google Scholar] [CrossRef]
- Gorczyca, W.; Sun, Z.Y.; Cronin, W.; Li, X.; Mau, S.; Tugulea, S. Immunophenotypic pattern of myeloid populations by flow cytometry analysis. Methods Cell Biol. 2011, 103, 221–266. [Google Scholar]
- Puig-Kröger, A.; Sanz-Rodríguez, F.; Longo, N.; Sánchez-Mateos, P.; Botella, L.; Teixidó, J.; Bernabéu, C.; Corbí, A.L. Maturation-dependent expression and function of the CD49d integrin on monocyte-derived human dendritic cells. J. Immunol. 2000, 165, 4338–4345. [Google Scholar] [CrossRef]
- Elghetany, M.T. Surface antigen changes during normal neutrophilic development: A critical review. Blood Cells Mol. Dis. 2002, 28, 260–274. [Google Scholar] [CrossRef] [PubMed]
- Mackey, J.B.G.; Coffelt, S.B.; Carlin, L.M. Neutrophil Maturity in Cancer. Front. Immunol. 2019, 10, 1912. [Google Scholar] [CrossRef] [PubMed]
- Ledderose, C.; Hashiguchi, N.; Valsami, E.A.; Rusu, C.; Junger, W.G. Optimized flow cytometry assays to monitor neutrophil activation in human and mouse whole blood samples. J. Immunol. Methods 2023, 512, 113403. [Google Scholar] [CrossRef]
- Ito, Y.; Nakahara, F.; Kagoya, Y.; Kurokawa, M. CD62L expression level determines the cell fate of myeloid progenitors. Stem Cell Rep. 2021, 16, 2871–2886. [Google Scholar] [CrossRef]
- Nauseef, W.M. Myeloperoxidase in human neutrophil host defence. Cell. Microbiol. 2014, 16, 1146–1155. [Google Scholar] [CrossRef]
- Edberg, J.C.; Salmon, J.E.; Kimberly, R.P. Functional capacity of Fc gamma receptor III (CD16) on human neutrophils. Immunol. Res. 1992, 11, 239–251. [Google Scholar] [CrossRef]
- Zhang, Q.; Bi, J.; Zheng, X.; Chen, Y.; Wang, H.; Wu, W.; Wang, Z.; Wu, Q.; Peng, H.; Wei, H.; et al. Blockade of the checkpoint receptor TIGIT prevents NK cell exhaustion and elicits potent anti-tumor immunity. Nat. Immunol. 2018, 19, 723–732. [Google Scholar] [CrossRef]
- Liu, G.; Zhang, Q.; Yang, J.; Li, X.; Xian, L.; Li, W.; Lin, T.; Cheng, J.; Lin, Q.; Xu, X.; et al. Increased TIGIT expressing NK cells with dysfunctional phenotype in AML patients correlated with poor prognosis. Cancer Immunol. Immunother. 2022, 71, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Hoechst, B.; Voigtlaender, T.; Ormandy, L.; Gamrekelashvili, J.; Zhao, F.; Wedemeyer, H.; Lehner, F.; Manns, M.P.; Greten, T.F.; Korangy, F. Myeloid derived suppressor cells inhibit natural killer cells in patients with hepatocellular carcinoma via the NKp30 receptor. Hepatology 2009, 50, 799–807. [Google Scholar] [CrossRef]
- Shibuya, A.; Shibuya, K. DNAM-1 versus TIGIT: Competitive roles in tumor immunity and inflammatory responses. Int. Immunol. 2021, 33, 687–692. [Google Scholar] [CrossRef]
- Kobecki, J.; Gajdzis, P.; Mazur, G.; Chabowski, M. Nectins and Nectin-like Molecules in Colorectal Cancer: Role in Diagnostics, Prognostic Values, and Emerging Treatment Options: A Literature Review. Diagnostics 2022, 12, 3076. [Google Scholar] [CrossRef]
- Sanchez-Correa, B.; Valhondo, I.; Hassouneh, F.; Lopez-Sejas, N.; Pera, A.; Bergua, J.M.; Arcos, M.J.; Bañas, H.; Casas-Avilés, I.; Durán, E.; et al. DNAM-1 and the TIGIT/PVRIG/TACTILE Axis: Novel Immune Checkpoints for Natural Killer Cell-Based Cancer Immunotherapy. Cancers 2019, 11, 877. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan Nair, V.; Saleh, R.; Toor, S.M.; Alajez, N.M.; Elkord, E. Transcriptomic Analyses of Myeloid-Derived Suppressor Cell Subsets in the Circulation of Colorectal Cancer Patients. Front. Oncol. 2020, 10, 1530. [Google Scholar] [CrossRef] [PubMed]
- Hossain, F.; Al-Khami, A.A.; Wyczechowska, D.; Hernandez, C.; Zheng, L.; Reiss, K.; Valle, L.D.; Trillo-Tinoco, J.; Maj, T.; Zou, W.; et al. Inhibition of Fatty Acid Oxidation Modulates Immunosuppressive Functions of Myeloid-Derived Suppressor Cells and Enhances Cancer Therapies. Cancer Immunol. Res. 2015, 3, 1236–1247. [Google Scholar] [CrossRef] [PubMed]
- Karakasheva, T.A.; Dominguez, G.A.; Hashimoto, A.; Lin, E.W.; Chiu, C.; Sasser, K.; Lee, J.W.; Beatty, G.L.; Gabrilovich, D.I.; Rustgi, A.K. CD38+ M-MDSC expansion characterizes a subset of advanced colorectal cancer patients. JCI Insight 2018, 3, e97022. [Google Scholar] [CrossRef] [PubMed]
- Mundy-Bosse, B.L.; Young, G.S.; Bauer, T.; Binkley, E.; Bloomston, M.; Bill, M.A.; Bekaii-Saab, T.; Carson, W.E., 3rd; Lesinski, G.B. Distinct myeloid suppressor cell subsets correlate with plasma IL-6 and IL-10 and reduced interferon-alpha signaling in CD4⁺ T cells from patients with GI malignancy. Cancer Immunol. Immunother. 2011, 60, 1269–1279. [Google Scholar] [CrossRef]
- Santegoets, S.J.; Stam, A.G.; Lougheed, S.M.; Gall, H.; Jooss, K.; Sacks, N.; Hege, K.; Lowy, I.; Scheper, R.J.; Gerritsen, W.R.; et al. Myeloid derived suppressor and dendritic cell subsets are related to clinical outcome in prostate cancer patients treated with prostate GVAX and ipilimumab. J. Immunother. Cancer 2014, 2, 31. [Google Scholar] [CrossRef] [PubMed]
- Meyer, C.; Cagnon, L.; Costa-Nunes, C.M.; Baumgaertner, P.; Montandon, N.; Leyvraz, L.; Michielin, O.; Romano, E.; Speiser, D.E. Frequencies of circulating MDSC correlate with clinical outcome of melanoma patients treated with ipilimumab. Cancer Immunol. Immunother. 2014, 63, 247–257. [Google Scholar] [CrossRef]
- Bi, J.; Tian, Z.N.K. Cell Dysfunction and Checkpoint Immunotherapy. Front. Immunol. 2019, 10, 1999. [Google Scholar] [CrossRef]
- Hasan, M.F.; Croom-Perez, T.J.; Oyer, J.L.; Dieffenthaller, T.A.; Robles-Carrillo, L.D.; Eloriaga, J.E.; Kumar, S.; Andersen, B.W.; Copik, A.J. TIGIT Expression on Activated NK Cells Correlates with Greater Anti-Tumor Activity but Promotes Functional Decline upon Lung Cancer Exposure: Implications for Adoptive Cell Therapy and TIGIT-Targeted Therapies. Cancers 2023, 15, 2712. [Google Scholar] [CrossRef]
- Ozbay Kurt, F.G.; Lasser, S.; Arkhypov, I.; Utikal, J.; Umansky, V. Enhancing immunotherapy response in melanoma: Myeloid-derived suppressor cells as a therapeutic target. J. Clin. Investig. 2023, 133, e170762. [Google Scholar] [CrossRef] [PubMed]
- Dar, A.; Kollet, O.; Lapidot, T. Mutual, reciprocal SDF-1/CXCR4 interactions between hematopoietic and bone marrow stromal cells regulate human stem cell migration and development in NOD/SCID chimeric mice. Exp. Hematol. 2006, 34, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Campana, S.; Di Carlo, E.; De Pasquale, C.; Barberi, C.; Oliveri, D.; Migliore, G.S.; Cannavò, S.P.; Galletti, B.; Pende, D.; Carrega, P.; et al. Dendritic cell recognition by group 3 innate lymphoid cells through DNAX accessory molecule 1 triggers proinflammatory reciprocal cell activation. J. Allergy Clin. Immunol. 2019, 144, 1118–1122. [Google Scholar] [CrossRef] [PubMed]
- Campana, S.; De Pasquale, C.; Barberi, C.; Oliveri, D.; Sidoti Migliore, G.; Galletti, B.; Guarneri, C.; Cannavò, S.P.; Ferlazzo, G. Circulating ILC precursors expressing CD62L exhibit a type 2 signature distinctly decreased in psoriatic patients. Eur. J. Immunol. 2021, 51, 1792–1798. [Google Scholar] [CrossRef] [PubMed]
- Sidoti Migliore, G.; Campana, S.; Barberi, C.; De Pasquale, C.; Pezzino, G.; Cavaliere, R.; Orecchia, P.; Ginestra, G.; Mandalari, G.; Del Zotto, G.; et al. Mechanical bacterial lysate enhances antimicrobial barrier mechanisms in human airway epithelial cells. J. Leukoc. Biol. 2023, 113, 535–540. [Google Scholar] [CrossRef] [PubMed]


| mAbs | Clone | Fluorochrome | Distributors |
|---|---|---|---|
| CD3 | UCHT1 | FITC | Beckman Coulter (Brea, CA, USA) |
| CD19 | J3.119 | FITC | Beckman Coulter |
| CD45 | J.33 | Krome Orange | Beckman Coulter |
| CD14 | RMO52 | APC-Alexa Fluor 700 | Beckman Coulter |
| CD11b | Bear1 | APC | Beckman Coulter |
| CD33 | D3HL60.251 | PC5 | Beckman Coulter |
| HLA-DR | Immu-357 | PC7 | Beckman Coulter |
| CD15 | 80H5 | FITC | Beckman Coulter |
| CD62L | DREG56 | PE | Beckman Coulter |
| CD66b | 80H3 | APC-Alex Fluor 750 | Beckman Coulter |
| MPO | 5B8 | PE | BD Biosciences |
| CD49d | HP2/1 | APC | Beckman Coulter |
| CD16 | 3G8 | Pacific Blue | Beckman Coulter |
| CD45RA | 2H4LDH11LDB9 (2H4) | ECD | Beckman Coulter |
| CD123 | SSDCLY107D2 | PC5.5 | Beckman Coulter |
| CD56 | N901 (NKH-1) | PC7 | Beckman Coulter |
| IFN-γ | 45 | FITC | Beckman Coulter |
| CD107a | H4-A3 | Pacific Blue | Beckman Coulter |
| TIGIT | 741182 | BUV395 | BD Biosciences |
| DNAM-1 | 11A8 | BV605 | BD Biosciences |
| CD34 | 581 | ECD | Beckman Coulter |
| CD38 | LS198-4-3 | APC-AlexaFluor 700 | Beckman Coulter |
| Dapi | n.a. | n.a. | ThermoFisher |
| NKp30 | Z25 | PE | Beckman Coulter |
| NKp46 | BAB281 | PC5 | Beckman Coulter |
| NKG2D | ON72 | APC | Beckman Coulter |
| KIR(KIR2DL2/DL3) | DX27 | FITC | Miltenyi Biotec |
| PD-1 | PD1.3 | PC7 | Beckman Coulter |
| Anti-IgG1 | n.a. | APC-AlexaFluor 488 | Invitrogen (Waltham, MA USA) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calabrò, A.; Drommi, F.; Sidoti Migliore, G.; Pezzino, G.; Vento, G.; Freni, J.; Costa, G.; Cavaliere, R.; Bonaccorsi, I.; Sionne, M.; et al. Neutrophil-like Monocytes Increase in Patients with Colon Cancer and Induce Dysfunctional TIGIT+ NK Cells. Int. J. Mol. Sci. 2024, 25, 8470. https://doi.org/10.3390/ijms25158470
Calabrò A, Drommi F, Sidoti Migliore G, Pezzino G, Vento G, Freni J, Costa G, Cavaliere R, Bonaccorsi I, Sionne M, et al. Neutrophil-like Monocytes Increase in Patients with Colon Cancer and Induce Dysfunctional TIGIT+ NK Cells. International Journal of Molecular Sciences. 2024; 25(15):8470. https://doi.org/10.3390/ijms25158470
Chicago/Turabian StyleCalabrò, Alessia, Fabiana Drommi, Giacomo Sidoti Migliore, Gaetana Pezzino, Grazia Vento, José Freni, Gregorio Costa, Riccardo Cavaliere, Irene Bonaccorsi, Mariagrazia Sionne, and et al. 2024. "Neutrophil-like Monocytes Increase in Patients with Colon Cancer and Induce Dysfunctional TIGIT+ NK Cells" International Journal of Molecular Sciences 25, no. 15: 8470. https://doi.org/10.3390/ijms25158470
APA StyleCalabrò, A., Drommi, F., Sidoti Migliore, G., Pezzino, G., Vento, G., Freni, J., Costa, G., Cavaliere, R., Bonaccorsi, I., Sionne, M., Nigro, S., Navarra, G., Ferlazzo, G., De Pasquale, C., & Campana, S. (2024). Neutrophil-like Monocytes Increase in Patients with Colon Cancer and Induce Dysfunctional TIGIT+ NK Cells. International Journal of Molecular Sciences, 25(15), 8470. https://doi.org/10.3390/ijms25158470






