Avian Influenza Virus A(H5Nx) and Prepandemic Candidate Vaccines: State of the Art
Abstract
1. A Brief History of Influenza Virus A(H5N1) 1996–2021
2. The H5N1 Enzootics since 2022
3. H5N1 Vaccines for Human Use
- On 14 November 2013, the FDA licensed the ID Biomedical Corporation’s (a Canadian subsidiary of Glaxo Smith Kline Biologicals) egg-based Influenza A (H5N1) Virus Monovalent Vaccine, including A/Indonesia/05/2005 (H5N1) and the AS03 adjuvant [91], but the vaccine was not commercially available: the US federal government purchased it for the NPIVS for as-needed distribution [92].
- On 31 January 2020, the US Food and Drug Administration (FDA) authorized the use of CSL Seqirus Inc.’s Audenz™ (Holly Springs, NC, USA) subunit influenza vaccine prepared from A/turkey/Turkey/1/2005 NIBRG-23 virus propagated in Madin Darby Canine Kidney (MDCK) cells, adjuvanted with MF59. The vaccine was manufactured by I.D. Biomedical Corporation.
- Aflunov™, which contains an A/turkey/Turkey/1/2005 (H5N1)-like strain (NIBRG-23) (clade 2.2.1) was submitted but the application was withdrawn on June 2008.
- Celldemic™ and Incellipan™ (zoonotic influenza vaccine (H5N1) (surface antigens, inactivated, adjuvanted, prepared in cell cultures) were authorized in April 2024.
| Target Influenza | Brand Name | Manufacturer | Used Lineage | Cell Substrate | Antigen | Adjuvant | Regulatory Status | Availability |
|---|---|---|---|---|---|---|---|---|
| Pandemic H1N1pdm09 | Pandemrix™ | GlaxoSmithKline Biologicals S.A. (Rixensart, Belgium) | A/California/7/2009 (H1N1)v-like strain X-179A (derived from A/Puerto Rico/8/1934) | 11-day-old fertilized hens’ eggs | Split virion | AS03 | Administered outside USA | EMA marketing authorization expired August 2015 |
| Arepanrix™ | Unknown | |||||||
| Focetria™ | Novartis Vaccines and Diagnostics S.r.l. (Siena, Italy) | A/California/7/2009 with HA1:N146D | Surface | MF59C.1 | Unknown | |||
| Celvapan™ | Nanotherapeutics Bohumil, s.r.o. (Jevany, Czech Republic) | A/California/7/2009 | Vero cells | HA | AS03 | Unknown | ||
| Panvax™ | CSL Biotherapies Ltd. (Melbourne, Australia) | A/California/7/2009 (H1N1)v-like | 11-day-old fertilized hens’ eggs | Split-virion | no | Approved by TGA (Australia), FDA, EMA, Singapore, Germany, and New Zealand [93] | Unknown | |
| Prepandemic H5N1 | none | ID Biomedical Corp (Vancouver, Canada), acquired by GSK in 2005 | A/Indonesia/05/2005 (H5N1) (clade 2) | 11-day-old fertilized hens’ eggs | Split-virion | AS03 | Approved by FDA | US stockpile only |
| Influenza Virus Vaccine, H5N1 | Sanofi Pasteur (Lyon, France) | A/Vietnam/1203/2004 (H5N1, clade 1) | Split-virion | No | Approved by FDA since 2007 for national stockpile | Unknown | ||
| Audenz™ | Seqirus Inc, USA, now part of CSL Seqirus (Melborne, Australia) | A/turkey/Turkey/1/2005 (H5N1)-like strain (NIBRG-23) (clade 2.2.1) | MDCK cells | Subunit | MF59 | Approved by FDA | Unknown | |
| Incellipan™ | Seqirus Netherlands B.V. (Amsterdam, The Netherlands) | HA and NA | MF59C.1 | Authorized by EMA | Unavailable | |||
| Celldemic™ | Authorized by EMA | Unknown | ||||||
| Aflunov™ | Novartis Vaccines and Diagnostics S.r.l. (Siena, Italy) | 11-day-old fertilized hens’ eggs | EMA request withdrawn in 2008 | Unknown | ||||
| Foclivia™ | Seqirus S.r.l. (Siena, Italy) | A/Vietnam/1194/2004 (H5N1) (clade 1) | Authorized by EMA | Unknown | ||||
| Pumarix™ | GSK Biologicals S.A. (Rixensart, Belgium) | A/Indonesia/05/2005 (H5N1) like strain used (PR8-IBCDC-RG2) (clade 2) | Split virion | AS03 | Authorization withdrawn by EMA | Unavailable | ||
| Prepandrix™ (formerly GSK1557484A [94,95]) | Authorization withdrawn by EMA | Unavailable | ||||||
| Adjupanrix™ (formerly GSK1562902A) [96,97,98,99] | A/VietNam/1194/2004 NIBRG 14 (clade 1) | Authorized by EMA | Unknown | |||||
| Panflu™ | Sinovac Biotech Ltd. (Beijing, China) | A/Vietnam/1194/2004-A/PR/8/34 (NIBRG-14) | Whole-virion | Yes | Approved in China | Unknown |
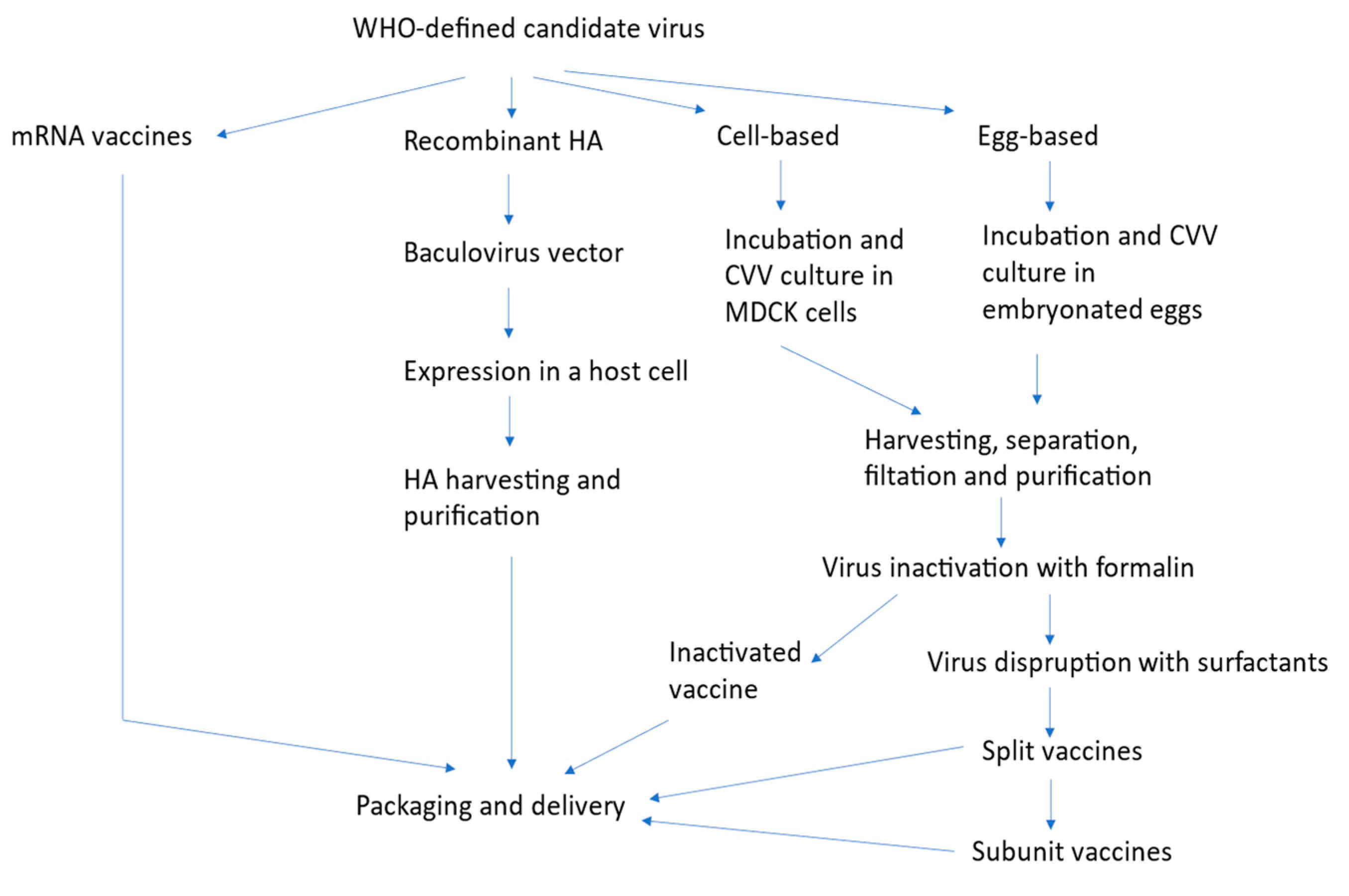
4. Production Technologies
5. Lessons Learnt from H5N1 Clinical Trials
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Beare, A.S.; Webster, R.G. Replication of avian influenza viruses in humans. Arch. Virol. 1991, 119, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Kandeil, A.; Patton, C.; Jones, J.C.; Jeevan, T.; Harrington, W.N.; Trifkovic, S.; Seiler, J.P.; Fabrizio, T.; Woodard, K.; Turner, J.C.; et al. Rapid evolution of A(H5N1) influenza viruses after intercontinental spread to North America. Nat. Commun. 2023, 14, 3082. [Google Scholar] [CrossRef] [PubMed]
- Evolution Working Group. Toward a unified nomenclature system for highly pathogenic avian influenza virus (H5N1). Emerg. Infect. Dis. 2008, 14, e1. [Google Scholar] [CrossRef] [PubMed]
- Lisovski, S.; Günther, A.; Dewar, M.; Ainley, D.; Aldunate, F.; Arce, R.; Ballard, G.; Bauer, S.; Belliure, J.; Banyard, A.C.; et al. Unexpected delayed incursion of highly pathogenic avian influenza H5N1 (clade 2.3.4.4b) in the Antarctic region. bioRxiv 2024. [Google Scholar] [CrossRef]
- Heo, G.; Kang, Y.; An, S.; Kim, Y.; Cha, R.; Jang, Y. Concurrent Infection with Clade 2.3.4.4b Highly Pathogenic Avian Influenza H5N6 and H5N1 Viruses, South Korea, 2023. Emerg. Infect. Dis. 2024, 30, 1223–1227. [Google Scholar] [CrossRef]
- Rosone, F.; Bonfante, F.; Sala, M.G.; Maniero, S.; Cersini, A.; Ricci, I.; Garofalo, L.; Caciolo, D.; Denisi, A.; Napolitan, A.; et al. Seroconversion of a Swine Herd in a Free-Range Rural Multi-Species Farm against HPAI H5N1 2.3.4.4b Clade Virus. Microorganisms 2023, 11, 1162. [Google Scholar] [CrossRef] [PubMed]
- Rijks, J.M.; Hesselink, H.; Lollinga, P.; Wesselman, R.; Prins, P.; Weesendorp, E.; Engelsma, M.; Heutink, R.; Harders, F.; Kik, M.; et al. Highly Pathogenic Avian Influenza A(H5N1) Virus in Wild Red Foxes, the Netherlands, 2021. Emerg. Infect. Dis. 2021, 27, 2960–2962. [Google Scholar] [CrossRef] [PubMed]
- Alkie, T.N.; Cox, S.; Embury-Hyatt, C.; Stevens, B.; Pople, N.; Pybus, M.J.; Xu, W.; Hisanaga, T.; Suderman, M.; Koziuk, J.; et al. Characterization of neurotropic HPAI H5N1 viruses with novel genome constellations and mammalian adaptive mutations in free-living mesocarnivores in Canada. Emerg. Microbes Infect. 2023, 12, 2186608. [Google Scholar] [CrossRef]
- Lair, S.; Quesnel, L.; Signore, A.; Delnatte, P.; Embury-Hyatt, C.; Nadeau, M.-S. Outbreak of highly pathogenic avian influenza A(H5N1) virus in seals, St. Lawrence Estuary, Quebec, Canada. Emerg. Infect. Dis. 2024, 30, 1133–1143. [Google Scholar] [CrossRef] [PubMed]
- Puryear, W.; Sawatzki, K.H.N.; Foss, A.; Stone, J.; Doughty, L. Highly Pathogenic Avian Influenza A(H5N1) Virus Outbreak in New England Seals, United States. Emerg. Infect. Dis. 2023, 29, 786–791. [Google Scholar] [CrossRef] [PubMed]
- Murawski, A.; Fabrizio, T.; Ossiboff, R.; Kackos, C.; Jeevan, T.; Jones, J.C.; Kandeil, A.; Walker, D.; Turner, J.C.M.; Patton, C.; et al. Highly pathogenic avian influenza A(H5N1) virus in a common bottlenose dolphin (Tursiops truncatus) in Florida. Commun. Biol. 2024, 7, 476. [Google Scholar] [CrossRef] [PubMed]
- Agüero, M.; Monne, I.; Sánchez, A.; Zecchin, B.; Fusaro, A.; Ruano, M.J.; del Valle Arrojo, M.; Fernández-Antonio, R.; Souto, A.M.; Tordable, P.; et al. Highly pathogenic avian influenza A(H5N1) virus infection in farmed minks, Spain, October 2022. Eurosurveillance 2023, 28, 2300001. [Google Scholar] [CrossRef] [PubMed]
- Pardo-Roa, C.; Nelson, M.I.; Ariyama, N.; Aguayo, C.; Almonacid, L.I.; Munoz, G.; Navarro, C.; Avila, C.; Ulloa, M.; Reyes, R.; et al. Cross-species transmission and PB2 mammalian adaptations of highly pathogenic avian influenza A/H5N1 viruses in Chile. bioRxiv 2023. [Google Scholar] [CrossRef]
- Uhart, M.; Vanstreels, R.E.T.; Nelson, M.I.; Olivera, V.; Campagna, J.; Zavattieri, V.; Lemey, P.; Campagna, C.; Falabella, V.; Rimondi, A. Massive outbreak of Influenza A H5N1 in elephant seals at Península Valdés, Argentina: Increased evidence for mammal-to-mammal transmission. bioRxiv 2024. [Google Scholar] [CrossRef]
- Gamarra-Toledo, V.; Plaza, P.I.; Gutiérrez, R.; Inga-Diaz, G.; Saravia-Guevara, P.; Pereyra-Meza, O.; Coronado-Flores, E.; Calderón-Cerrón, A.; Quiroz-Jiménez, G.; Martinez, P.; et al. Mass Mortality of Marine Mammals Associated to Highly Pathogenic Influenza Virus (H5N1) in South America. bioRxiv 2023. [Google Scholar] [CrossRef]
- Leguia, M.; Garcia-Glaessner, A.; Muñoz-Saavedra, B.; Juarez, D.; Barrera, P.; Calvo-Mac, C.; Jara, J.; Silva, W.; Ploog, K.; Amaro, L.; et al. Highly pathogenic avian influenza A (H5N1) in marine mammals and seabirds in Peru. Nat. Comm. 2023, 14, 5489. [Google Scholar] [CrossRef] [PubMed]
- Rabalski, L.; Milewska, A.; Pohlmann, A.; Gackowska, K.; Lepionka, T.; Szczepaniak, K.; Swiatalska, A.; Sieminska, I.; Arent, Z.; Beer, M.; et al. Emergence and potential transmission route of avian influenza A (H5N1) virus in domestic cats in Poland, June 2023. Eurosurveillance 2023, 28, 2300390. [Google Scholar] [CrossRef] [PubMed]
- Domańska-Blicharz, K.; Świętoń, E.; Świątalska, A.; Monne, I.; Fusaro, A.; Tarasiuk, K.; Wyrostek, K.; Styś-Fijoł, N.; Giza, A.; Pietruk, M.; et al. Outbreak of highly pathogenic avian influenza A(H5N1) clade 2.3.4.4b virus in cats, Poland, June to July 2023. Eurosurveillance 2023, 28, 2300366. [Google Scholar] [CrossRef] [PubMed]
- Lindh, E.; Lounela, H.; Ikonen, N.; Kantala, T.; Savolainen-Kopra, C.; Kauppinen, A.; Österlund, P.; Kareinen, L.; Katz, A.; Nokireki, T.; et al. Highly pathogenic avian influenza A(H5N1) virus infection on multiple fur farms in the South and Central Ostrobothnia regions of Finland, July 2023. Eurosurveillance 2023, 28, 2300400. [Google Scholar] [CrossRef]
- Lee, K.; Yeom, M.; Vu, T.T.H.; Do, H.-Q.; Na, W.; Lee, M.; Jeong, D.G.; Cheon, D.-S.; Song, D. Characterization of highly pathogenic avian influenza A (H5N1) viruses isolated from cats in South Korea, 2023. Emerg. Microbes Infect. 2024, 13, 2290835. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.; Black, A.; Haman, K.; Diel, D.; Ramirez, V.; Ziejka, R. Antibodies to Influenza A(H5N1) Virus in Hunting Dogs Retrieving Wild Fowl, Washington, USA. Emerg. Infect. Dis. 2024, 30, 1271–1274. [Google Scholar] [CrossRef] [PubMed]
- Falchieri, M.; Reid, S.M.; Dastderji, A.; Cracknell, J.; Warren, C.J.; Mollett, B.C.; Peers-Dent, J.; Schlachter, A.-L.D.; Mcginn, N.; Hepple, R.; et al. Rapid mortality in captive bush dogs (Speothos venaticus) caused by influenza A of avian origin (H5N1) at a wildlife collection in the United Kingdom. bioRxiv 2024. [Google Scholar] [CrossRef] [PubMed]
- Pulit-Penaloza, J.A.; Belser, J.A.; Brock, N.; Thakur, P.B.; Tumpey, T.M.; Maines, T.R. Pathogenesis and Transmissibility of North American Highly Pathogenic Avian Influenza A(H5N1) Virus in Ferrets. Emerg. Infect. Dis. 2022, 28, 1913–1915. [Google Scholar] [CrossRef] [PubMed]
- Pulit-Penaloza, J.A.; Brock, N.; Belser, J.A.; Sun, X.; Pappas, C.; Kieran, T.J.; Thakur, P.B.; Zeng, H.; Cui, D.; Frederick, J.; et al. Highly pathogenic avian influenza A(H5N1) virus of clade 2.3.4.4b isolated from a human case in Chile causes fatal disease and transmits between co-housed ferrets. Emerg. Microbes Infect. 2024, 13, 2332667. [Google Scholar] [CrossRef] [PubMed]
- Restori, K.H.; Septer, K.M.; Field, C.J.; Patel, D.R.; VanInsberghe, D.; Raghunathan, V.; Lowen, A.C.; Sutton, T.C. Risk assessment of a highly pathogenic H5N1 influenza virus from mink. Nat. Commun. 2024, 15, 4112. [Google Scholar] [CrossRef] [PubMed]
- Vairo, F.; Haider, N.; Kock, R.; Ntoumi, F.; Ippolito, G.; Zumla, A. Chikungunya: Epidemiology, Pathogenesis, Clinical Features, Management, and Prevention. Infect. Dis. Clin. N. Am. 2019, 33, 1003–1025. [Google Scholar] [CrossRef] [PubMed]
- Klein, B.; Kraemer, M.; Scarpino, S. Timeline for H5N1 in the USA during the 2024 Outbreak. 2024. Available online: https://github.com/Emergent-Epidemics/H5N1_US2024_timeline (accessed on 29 April 2024).
- Nguyen, T.-Q.; Hutter, C.; Markin, A.; Thomas, M.N.; Lantz, K.; Killian, M.L.; Janzen, G.M.; Vijendran, S.; Wagle, S.; Inderski, B.; et al. Emergence and interstate spread of highly pathogenic avian influenza A(H5N1) in dairy cattle. bioRxiv 2024. [Google Scholar] [CrossRef]
- Stone, H.; Jindal, M.; Lim, S.; Dawson, R.; Quigley, A.; Scotch, M.; MacIntyre, C.R. Potential Pathways of Spread of Highly Pathogenic Avian Influenza A/H5N1 Clade 2.3.4.4b Across Dairy Farms in the United States. medRxiv 2024. [Google Scholar] [CrossRef]
- Caserta, L.C.; Frye, E.A.; Butt, S.L.; Laverack, M.; Nooruzzaman, M.; Covaleda, L.M.; Thompson, A.C.; Koscielny, M.P.; Cronk, B.; Johnson, A.; et al. Spillover of highly pathogenic avian influenza H5N1 virus to dairy cattle. Nature 2024. Epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Kalthoff, D.; Hoffmann, B.; Harder, T.; Durban, M.; Beer, M. Experimental infection of cattle with highly pathogenic avian influenza virus (H5N1). Emerg. Infect. Dis. 2008, 14, 1132–1134. [Google Scholar] [CrossRef] [PubMed]
- Gangavarapu, K.; Pekar, J.; Débarre, F.; Andersen, K.; Dudas, G.; Goldhill, D.; Hughes, J.; Ji, X.; Joy, J.; Kraemer, M.; et al. Note about Availability of H5N1 2.3.4.4b Consensus Sequences from Cattle and Other Species. Available online: https://virological.org/t/note-about-availability-of-h5n1-2-3-4-4b-consensus-sequences-from-cattle-and-other-species/967 (accessed on 29 April 2024).
- Moncla, L. Real-Time Tracking of Influenza A/H5N1 Virus Evolution. 2024. Available online: https://nextstrain.org/avian-flu/h5n1/ha/2y?dmax=2023-06-19&dmin=2020-08-04 (accessed on 29 April 2024).
- Hu, X.; Saxena, A.; Magstadt, D.R.; Gauger, P.C.; Burrough, E.; Zhang, J.; Siepker, C.; Mainenti, M.; Gorden, P.J.; Plummer, P.; et al. Highly Pathogenic Avian Influenza A (H5N1) clade 2.3.4.4b Virus detected in dairy cattle. Emerg. Microbes Infect. 2024, 13, 1335–1343. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Suzuki, Y.; Suzuki, T.; Le, M.Q.; Nidom, C.A.; Sakai-Tagawa, Y.; Muramoto, Y.; Ito, M.; Kiso, M.; Horimoto, T.; et al. Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors. Nature 2006, 444, 378–382. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhang, Y.; Shinya, K.; Deng, G.; Jiang, Y.; Li, Z.; Guan, Y.; Tian, G.; Li, Y.; Shi, J.; et al. Identification of amino acids in HA and PB2 critical for the transmission of H5N1 avian influenza viruses in a mammalian host. PLoS Pathog. 2009, 5, e1000709. [Google Scholar] [CrossRef] [PubMed]
- Van Riel, D.; den Bakker, M.A.; Leijten, L.M.E.; Chutinimitkul, S.; Munster, V.J.; de Wit, E.; Rimmelzwaan, G.F.; Fouchier, R.A.M.; Osterhaus, A.D.M.E.; Kuiken, T. Seasonal and Pandemic Human Influenza Viruses Attach Better to Human Upper Respiratory Tract Epithelium than Avian Influenza Viruses. Am. J. Pathol. 2010, 176, 1614–1618. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Deng, G.; Song, J.; Tian, G.; Suo, Y.; Jiang, Y.; Guan, Y.; Bu, Z.; Kawaoka, Y.; Chen, H. Two amino acid residues in the matrix protein M1 contribute to the virulence difference of H5N1 avian influenza viruses in mice. Virology 2009, 384, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Suttie, A.; Deng, Y.M.; Greenhill, A.R.; Dussart, P.; Horwood, P.F.; Karlsson, E.A. Inventory of molecular markers affecting biological characteristics of avian influenza A viruses. Virus Genes 2019, 55, 739–768. [Google Scholar] [CrossRef] [PubMed]
- Nao, N.; Kajihara, M.; Manzoor, R.; Maruyama, J.; Yoshida, R.; Muramatsu, M.; Miyamoto, H.; Igarashi, M.; Eguchi, N.; Sato, M.; et al. A Single Amino Acid in the M1 Protein Responsible for the Different Pathogenic Potentials of H5N1 Highly Pathogenic Avian Influenza Virus Strains. PLoS ONE 2015, 10, e0137989. [Google Scholar] [CrossRef] [PubMed]
- Bordes, L.; Vreman, S.; Heutink, R.; Roose, M.; Venema, S.; Pritz-Verschuren, S.B.E.; Rijks, J.M.; Gonzales, J.L.; Germeraad, E.A.; Engelsma, M.; et al. Highly Pathogenic Avian Influenza H5N1 Virus Infections in Wild Red Foxes (Vulpes vulpes) Show Neurotropism and Adaptive Virus Mutations. Microbiol. Spectr. 2023, 11, e0286722. [Google Scholar] [CrossRef] [PubMed]
- Hatta, M.; Hatta, Y.; Kim, J.H.; Watanabe, S.; Shinya, K.; Nguyen, T.; Lien, P.S.; Le, Q.M.; Kawaoka, Y. Growth of H5N1 influenza A viruses in the upper respiratory tracts of mice. PLoS Pathog. 2007, 3, 1374–1379. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Hatta, M.; Staker, B.L.; Watanabe, S.; Imai, M.; Shinya, K.; Sakai-Tagawa, Y.; Ito, M.; Ozawa, M.; Watanabe, T.; et al. Biological and structural characterization of a host-adapting amino acid in influenza virus. PLoS Pathog. 2010, 6, e1001034. [Google Scholar] [CrossRef] [PubMed]
- Lina, L.; Saijuan, C.; Chengyu, W.; Yuefeng, L.; Shishan, D.; Ligong, C.; Kangkang, G.; Zhendong, G.; Jiakai, L.; Jianhui, Z.; et al. Adaptive amino acid substitutions enable transmission of an H9N2 avian influenza virus in guinea pigs. Sci. Rep. 2019, 9, 19734. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, R.; Yamada, S.; Le, M.Q.; Li, C.; Chen, H.; Qurnianingsih, E.; Nidom, C.A.; Ito, M.; Sakai-Tagawa, Y.; Kawaoka, Y. Identification of PB2 Mutations Responsible for the Efficient Replication of H5N1 Influenza Viruses in Human Lung Epithelial Cells. J. Virol. 2015, 89, 3947–3956. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Trujillo, J.; McDowell, C. Detection and characterization of H5N1 HPAIV in environmental samples from a dairy farm. Virus Genes 2024. [Google Scholar] [CrossRef] [PubMed]
- Burrough, E.R.; Magstadt, D.R.; Petersen, B.; Timmermans, S.J.; Gauger, P.C.; Zhang, J.; Siepker, C.; Mainenti, M.; Li, G.; Thompson, A.C.; et al. Highly Pathogenic Avian Influenza A(H5N1) Clade 2.3.4.4b Virus Infection in Domestic Dairy Cattle and Cats, United States, 2024. Emerg. Infect. Dis. 2024, 30, 1335–1343. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wu, X.; Cheng, Y.; An, Y.; Ning, Z. Tissue distribution of human and avian type sialic acid influenza virus receptors in domestic cat. Acta Vet. Hung. 2013, 61, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Rimmelzwaan, G.F.; van Riel, D.; Baars, M.; Bestebroer, T.M.; van Amerongen, G.; Fouchier, R.A.; Osterhaus, A.D.; Kuiken, T. Influenza A virus (H5N1) infection in cats causes systemic disease with potential novel routes of virus spread within and between hosts. Am. J. Pathol. 2006, 168, 176–183, quiz 364. [Google Scholar] [CrossRef] [PubMed]
- Kuchipudi, S.V.; Nelli, R.K.; Gontu, A.; Satyakumar, R.; Surendran Nair, M.; Subbiah, M. Sialic Acid Receptors: The Key to Solving the Enigma of Zoonotic Virus Spillover. Viruses 2021, 13, 262. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, C.A.; Walker, R.V.; Bannister, G.L. Preliminary Experiments Relating to the Propagation of Viruses in the Bovine Mammary Gland. Can. J. Comp. Med. Vet. Sci. 1953, 17, 97–104. [Google Scholar] [PubMed]
- Paquette, S.G.; Banner, D.; Huang, S.S.; Almansa, R.; Leon, A.; Xu, L.; Bartoszko, J.; Kelvin, D.J.; Kelvin, A.A. Influenza Transmission in the Mother-Infant Dyad Leads to Severe Disease, Mammary Gland Infection, and Pathogenesis by Regulating Host Responses. PLoS Pathog. 2015, 11, e1005173. [Google Scholar] [CrossRef] [PubMed]
- Le Sage, V.; Campbell, A.J.; Reed, D.; Duprex, W.P.; Lakdawala, S. Influenza H5N1 and H1N1 viruses remain infectious in unpasteurized milk on milking machinery surfaces. medRxiv 2024. [Google Scholar] [CrossRef]
- Guan, L.; Eisfeld, A.J.; Pattinson, D.; Gu, C.; Biswas, A.; Maemura, T.; Trifkovic, S.; Babujee, L.; Presler, R.; Dahn, R.; et al. Cow’s Milk Containing Avian Influenza A(H5N1) Virus—Heat Inactivation and Infectivity in Mice. N. Engl. J. Med. 2024, 391, 87–90. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J. Bird Flu May be Spreading in Cows via Milking and Herd Transport. Science Insider News 2024. [Google Scholar] [CrossRef]
- Ssematimba, A.; Hagenaars, T.J.; de Jong, M.C. Modelling the wind-borne spread of highly pathogenic avian influenza virus between farms. PLoS ONE 2012, 7, e31114. [Google Scholar] [CrossRef] [PubMed]
- Wanaratana, S.; Panyim, S.; Pakpinyo, S. The potential of house flies to act as a vector of avian influenza subtype H5N1 under experimental conditions. Med. Vet. Entomol. 2011, 25, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Kupferschmidt, K. To combat cow flu outbreak, scientists plan to infect cattle with influenza in high-security labs. Sci. Insid. News 2024. [Google Scholar] [CrossRef]
- Baker, A.L.; Arruda, B.; Palmer, M.V.; Boggiatto, P.; Sarlo Davila, K.; Buckley, A.; Ciacci Zanella, G.; Snyder, C.A.; Anderson, T.K.; Hutter, C.; et al. Experimental reproduction of viral replication and disease in dairy calves and lactating cows inoculated with highly pathogenic avian influenza H5N1 clade 2.3.4.4b. bioRxiv 2024. [Google Scholar] [CrossRef]
- Kristensen, C.; Jensen, H.E.; Trebbien, R.; Webby, R.J.; Larsen, L.E. The avian and human influenza A virus receptors sialic acid (SA)-α2,3 and SA-α2,6 are widely expressed in the bovine mammary gland. bioRxiv 2024. [Google Scholar] [CrossRef]
- Nwosu, C.C.; Aldredge, D.L.; Lee, H.; Lerno, L.A.; Zivkovic, A.M.; German, J.B.; Lebrilla, C.B. Comparison of the human and bovine milk N-glycome via high-performance microfluidic chip liquid chromatography and tandem mass spectrometry. J. Proteome Res. 2012, 11, 2912–2924. [Google Scholar] [CrossRef] [PubMed]
- Takimori, S.; Shimaoka, H.; Furukawa, J.; Yamashita, T.; Amano, M.; Fujitani, N.; Takegawa, Y.; Hammarström, L.; Kacskovics, I.; Shinohara, Y.; et al. Alteration of the N-glycome of bovine milk glycoproteins during early lactation. FEBS J. 2011, 278, 3769–3781. [Google Scholar] [CrossRef] [PubMed]
- Tao, N.; DePeters, E.J.; German, J.B.; Grimm, R.; Lebrilla, C.B. Variations in bovine milk oligosaccharides during early and middle lactation stages analyzed by high-performance liquid chromatography-chip/mass spectrometry. J. Dairy Sci. 2009, 92, 2991–3001. [Google Scholar] [CrossRef] [PubMed]
- Rios Carrasco, M.; Grone, A.; van den Brand, J.M.A.; de Vries, R.P. The mammary glands of cows abundantly display receptors for circulating avian H5 viruses. bioRxiv 2024. [Google Scholar] [CrossRef]
- Eisfeld, A.J.; Biswas, A.; Guan, L.; Gu, C.; Maemura, T.; Trifkovic, S.; Wang, T.; Babujee, L.; Dahn, R.; Halfmann, P.J.; et al. Pathogenicity and transmissibility of bovine H5N1 influenza virus. Nature 2024. Epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Pitino, M.A.; O’Connor, D.L.; McGeer, A.J.; Unger, S. The impact of thermal pasteurization on viral load and detectable live viruses in human milk and other matrices: A rapid review. Appl. Physiol. Nutr. Metab. 2021, 46, 10–26. [Google Scholar] [CrossRef] [PubMed]
- Jay, J.M.; Loessner, M.J.; Golden, D.A. (Eds.) Food Protection with High Temperatures, and Characteristics of Thermophilic Microorganisms. In Modern Food Microbiology; Springer: Boston, MA, USA, 2005; pp. 415–441. [Google Scholar]
- Chmielewski, R.A.; Beck, J.R.; Swayne, D.E. Thermal Inactivation of Avian Influenza Virus and Newcastle Disease Virus in a Fat-Free Egg Product. J. Food Prot. 2011, 74, 1161–1169. [Google Scholar] [CrossRef]
- Chmielewski, R.A.; Beck, J.R.; Juneja, V.K.; Swayne, D.E. Inactivation of low pathogenicity notifiable avian influenza virus and lentogenic Newcastle disease virus following pasteurization in liquid egg products. LWT-Food Sci. Technol. 2013, 52, 27–30. [Google Scholar] [CrossRef]
- Chmielewski, R.A.; Beck, J.R.; Swayne, D.E. Evaluation of the U.S. Department of Agriculture’s Egg Pasteurization Processes on the Inactivation of High-Pathogenicity Avian Influenza Virus and Velogenic Newcastle Disease Virus in Processed Egg Products. J. Food Prot. 2013, 76, 640–645. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Palme, D.I.; Lang, J.; Helke, D.; Kuryshko, M.; Abdelwhab, E.M. Strain-dependent variations in replication of European clade 2.3.4.4b influenza A(H5N1) viruses in bovine cells and thermal inactivation in semi-skimmed or whole milk. Eurosurveillance 2024, 29, 2400436. [Google Scholar] [CrossRef] [PubMed]
- Wallace, H.L.; Wight, J.; Dowding, B.; Etchebarne, M.B.; Flamand, L.; Hobman, T.; Jean, F.; Joy, J.B.; Lang, A.S.; McCormick, C.; et al. Longitudinal Influenza A Virus Screening of Retail Milk from Canadian Provinces (Rolling Updates). medRxiv 2024. [Google Scholar] [CrossRef]
- Blais-Savoie, J.; Yim, W.; Kotwa, J.D.; Yip, L.; Kozak, R.; McGeer, A.; Mubareka, S.D. Screening of retail milk in Ontario for the presence of influenza A. medRxiv 2024. [Google Scholar] [CrossRef]
- Uyeki, T.M.; Milton, S.; Hamid, C.A.; Webb, C.R.; Presley, S.M.; Shetty, V.; Rollo, S.N.; Martinez, D.L.; Rai, S.; Gonzales, E.R.; et al. Highly Pathogenic Avian Influenza A(H5N1) Virus Infection in a Dairy Farm Worker. N. Engl. J. Med. 2024, 390, 2028–2029. [Google Scholar] [CrossRef] [PubMed]
- Shittu, I.; Silva, D.; Oguzie, J.U.; Marushchak, L.V.; Olinger, G.G.; Lednicky, J.A.; Trujillo-Vargas, C.M.; Schneider, N.E.; Hao, H.; Gray, G.C. A One Health Investigation into H5N1 Avian Influenza Virus Epizootics on Two Dairy Farms. medRxiv 2024. [Google Scholar] [CrossRef]
- Focosi, D.; Spezia, P.G.; Maggi, F. Online dashboards for SARS-CoV-2 wastewater-based epidemiology. Future Microbiol. 2024, 19, 761–769. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, M.K.; Duong, D.; Shelden, B.; Chan, E.M.G.; Chan-Herur, V.; Hilton, S.; Paulos, A.H.; Zulli, A.; White, B.; Boehm, A. Detection of hemagglutinin H5 influenza A virus sequence in municipal wastewater solids at wastewater treatment plants with increases in influenza A in spring, 2024. Environ. Sci. Technol. Lett. 2024, 11, 526–532. [Google Scholar] [CrossRef]
- Tisza, M.J.; Hanson, B.; Clark, J.R.; Wang, L.; Payne, K.; Ross, M.C.; Mena, K.D.; Gitter, A.; Cregeen, S.J.J.; Cormier, J.J.; et al. Virome Sequencing Identifies H5N1 Avian Influenza in Wastewater from Nine Cities. medRxiv 2024. [Google Scholar] [CrossRef]
- Focosi, D.; Franchini, M.; Senefeld, J.W.; Joyner, M.J.; Sullivan, D.J.; Pekosz, A.; Maggi, F.; Casadevall, A. Passive immunotherapies for the next influenza pandemic. Rev. Med. Virol. 2024, 34, e2533. [Google Scholar] [CrossRef]
- Tan, S.K.; Cebrik, D.; Plotnik, D.; Agostini, M.L.; Boundy, K.; Hebner, C.M.; Yeh, W.W.; Pang, P.S.; Moya, J.; Fogarty, C.; et al. A Randomized, Placebo-Controlled Trial to Evaluate the Safety and Efficacy of VIR-2482 in Healthy Adults for Prevention of Influenza A Illness (PENINSULA). medRxiv 2024. [Google Scholar] [CrossRef] [PubMed]
- Belshe, R.B.; Frey, S.E.; Graham, I.; Mulligan, M.J.; Edupuganti, S.; Jackson, L.A.; Wald, A.; Poland, G.; Jacobson, R.; Keyserling, H.L.; et al. Safety and immunogenicity of influenza A H5 subunit vaccines: Effect of vaccine schedule and antigenic variant. J. Infect. Dis. 2011, 203, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Jennings, L.C.; Monto, A.S.; Chan, P.K.; Szucs, T.D.; Nicholson, K.G. Stockpiling prepandemic influenza vaccines: A new cornerstone of pandemic preparedness plans. Lancet Infect. Dis. 2008, 8, 650–658. [Google Scholar] [CrossRef] [PubMed]
- Treanor, J.J.; Campbell, J.D.; Zangwill, K.M.; Rowe, T.; Wolff, M. Safety and immunogenicity of an inactivated subvirion influenza A (H5N1) vaccine. New Engl. J. Med. 2006, 354, 1343–1351. [Google Scholar] [CrossRef] [PubMed]
- Treanor, J.J.; Wilkinson, B.E.; Masseoud, F.; Hu-Primmer, J.; Battaglia, R.; O’Brien, D.; Wolff, M.; Rabinovich, G.; Blackwelder, W.; Katz, J.M. Safety and immunogenicity of a recombinant hemagglutinin vaccine for H5 influenza in humans. Vaccine 2001, 19, 1732–1737. [Google Scholar] [CrossRef] [PubMed]
- Wimmers, F.; Donato, M.; Kuo, A.; Ashuach, T.; Gupta, S.; Li, C.; Dvorak, M.; Foecke, M.H.; Chang, S.E.; Hagan, T.; et al. The single-cell epigenomic and transcriptional landscape of immunity to influenza vaccination. Cell 2021, 184, 3915–3935.e3921. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.S.; Montomoli, E.; Pasini, F.L.; Steinman, L. The Safety of Adjuvanted Vaccines Revisited: Vaccine-Induced Narcolepsy. Isr. Med. Assoc. J. 2016, 18, 216–220. [Google Scholar] [PubMed]
- Ahmed, S.S.; Volkmuth, W.; Duca, J.; Corti, L.; Pallaoro, M.; Pezzicoli, A.; Karle, A.; Rigat, F.; Rappuoli, R.; Narasimhan, V.; et al. Antibodies to influenza nucleoprotein cross-react with human hypocretin receptor 2. Sci. Transl. Med. 2015, 7, 294ra105. [Google Scholar] [CrossRef]
- Luo, G.; Ambati, A.; Lin, L.; Bonvalet, M.; Partinen, M.; Ji, X.; Maecker, H.T.; Mignot, E.J. Autoimmunity to hypocretin and molecular mimicry to flu in type 1 narcolepsy. Proc. Natl. Acad. Sci. USA 2018, 115, E12323–E12332. [Google Scholar] [CrossRef] [PubMed]
- Vaarala, O.; Vuorela, A.; Partinen, M.; Baumann, M.; Freitag, T.L.; Meri, S.; Saavalainen, P.; Jauhiainen, M.; Soliymani, R.; Kirjavainen, T.; et al. Antigenic differences between AS03 adjuvanted influenza A (H1N1) pandemic vaccines: Implications for pandemrix-associated narcolepsy risk. PLoS ONE 2014, 9, e114361. [Google Scholar] [CrossRef] [PubMed]
- Vogel, O.A.; Manicassamy, B. Broadly Protective Strategies Against Influenza Viruses: Universal Vaccines and Therapeutics. Front. Microbiol. 2020, 11, 135. [Google Scholar] [CrossRef] [PubMed]
- Booth, C.M.; Matukas, L.M.; Tomlinson, G.A.; Rachlis, A.R.; Rose, D.B.; Dwosh, H.A.; Walmsley, S.L.; Mazzulli, T.; Avendano, M.; Derkach, P.; et al. Clinical features and short-term outcomes of 144 patients with SARS in the greater Toronto area. JAMA 2003, 289, 2801–2809. [Google Scholar] [CrossRef] [PubMed]
- Bermingham, A.; Chand, M.A.; Brown, C.S.; Aarons, E.; Tong, C.; Langrish, C.; Hoschler, K.; Brown, K.; Galiano, M.; Myers, R.; et al. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill. 2012, 17, 20290. [Google Scholar] [CrossRef] [PubMed]
- McVernon, J.; Nolan, T. Panvax(®): A monovalent inactivated unadjuvanted vaccine against pandemic influenza A (H1N1) 2009. Expert Rev. Vaccines 2011, 10, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Kosalaraksa, P.; Jeanfreau, R.; Frenette, L.; Drame, M.; Madariaga, M.; Innis, B.L.; Godeaux, O.; Izurieta, P.; Vaughn, D.W. AS03B-adjuvanted H5N1 influenza vaccine in children 6 months through 17 years of age: A phase 2/3 randomized, placebo-controlled, observer-blinded trial. J. Infect. Dis. 2015, 211, 801–810. [Google Scholar] [CrossRef]
- Standaert, B.; Dort, T.; Linden, J.; Madan, A.; Bart, S.; Chu, L.; Hayney, M.S.; Kosinski, M.; Kroll, R.; Malak, J.; et al. Usability of daily SF36 questionnaires to capture the QALD variation experienced after vaccination with AS03(A)-adjuvanted monovalent influenza A (H5N1) vaccine in a safety and tolerability study. Health Qual. Life Outcomes 2019, 17, 80. [Google Scholar] [CrossRef] [PubMed]
- Gillard, P.; Chu, D.W.; Hwang, S.J.; Yang, P.C.; Thongcharoen, P.; Lim, F.S.; Dramé, M.; Walravens, K.; Roman, F. Long-term booster schedules with AS03A-adjuvanted heterologous H5N1 vaccines induces rapid and broad immune responses in Asian adults. BMC Infect. Dis. 2014, 14, 142. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.J.; Chang, S.C.; Yu, C.J.; Chan, Y.J.; Chen, T.J.; Hsieh, S.L.; Lai, H.Y.; Lin, M.H.; Liu, J.Y.; Ong, G.; et al. Immunogenicity and safety of an AS03(A)-adjuvanted H5N1 influenza vaccine in a Taiwanese population. J. Formos. Med. Assoc. 2011, 110, 780–786. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Leroux-Roels, I.; Borkowski, A.; Vanwolleghem, T.; Dramé, M.; Clement, F.; Hons, E.; Devaster, J.-M.; Leroux-Roels, G. Antigen sparing and cross-reactive immunity with an adjuvanted rH5N1 prototype pandemic influenza vaccine: A randomised controlled trial. Lancet 2007, 370, 580–589. [Google Scholar] [CrossRef] [PubMed]
- Leroux-Roels, I.; Bernhard, R.; Gérard, P.; Dramé, M.; Hanon, E.; Leroux-Roels, G. Broad Clade 2 Cross-Reactive Immunity Induced by an Adjuvanted Clade 1 rH5N1 Pandemic Influenza Vaccine. PLoS ONE 2008, 3, e1665. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.H.; Jackson, L.A.; Edwards, K.M.; Keitel, W.A.; Hill, H.; Noah, D.L.; Creech, C.B.; Patel, S.M.; Mangal, B.; Kotloff, K.L. Safety, Reactogenicity, and Immunogenicity of Inactivated Monovalent Influenza A(H5N1) Virus Vaccine Administered with or Without AS03 Adjuvant. Open Forum Infect. Dis. 2014, 1, ofu091. [Google Scholar] [CrossRef] [PubMed]
- Nuwarda, R.F.; Alharbi, A.A.; Kayser, V. An Overview of Influenza Viruses and Vaccines. Vaccines 2021, 9, 1032. [Google Scholar] [CrossRef] [PubMed]
- Yamayoshi, S.; Kawaoka, Y. Current and future influenza vaccines. Nat. Med. 2019, 25, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Gunjan, K.; Pandey, R.P.; Himanshu; Mukherjee, R.; Chang, C.-M. Navigating the global egg shortage: A comprehensive study of interconnected challenges. medRxiv 2024. [Google Scholar] [CrossRef]
- Focosi, D. An inactivated subvirion influenza A (H5N1) vaccine. N. Engl. J. Med. 2006, 354, 2724–2725. [Google Scholar] [PubMed]
- Zhou, F.; Hansen, L.; Pedersen, G.; Grødeland, G.; Cox, R. Matrix M Adjuvanted H5N1 Vaccine Elicits Broadly Neutralizing Antibodies and Neuraminidase Inhibiting Antibodies in Humans That Correlate With In Vivo Protection. Front. Immunol. 2021, 12, 747774. [Google Scholar] [CrossRef] [PubMed]
- Endo, M.; Tanishima, M.; Ibaragi, K.; Hayashida, K.; Fukuda, T.; Tanabe, T.; Naruse, T.; Kino, Y.; Ueda, K. Clinical phase II and III studies of an AS03-adjuvanted H5N1 influenza vaccine produced in an EB66(®) cell culture platform. Influenza Other Respir. Viruses 2020, 14, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Chanthavanich, P.; Versage, E.; Van Twuijver, E.; Hohenboken, M. Antibody responses against heterologous A/H5N1 strains for an MF59-adjuvanted cell culture-derived A/H5N1 (aH5N1c) influenza vaccine in healthy pediatric subjects. Vaccine 2021, 39, 6930–6935. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, K.; Migueles, S.A.; Huang, J.; Bolkhovitinov, L.; Stuccio, S.; Griesman, T.; Pullano, A.A.; Kang, B.H.; Ishida, E.; Zimmerman, M.; et al. A replication-competent adenovirus-vectored influenza vaccine induces durable systemic and mucosal immunity. J. Clin. Investig. 2021, 131, e140794. [Google Scholar] [CrossRef]
- Nicolodi, C.; Groiss, F.; Kiselev, O.; Wolschek, M.; Seipelt, J.; Muster, T. Safety and immunogenicity of a replication-deficient H5N1 influenza virus vaccine lacking NS1. Vaccine 2019, 37, 3722–3729. [Google Scholar] [CrossRef] [PubMed]
- Talaat, K.R.; Luke, C.J.; Khurana, S.; Manischewitz, J.; King, L.R.; McMahon, B.A.; Karron, R.A.; Lewis, K.D.; Qin, J.; Follmann, D.A.; et al. A live attenuated influenza A(H5N1) vaccine induces long-term immunity in the absence of a primary antibody response. J. Infect. Dis. 2014, 209, 1860–1869. [Google Scholar] [CrossRef] [PubMed]
- Daulagala, P.; Cheng, S.M.S.; Chin, A.; Luk, L.L.H.; Leung, K.; Wu, J.T.; Poon, L.L.M.; Peiris, M.; Yen, H.L. Avian Influenza A(H5N1) Neuraminidase Inhibition Antibodies in Healthy Adults after Exposure to Influenza A(H1N1)pdm09. Emerg. Infect. Dis. 2024, 30, 168–171. [Google Scholar] [CrossRef] [PubMed]
- Koutsakos, M.; Rockman, S.; Krammer, F. Is eradication of influenza B viruses possible? Lancet Infect. Dis. 2024, 24, 451–453. [Google Scholar] [CrossRef] [PubMed]
- Arevalo, C.P.; Bolton, M.J.; Le Sage, V.; Ye, N.; Furey, C.; Muramatsu, H.; Alameh, M.G.; Pardi, N.; Drapeau, E.M.; Parkhouse, K.; et al. A multivalent nucleoside-modified mRNA vaccine against all known influenza virus subtypes. Science 2022, 378, 899–904. [Google Scholar] [CrossRef] [PubMed]
- Widge, A.T.; Hofstetter, A.R.; Houser, K.V.; Awan, S.F.; Chen, G.L.; Burgos Florez, M.C.; Berkowitz, N.M.; Mendoza, F.; Hendel, C.S.; Holman, L.A.; et al. An influenza hemagglutinin stem nanoparticle vaccine induces cross-group 1 neutralizing antibodies in healthy adults. Sci. Transl. Med. 2023, 15, eade4790. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S.F.; Cominsky, L.Y.; Shimberg, G.D.; Gillespie, R.A.; Gorman, J.; Raab, J.E.; Brand, J.; Creanga, A.; Gajjala, S.R.; Narpala, S.; et al. An influenza H1 hemagglutinin stem-only immunogen elicits a broadly cross-reactive B cell response in humans. Sci. Transl. Med. 2023, 15, eade4976. [Google Scholar] [CrossRef] [PubMed]
- Dolgin, E. Self-copying RNA vaccine wins first full approval: What’s next? Nature 2023, 624, 236–237. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.; Music, N.; Cheung, M.; Rossignol, E.; Bedi, S.; Patel, H.; Safari, M.; Lee, C.; Otten, G.R.; Settembre, E.C.; et al. Self-amplifying mRNA bicistronic influenza vaccines raise cross-reactive immune responses in mice and prevent infection in ferrets. Mol. Ther.-Methods Clin. Dev. 2022, 27, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Feldman, R.A.; Fuhr, R.; Smolenov, I.; Mick Ribeiro, A.; Panther, L.; Watson, M.; Senn, J.J.; Smith, M.; Almarsson, Ö.; Pujar, H.S.; et al. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine 2019, 37, 3326–3334. [Google Scholar] [CrossRef] [PubMed]
- Baj, A.; Gasperina, D.D.; Focosi, D.; Forlani, G.; Ferrante, F.D.; Novazzi, F.; Azzi, L.; Maggi, F. Safety and immunogenicity of synchronous COVID19 and influenza vaccination. J. Clin. Virol. Plus 2022, 2, 100082. [Google Scholar] [CrossRef] [PubMed]
- Vistoli, F.; Focosi, D.; De Donno, M.; Giannecchini, S.; Mariotti, M.L.; Occhipinti, M.; Barsotti, M.; Duquesnoy, R.; Marchetti, P.; Scatena, F.; et al. Pancreas rejection after pandemic influenzavirus A(H1N1) vaccination or infection: A report of two cases. Transpl. Int. Off. J. Eur. Soc. Organ Transplant. 2011, 24, e28–e29. [Google Scholar] [CrossRef]
- Focosi, D.; Caracciolo, F.; Galimberti, S.; Papineschi, F.; Petrini, M. False positive PET scanning caused by inactivated influenza virus vaccination during complete remission from anaplastic T-cell lymphoma. Ann. Hematol. 2008, 87, 343–344. [Google Scholar] [CrossRef] [PubMed]
- Zeller, M.A.; Ma, J.; Wong, F.Y.; Tum, S.; Hidano, A.; Holt, H.; Chhay, T.; Sorn, S.; Koeut, D.; Seng, B.; et al. The genomic landscape of swine influenza A viruses in Southeast Asia. Proc. Natl. Acad. Sci. USA 2023, 120, e2301926120. [Google Scholar] [CrossRef] [PubMed]
- Coleman, K.K.; Bemis, I.G. Avian Influenza Virus Infections in Felines: A Systematic Review of Two Decades of Literature. medRxiv 2024. [Google Scholar] [CrossRef]
- Chen, Y.; Zhong, G.; Wang, G.; Deng, G.; Li, Y.; Shi, J.; Zhang, Z.; Guan, Y.; Jiang, Y.; Bu, Z.; et al. Dogs are highly susceptible to H5N1 avian influenza virus. Virology 2010, 405, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Pearson, H. Cows could foster flu pandemics. Nature News 2002. [Google Scholar] [CrossRef]
- Kwon, T.; Trujillo, J.D.; Carossino, M.; Lyoo, E.L.; McDowell, C.D.; Cool, K.; Matias-Ferreyra, F.S.; Jeevan, T.; Morozov, I.; Gaudreault, N.N.; et al. Pigs are highly susceptible to but do not transmit mink-derived highly pathogenic avian influenza virus H5N1 clade 2.3.4.4b. Emerg. Microbes Infect. 2024, 13, 2353292. [Google Scholar] [CrossRef] [PubMed]
- WHO Disease Outbreak News. Avian Influenza A (H5N2)—Mexico. Available online: https://www.who.int/emergencies/disease-outbreak-news/item/2024-DON520 (accessed on 6 June 2024).
- Staller, E.; Carrique, L.; Swann, O.C.; Fan, H.; Keown, J.R.; Sheppard, C.M.; Barclay, W.S.; Grimes, J.M.; Fodor, E. Structures of H5N1 influenza polymerase with ANP32B reveal mechanisms of genome replication and host adaptation. Nat. Commun. 2024, 15, 4123. [Google Scholar] [CrossRef]
- Briggs, K.; Kapczynski, D.R. Comparative analysis of PB2 residue 627E/K/V in H5 subtypes of avian influenza viruses isolated from birds and mammals. Front. Vet. Sci. 2023, 10, 1250952. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, C.M.; Goldhill, D.H.; Swann, O.C.; Staller, E.; Penn, R.; Platt, O.K.; Sukhova, K.; Baillon, L.; Frise, R.; Peacock, T.P.; et al. An Influenza A virus can evolve to use human ANP32E through altering polymerase dimerization. Nat. Commun. 2023, 14, 6135. [Google Scholar] [CrossRef] [PubMed]
- Dadonaite, B.; Ahn, J.J.; Ort, J.T.; Yu, J.; Furey, C.; Dosey, A.; Hannon, W.W.; Baker, A.V.; Webby, R.J.; King, N.P.; et al. Deep mutational scanning of H5 hemagglutinin to inform influenza virus surveillance. bioRxiv 2024. [Google Scholar] [CrossRef]
- Teo, Q.W.; Wang, Y.; Lv, H.; Mao, K.J.; Tan, T.J.C.; Huan, Y.W.; Rivera-Cardona, J.; Shao, E.K.; Choi, D.; Dargani, Z.T.; et al. Deep mutational scanning of influenza A virus NEP reveals pleiotropic mutations in its N-terminal domain. bioRxiv 2024. [Google Scholar] [CrossRef]
- Long, J.S.; Mistry, B.; Haslam, S.M.; Barclay, W.S. Host and viral determinants of influenza A virus species specificity. Nat. Rev. Microbiol. 2019, 17, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Pinto, R.M.; Bakshi, S.; Lytras, S.; Zakaria, M.K.; Swingler, S.; Worrell, J.C.; Herder, V.; Hargrave, K.E.; Varjak, M.; Cameron-Ruiz, N.; et al. BTN3A3 evasion promotes the zoonotic potential of influenza A viruses. Nature 2023, 619, 338–347. [Google Scholar] [CrossRef]
- Zúñiga, S.; Sola, I.; Moreno, J.L.; Sabella, P.; Plana-Durán, J.; Enjuanes, L. Coronavirus nucleocapsid protein is an RNA chaperone. Virology 2007, 357, 215–227. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Focosi, D.; Maggi, F. Avian Influenza Virus A(H5Nx) and Prepandemic Candidate Vaccines: State of the Art. Int. J. Mol. Sci. 2024, 25, 8550. https://doi.org/10.3390/ijms25158550
Focosi D, Maggi F. Avian Influenza Virus A(H5Nx) and Prepandemic Candidate Vaccines: State of the Art. International Journal of Molecular Sciences. 2024; 25(15):8550. https://doi.org/10.3390/ijms25158550
Chicago/Turabian StyleFocosi, Daniele, and Fabrizio Maggi. 2024. "Avian Influenza Virus A(H5Nx) and Prepandemic Candidate Vaccines: State of the Art" International Journal of Molecular Sciences 25, no. 15: 8550. https://doi.org/10.3390/ijms25158550
APA StyleFocosi, D., & Maggi, F. (2024). Avian Influenza Virus A(H5Nx) and Prepandemic Candidate Vaccines: State of the Art. International Journal of Molecular Sciences, 25(15), 8550. https://doi.org/10.3390/ijms25158550







