Genome-Wide Identification and Expression Pattern Analysis of the WNK Gene Family in Apple under Abiotic Stress and Colletotrichum siamense Infection
Abstract
1. Introduction
2. Results
2.1. Identification and Characterization of WNK Gene Family in Apple
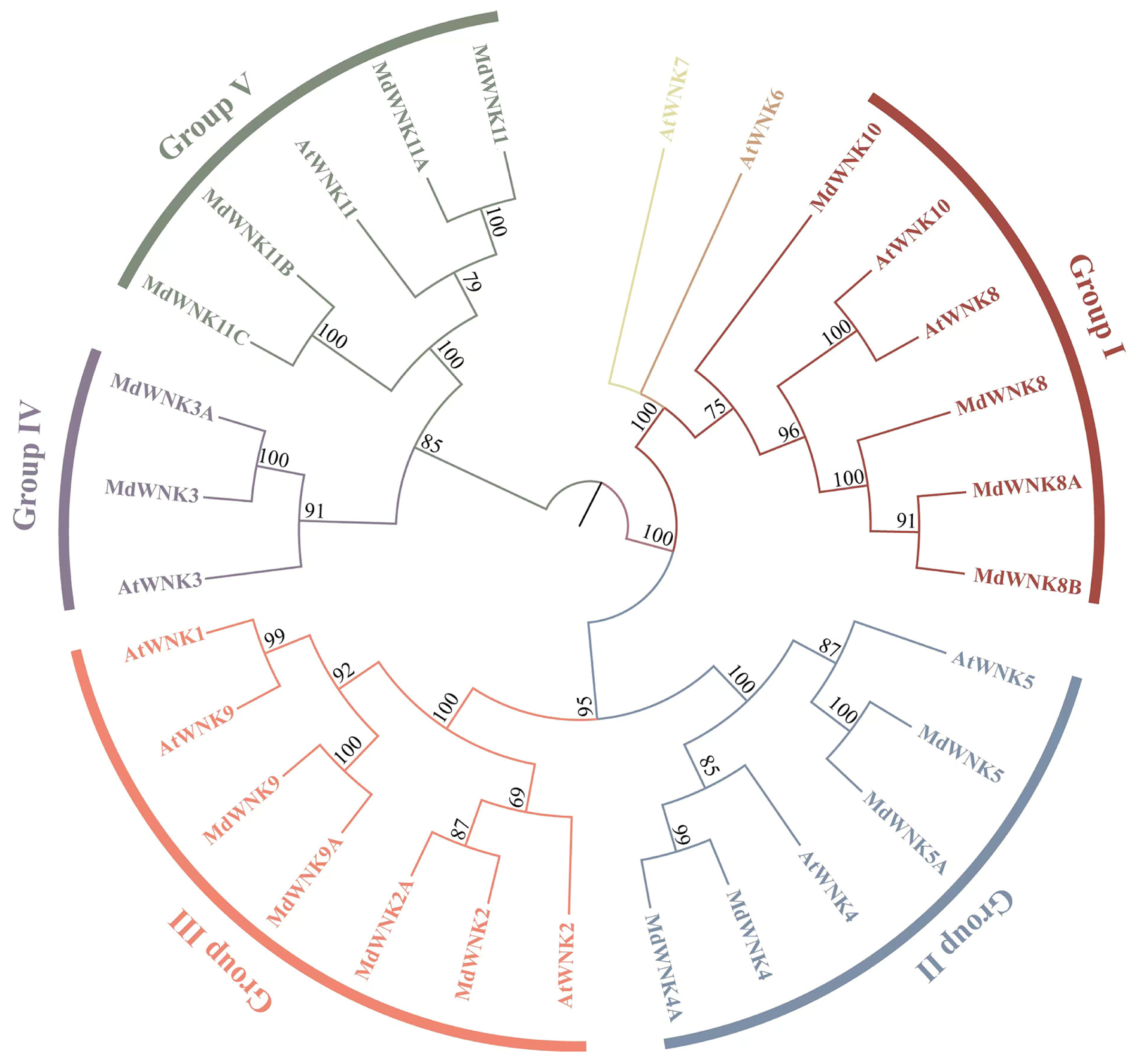
2.2. Phylogenetic Analysis of the WNK Family
2.3. Structure Prediction of MdWNK Proteins
2.4. Gene Structure, Conserved Domain, and Motif Analysis of MdWNKs
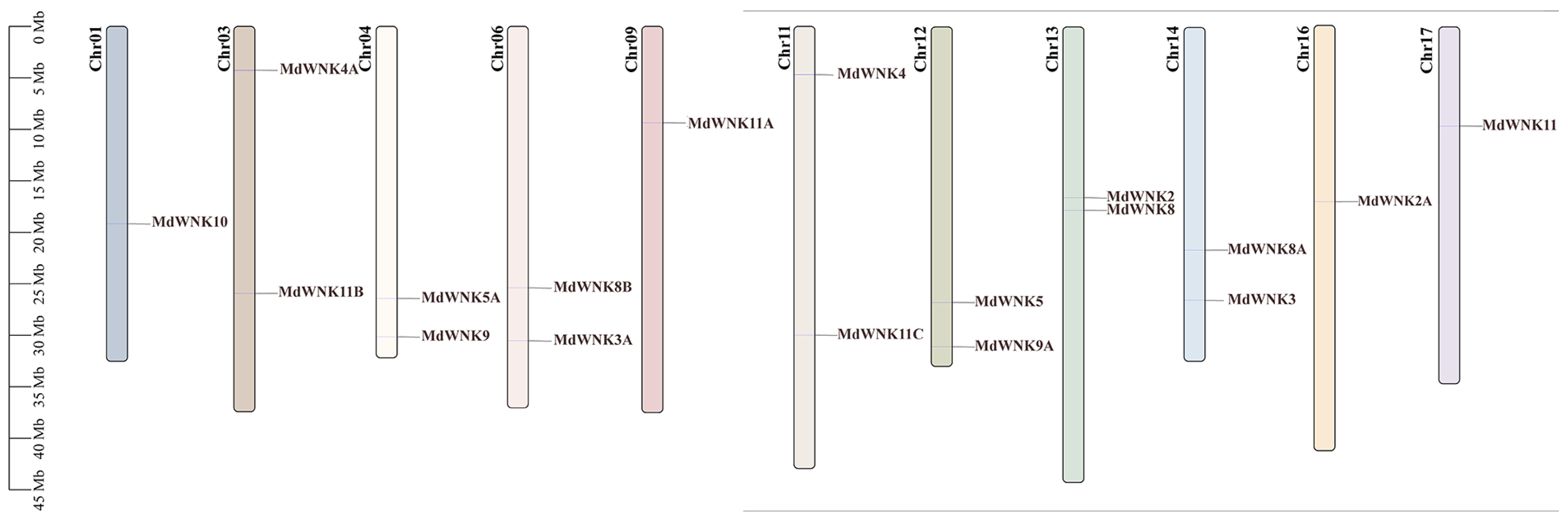
2.5. Chromosomal Localization and Synteny Analysis of MdWNKs
2.6. Promoter Cis-Acting Element Analysis of MdWNK Genes
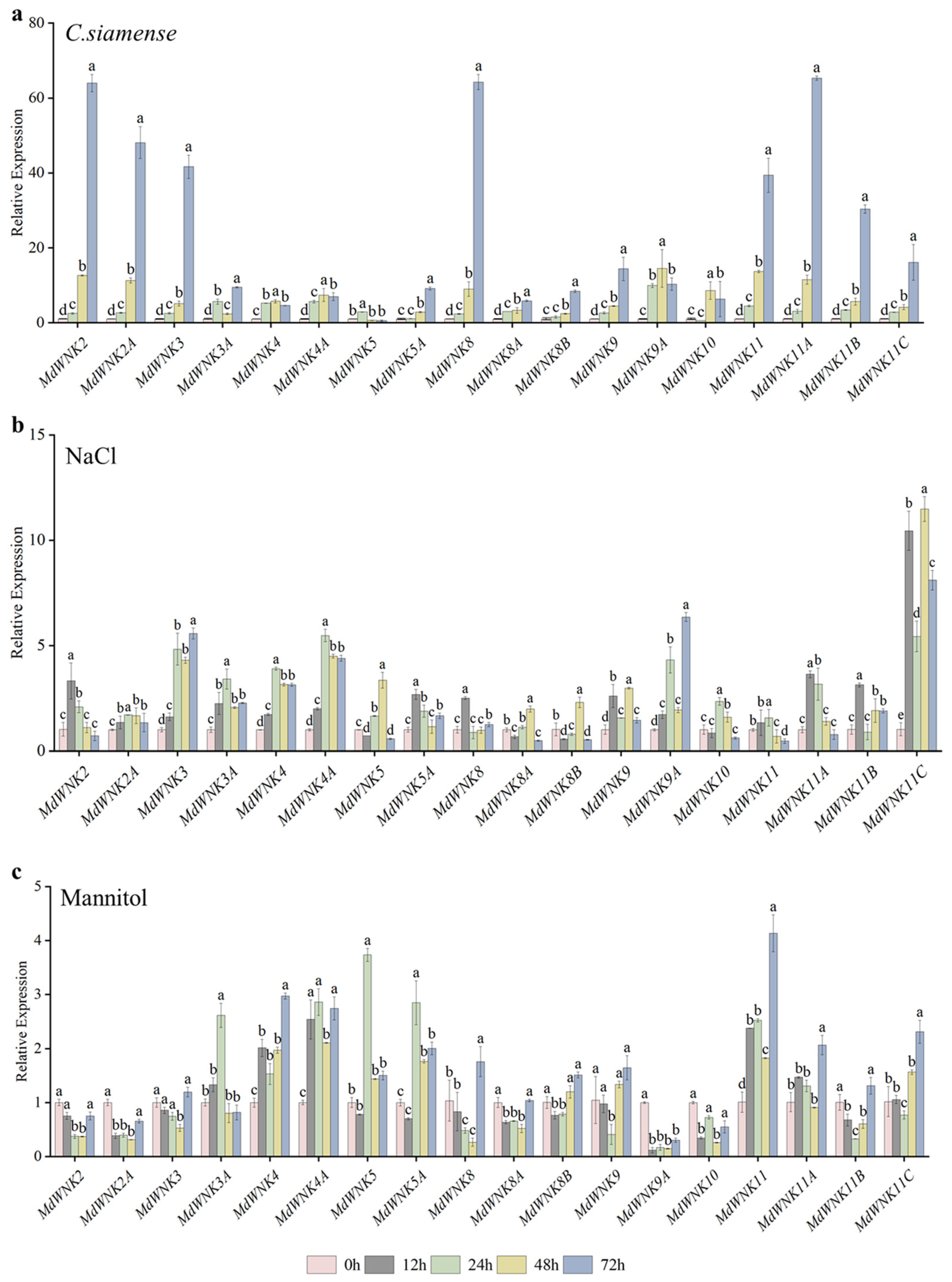
2.7. Expression Analysis of MdWNK Genes under Different Stress Treatments
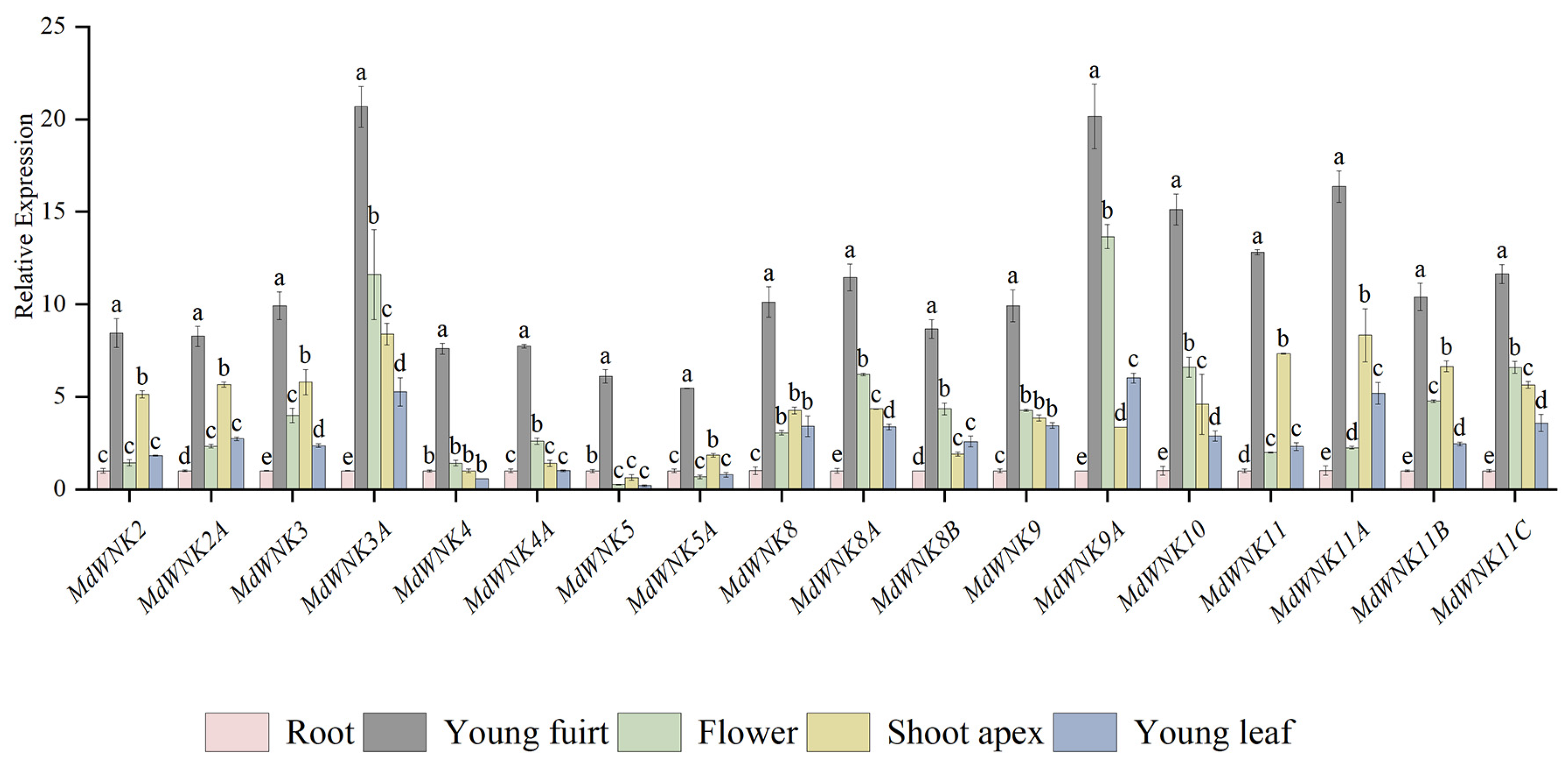
2.8. Tissue-Specific Expression Analysis of MdWNK Genes
3. Discussion
4. Materials and Methods
4.1. Identification of WNK Family Members in Apple
4.2. Phylogenetic Analysis
4.3. Sequence Analysis of MdWNK Family Members
4.4. Promoter Cis-Acting Element Analysis
4.5. Chromosomal Localization and Synteny Analysis
4.6. Plant Material and Stress Treatment
4.7. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hunter, T. Signaling-2000 and beyond. Cell 2000, 100, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Hanks, S.K.; Quinn, A.M. Protein kinase catalytic domain sequence database: Identification of conserved features of primary structure and classification of family members. Meth. Enzymol. 1991, 200, 38–61. [Google Scholar]
- Xu, B.; English, J.M.; Wilsbacher, J.L.; Stippec, S.; Goldsmith, E.J.; Cobb, M.H. WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II. J. Biol. Chem. 2000, 275, 16795–16801. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Min, X.; Stippec, S.; Lee, B.H.; Goldsmith, E.J.; Cobb, M.H. Regulation of WNK1 by an autoinhibitory domain and autophosphorylation. J. Biol. Chem. 2002, 277, 48456–48462. [Google Scholar] [CrossRef]
- Wilson, F.H.; Disse-Nicodème, S.; Choate, K.A.; Ishikawa, K.; Nelson-Williams, C.; Desitter, I.; Gunel, M.; Milford, D.V.; Lipkin, G.W.; Achard, J.M.; et al. Human hypertension caused by mutations in WNK kinases. Science 2001, 293, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Veríssimo, F.; Jordan, P. WNK kinases, a novel protein kinase subfamily in multi-cellular organisms. Oncogene 2001, 20, 5562–5569. [Google Scholar] [CrossRef]
- Liu, R.; Vasupalli, N.; Hou, D.; Stalin, A.; Wei, H.; Zhang, H.; Lin, X. Genome-wide identification and evolution of WNK kinases in Bambusoideae and transcriptional profiling during abiotic stress in Phyllostachys edulis. PeerJ 2022, 10, e12718. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, C.D.; Pan, Z.Y.; Lin, H.R.; Li, Z.B.; Hou, X.H.; Liu, J.S.; Nie, X.H.; Wu, Y.L. Genome-wide identification and analysis of the WNK kinase gene family in upland cotton. Plants 2023, 12, 4036. [Google Scholar] [CrossRef]
- Saddhe, A.A.; Karle, S.B.; Aftab, T.; Kumar, K. With no lysine kinases: The key regulatory networks and phytohormone cross talk in plant growth, development and stress response. Plant Cell Rep. 2021, 40, 2097–2109. [Google Scholar] [CrossRef]
- Zhang, B.G.; Liu, K.D.; Zheng, Y.; Wang, Y.X.; Wang, J.X.; Liao, H. Disruption of AtWNK8 enhances tolerance of Arabidopsis to salt and osmotic stresses via modulating proline content and activities of catalase and peroxidase. Int. J. Mol. Sci. 2013, 14, 7032–7047. [Google Scholar] [CrossRef]
- Xie, M.M.; Wu, D.; Duan, G.F.; Wang, L.Q.; He, R.Q.; Li, X.S.; Tang, D.Y.; Zhao, X.Y.; Liu, X.M. AtWNK9 is regulated by ABA and dehydration and is involved in drought tolerance in Arabidopsis. Plant Physiol. Biochem. 2014, 77, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Manuka, R.; Saddhe, A.A.; Kumar, K. Genome-wide identification and expression analysis of WNK kinase gene family in rice. Comput. Biol. Chem. 2015, 59, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Hao, P.; Shu, W.; Wang, G.; Xie, Z.; Gu, C.; Zhang, S. Phylogenetic and expression analyses of with-no-lysine kinase genes reveal novel gene family diversity in fruit trees. Hortic. Plant J. 2019, 5, 5–16. [Google Scholar] [CrossRef]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Rannala, B. Molecular phylogenetics: Principles and practice. Nat. Rev. Genet. 2012, 13, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Hemmer, W.; McGlone, M.; Tsigelny, I.; Taylor, S.S. Role of the glycine triad in the ATP-binding site of cAMP-dependent protein kinase. J. Biol. Chem. 1997, 272, 16946–16954. [Google Scholar] [CrossRef] [PubMed]
- Hanks, S.K.; Hunter, T. Protein kinases 6. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. FASEB J. 1995, 9, 576–596. [Google Scholar] [CrossRef] [PubMed]
- Cao-Pham, A.H.; Urano, D.; Ross-Elliott, T.J.; Jones, A.M. Nudge-nudge, WNK-WNK (kinases), say no more? New Phytol. 2018, 220, 35–48. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef]
- Nakamichi, N.; Murakami-Kojima, M.; Sato, E.; Kishi, Y.; Yamashino, T.; Mizuno, T. Compilation and characterization of a novel WNK family of protein kinases in Arabidopsis thaliana with reference to circadian rhythms. Biosci. Biotechnol. Biochem. 2002, 66, 2429–2436. [Google Scholar] [CrossRef]
- Kumar, K.; Rao, K.P.; Biswas, D.K.; Sinha, A.K. Rice WNK1 is regulated by abiotic stress and involved in internal circadian rhythm. Plant Signal. Behav. 2011, 6, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Geng, D.L.; Chen, P.X.; Shen, X.X.; Zhang, Y.; Li, X.W.; Jiang, L.J.; Xie, Y.P.; Niu, C.D.; Zhang, J.; Huang, X.H.; et al. MdMYB88 and MdMYB124 enhance drought tolerance by modulating root vessels and cell walls in apple. Plant Physiol. 2018, 178, 1296–1309. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Chen, W.J.; Guo, N.; Zang, J.; Liu, L.F.; Zhang, Z.H.; Dai, H.Y. MdWRKY61 positively regulates resistance to Colletotrichum siamense in apple (Malus domestica). Physiol. Mol. Plant Pathol. 2022, 117, 101776. [Google Scholar] [CrossRef]
- Wang, Y.X.; Suo, H.C.; Zhuang, C.X.; Ma, H.; Yan, X.L. Overexpression of the soybean GmWNK1 altered the sensitivity to salt and osmotic stress in Arabidopsis. J. Plant Physiol. 2011, 168, 2260–2267. [Google Scholar] [CrossRef] [PubMed]
- Manuka, R.; Saddhe, A.A.; Kumar, K. Expression of OsWNK9 in Arabidopsis conferred tolerance to salt and drought stress. Plant Sci. 2018, 270, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.X.; Suo, H.C.; Zheng, Y.; Liu, K.D.; Zhuang, C.X.; Kahle, K.T.; Ma, H.; Yan, X.L. The soybean root-specific protein kinase GmWNK1 regulates stress-responsive ABA signaling on the root system architecture. Plant J. 2010, 64, 230–242. [Google Scholar] [CrossRef] [PubMed]
- Negi, Y.; Kumar, K. Cloning, homology modelling and expression analysis of Oryza sativa WNK gene family. Int. J. Biol. Macromol. 2023, 229, 994–1008. [Google Scholar] [CrossRef] [PubMed]
- Dunker, F.; Trutzenberg, A.; Rothenpieler, J.S.; Kuhn, S.; Pröls, R.; Schreiber, T.; Tissier, A.; Kemen, A.; Kemen, E.; Hückelhoven, R.; et al. Oomycete small RNAs bind to the plant RNA-induced silencing complex for virulence. eLife 2020, 9, e56096. [Google Scholar] [CrossRef] [PubMed]
- Lumba, S.; Toh, S.; Handfield, L.F.; Swan, M.; Liu, R.; Youn, J.Y.; Cutler, S.R.; Subramaniam, R.; Provart, N.; Moses, A.; et al. A mesoscale abscisic acid hormone interactome reveals a dynamic signaling landscape in Arabidopsis. Dev. Cell 2014, 29, 360–372. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.P.; Tang, H.B.; Debarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Xia, R. A painless way to customize circos plot: From datapreparation to visualization using TBtools. iMeta 2022, 1, e35. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.P.; Zhang, Y.B.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinf. 2010, 8, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Zhang, Z.; Yang, H.; Li, H.; Dai, H. Detection of strawberry RNA and DNA viruses by RT-PCR using total nucleic acid as a template. J. Phytopathol. 2007, 155, 431–436. [Google Scholar] [CrossRef]
- Chen, K.Q.; Song, M.R.; Guo, Y.N.; Liu, L.F.; Xue, H.; Dai, H.Y.; Zhang, Z.H. MdMYB46 could enhance salt and osmotic stress tolerance in apple by directly activating stress-responsive signals. Plant Biotechnol. J. 2019, 17, 2341–2355. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]




| Gene Name | Gene ID | Protein Length (aa) | MW (kDa) | pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity |
|---|---|---|---|---|---|---|---|
| MdWNK2 | MD13G1192500 | 737 | 84.10 | 5.35 | 39.67 | 76.61 | −0.636 |
| MdWNK2A | MD16G1193000 | 738 | 83.94 | 5.24 | 41.25 | 76.12 | −0.604 |
| MdWNK3 | MD14G1170800 | 632 | 70.65 | 4.87 | 48.31 | 79.18 | −0.618 |
| MdWNK3A | MD06G1165300 | 632 | 70.87 | 4.8 | 47.1 | 80.08 | −0.633 |
| MdWNK4 | MD11G1055100 | 588 | 66.47 | 6.22 | 39 | 79.59 | −0.464 |
| MdWNK4A | MD03G1053500 | 585 | 66.38 | 5.99 | 41.31 | 80.17 | −0.48 |
| MdWNK5 | MD12G1186000 | 604 | 68.82 | 5.45 | 51.93 | 78.11 | −0.559 |
| MdWNK5A | MD04G1172700 | 603 | 68.91 | 5.86 | 52.58 | 74.53 | −0.621 |
| MdWNK8 | MD13G1200200 | 409 | 46.34 | 5.21 | 34.43 | 93.37 | −0.267 |
| MdWNK8A | MD14G1136200 | 606 | 67.62 | 5.14 | 51.91 | 80.1 | −0.416 |
| MdWNK8B | MD06G1114800 | 605 | 67.62 | 5.19 | 44.31 | 82.63 | −0.44 |
| MdWNK9 | MD04G1220500 | 745 | 85.13 | 5.08 | 43.15 | 77.99 | −0.592 |
| MdWNK9A | MD12G1236900 | 593 | 67.85 | 5.31 | 39.43 | 77.22 | −0.475 |
| MdWNK10 | MD01G1085200 | 655 | 73.37 | 5.49 | 39.88 | 84.52 | −0.382 |
| MdWNK11 | MD17G1112400 | 350 | 39.96 | 8.64 | 43.63 | 91.63 | −0.33 |
| MdWNK11A | MD09G1121400 | 289 | 32.92 | 5.89 | 30.13 | 87.68 | −0.381 |
| MdWNK11B | MD03G1189000 | 295 | 33.73 | 5.47 | 37.06 | 83.9 | −0.465 |
| MdWNK11C | MD11G1205200 | 295 | 33.85 | 5.4 | 37.59 | 82.54 | −0.466 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, Z.; Wu, Z.-C.; Zang, J.; Zhao, D.; Guo, W.; Dai, H. Genome-Wide Identification and Expression Pattern Analysis of the WNK Gene Family in Apple under Abiotic Stress and Colletotrichum siamense Infection. Int. J. Mol. Sci. 2024, 25, 8528. https://doi.org/10.3390/ijms25158528
Wei Z, Wu Z-C, Zang J, Zhao D, Guo W, Dai H. Genome-Wide Identification and Expression Pattern Analysis of the WNK Gene Family in Apple under Abiotic Stress and Colletotrichum siamense Infection. International Journal of Molecular Sciences. 2024; 25(15):8528. https://doi.org/10.3390/ijms25158528
Chicago/Turabian StyleWei, Ziwen, Zheng-Chao Wu, Jian Zang, Di Zhao, Wei Guo, and Hongyan Dai. 2024. "Genome-Wide Identification and Expression Pattern Analysis of the WNK Gene Family in Apple under Abiotic Stress and Colletotrichum siamense Infection" International Journal of Molecular Sciences 25, no. 15: 8528. https://doi.org/10.3390/ijms25158528
APA StyleWei, Z., Wu, Z.-C., Zang, J., Zhao, D., Guo, W., & Dai, H. (2024). Genome-Wide Identification and Expression Pattern Analysis of the WNK Gene Family in Apple under Abiotic Stress and Colletotrichum siamense Infection. International Journal of Molecular Sciences, 25(15), 8528. https://doi.org/10.3390/ijms25158528






