Transcriptional Profiling of Staphylococcus aureus during the Transition from Asymptomatic Nasal Colonization to Skin Colonization/Infection in Patients with Atopic Dermatitis
Abstract
1. Introduction
2. Results and Discussion
2.1. Characteristics of Nasal and Skin Specimens from Patients with AD
2.2. In Vivo Transcriptional Profiling of S. aureus during Asymptomatic Nasal Colonization to Skin Colonization/Infection in Patients with AD
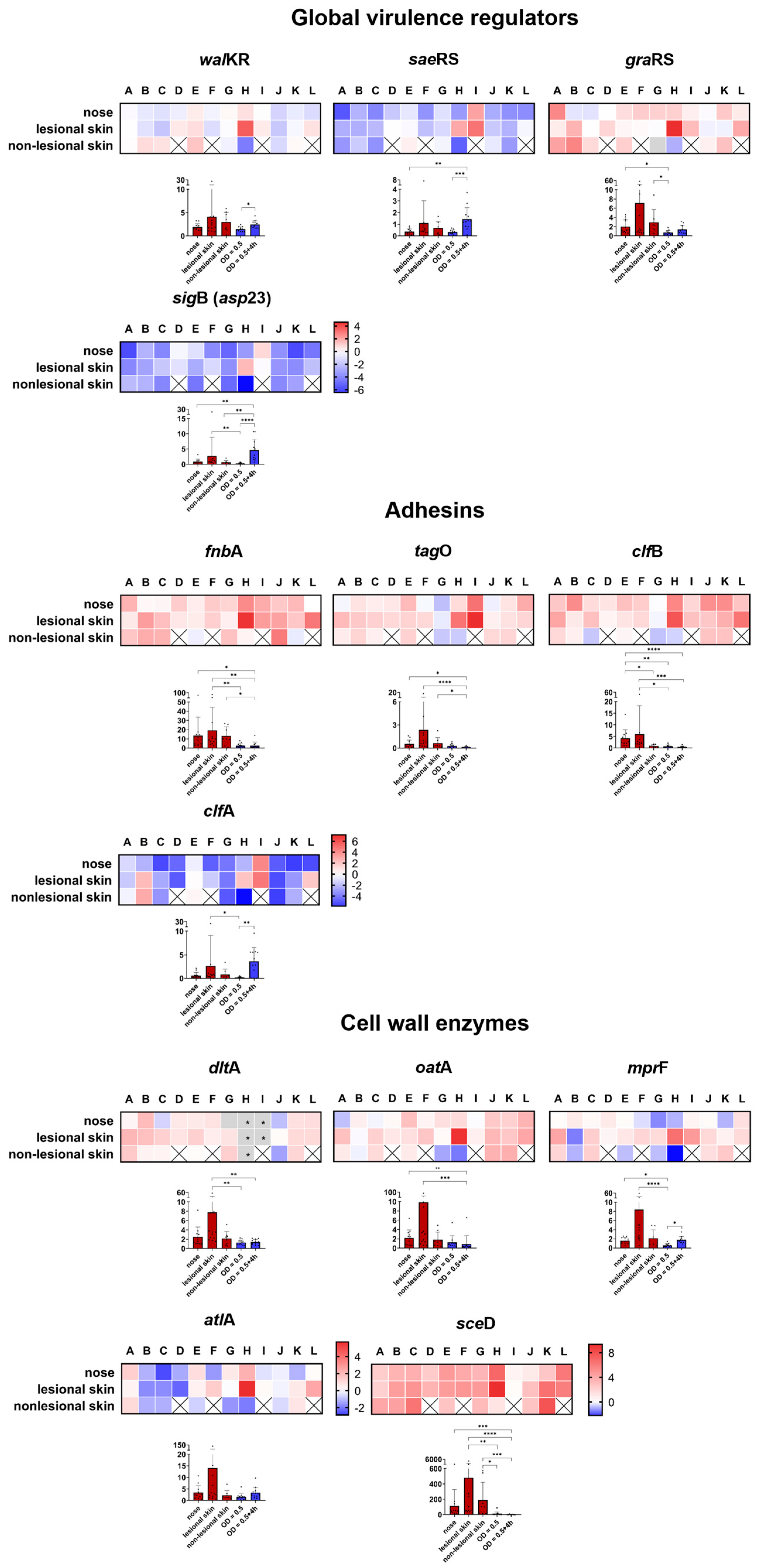
2.2.1. S. aureus Agr Activity Is Increased in the Skin Samples of Some Patients with AD
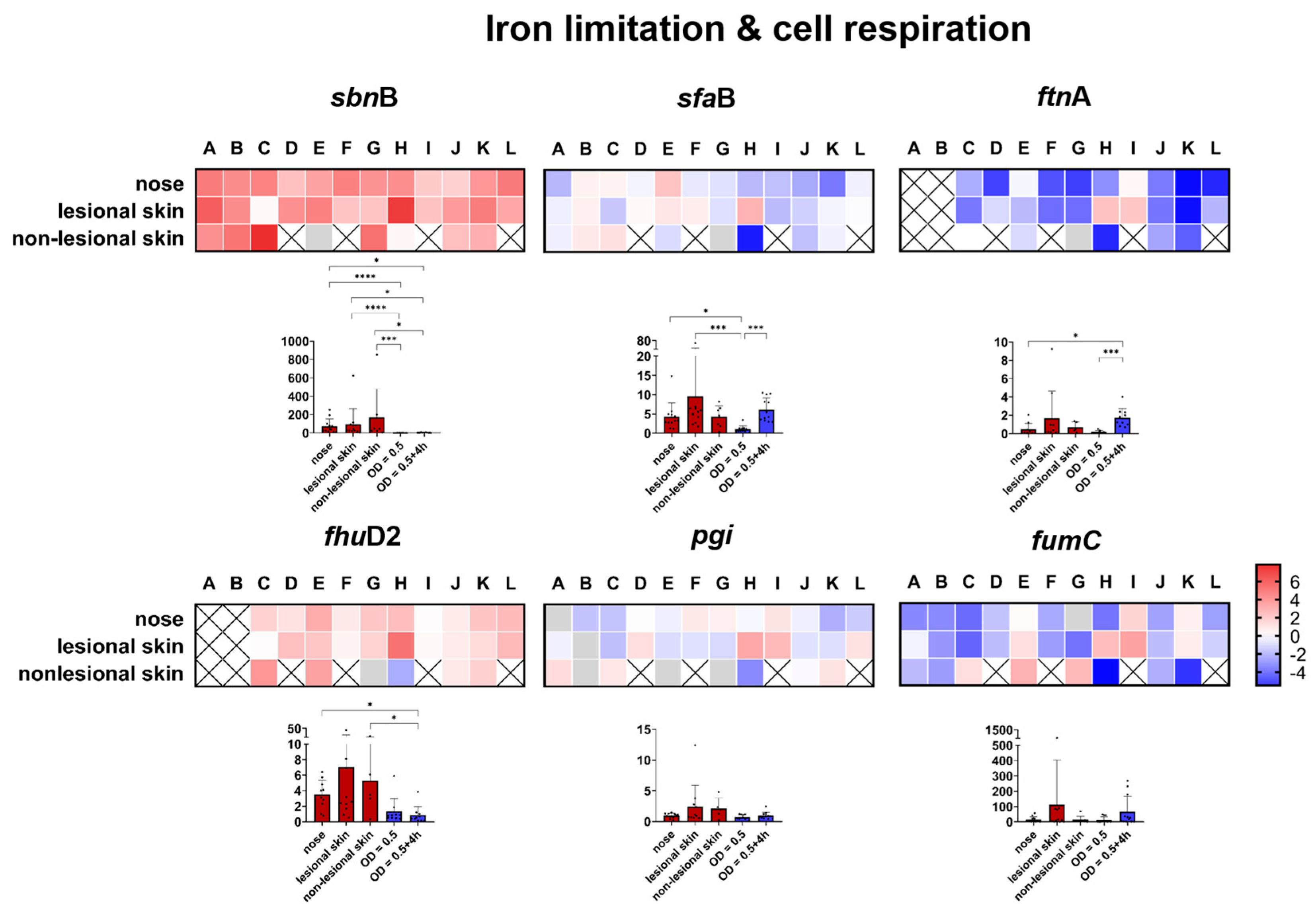
2.2.2. Gene Expression in the Nasal Cavity and Skin Environment Appears Similar
Global Transcriptional Regulators
Adhesins
Cell Wall Modification Enzymes
Immune Evasion
Proteases
Genes Involved in Iron Limitation and Cell Respiration
3. Materials and Methods
3.1. Ethics Statement
3.2. Study Population
3.3. Sampling, Bacteriological Analysis, and Growth of S. aureus Strains
3.4. RNA Isolation, Reverse Transcription, and Quantitative Real-Time PCR
3.5. Spa Genotyping
3.6. Data Visualization and Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wertheim, H.F.; Melles, D.C.; Vos, M.C.; van Leeuwen, W.; van Belkum, A.; Verbrugh, H.A.; Nouwen, J.L. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 2005, 5, 751–762. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.E. Healthy carriage of Staphylococcus aureus: Its prevalence and importance. Bacteriol. Rev. 1963, 27, 56–71. [Google Scholar] [CrossRef] [PubMed]
- van Belkum, A.; Verkaik, N.J.; de Vogel, C.P.; Boelens, H.A.; Verveer, J.; Nouwen, J.L.; Verbrugh, H.A.; Wertheim, H.F. Reclassification of Staphylococcus aureus nasal carriage types. J. Infect. Dis. 2009, 199, 1820–1826. [Google Scholar] [CrossRef]
- von Eiff, C.; Becker, K.; Machka, K.; Stammer, H.; Peters, G. Nasal carriage as a source of Staphylococcus aureus bacteremia. Study Group. N. Engl. J. Med. 2001, 344, 11–16. [Google Scholar] [CrossRef]
- Wertheim, H.F.; Vos, M.C.; Ott, A.; van Belkum, A.; Voss, A.; Kluytmans, J.A.; van Keulen, P.H.; Vandenbroucke-Grauls, C.M.; Meester, M.H.; Verbrugh, H.A. Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 2004, 364, 703–705. [Google Scholar] [CrossRef]
- Matsui, K.; Nishikawa, A.; Suto, H.; Tsuboi, R.; Ogawa, H. Comparative study of Staphylococcus aureus isolated from lesional and non-lesional skin of atopic dermatitis patients. Microbiol. Immunol. 2000, 44, 945–947. [Google Scholar] [CrossRef] [PubMed]
- Totte, J.E.; van der Feltz, W.T.; Hennekam, M.; van Belkum, A.; van Zuuren, E.J.; Pasmans, S.G. Prevalence and odds of Staphylococcus aureus carriage in atopic dermatitis: A systematic review and meta-analysis. Br. J. Dermatol. 2016, 175, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.H.; Oh, J.; Deming, C.; Conlan, S.; Grice, E.A.; Beatson, M.A.; Nomicos, E.; Polley, E.C.; Komarow, H.D.; Program, N.C.S.; et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012, 22, 850–859. [Google Scholar] [CrossRef]
- Byrd, A.L.; Deming, C.; Cassidy, S.K.B.; Harrison, O.J.; Ng, W.I.; Conlan, S.; Program, N.C.S.; Belkaid, Y.; Segre, J.A.; Kong, H.H. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci. Transl. Med. 2017, 9, eaal4651. [Google Scholar] [CrossRef]
- Tauber, M.; Balica, S.; Hsu, C.Y.; Jean-Decoster, C.; Lauze, C.; Redoules, D.; Viode, C.; Schmitt, A.M.; Serre, G.; Simon, M.; et al. Staphylococcus aureus density on lesional and nonlesional skin is strongly associated with disease severity in atopic dermatitis. J. Allergy Clin. Immunol. 2016, 137, 1272–1274.e3. [Google Scholar] [CrossRef]
- Langan, S.M.; Irvine, A.D. Childhood eczema and the importance of the physical environment. J. Investig. Dermatol. 2013, 133, 1706–1709. [Google Scholar] [CrossRef]
- Irvine, A.D.; McLean, W.H.; Leung, D.Y. Filaggrin mutations associated with skin and allergic diseases. N. Engl. J. Med. 2011, 365, 1315–1327. [Google Scholar] [CrossRef]
- Salimi, M.; Barlow, J.L.; Saunders, S.P.; Xue, L.; Gutowska-Owsiak, D.; Wang, X.; Huang, L.C.; Johnson, D.; Scanlon, S.T.; McKenzie, A.N.; et al. A role for IL-25 and IL-33-driven type-2 innate lymphoid cells in atopic dermatitis. J. Exp. Med. 2013, 210, 2939–2950. [Google Scholar] [CrossRef]
- Furue, M.; Chiba, T.; Tsuji, G.; Ulzii, D.; Kido-Nakahara, M.; Nakahara, T.; Kadono, T. Atopic dermatitis: Immune deviation, barrier dysfunction, IgE autoreactivity and new therapies. Allergol. Int. 2017, 66, 398–403. [Google Scholar] [CrossRef]
- Bieber, T. Atopic dermatitis. N. Engl. J. Med. 2008, 358, 1483–1494. [Google Scholar] [CrossRef] [PubMed]
- Weidinger, S.; Novak, N. Atopic dermatitis. Lancet 2016, 387, 1109–1122. [Google Scholar] [CrossRef]
- Silverberg, J.I. Public Health Burden and Epidemiology of Atopic Dermatitis. Dermatol. Clin. 2017, 35, 283–289. [Google Scholar] [CrossRef]
- Jenul, C.; Horswill, A.R. Regulation of Staphylococcus aureus Virulence. Microbiol. Spectr. 2019, 7, 1–21. [Google Scholar] [CrossRef]
- Bleul, L.; Francois, P.; Wolz, C. Two-Component Systems of S. aureus: Signaling and Sensing Mechanisms. Genes 2021, 13, 34. [Google Scholar] [CrossRef]
- Thoendel, M.; Kavanaugh, J.S.; Flack, C.E.; Horswill, A.R. Peptide signaling in the staphylococci. Chem. Rev. 2011, 111, 117–151. [Google Scholar] [CrossRef]
- Cheung, A.L.; Bayer, A.S.; Zhang, G.; Gresham, H.; Xiong, Y.Q. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 2004, 40, 1–9. [Google Scholar] [CrossRef]
- Novick, R.P. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 2003, 48, 1429–1449. [Google Scholar] [CrossRef] [PubMed]
- Brauweiler, A.M.; Goleva, E.; Leung, D.Y.M. Th2 cytokines increase Staphylococcus aureus alpha toxin-induced keratinocyte death through the signal transducer and activator of transcription 6 (STAT6). J. Investig. Dermatol. 2014, 134, 2114–2121. [Google Scholar] [CrossRef] [PubMed]
- Brauweiler, A.M.; Bin, L.; Kim, B.E.; Oyoshi, M.K.; Geha, R.S.; Goleva, E.; Leung, D.Y. Filaggrin-dependent secretion of sphingomyelinase protects against staphylococcal alpha-toxin-induced keratinocyte death. J. Allergy Clin. Immunol. 2013, 131, 421–427.e2. [Google Scholar] [CrossRef]
- Breuer, K.; Wittmann, M.; Kempe, K.; Kapp, A.; Mai, U.; Dittrich-Breiholz, O.; Kracht, M.; Mrabet-Dahbi, S.; Werfel, T. Alpha-toxin is produced by skin colonizing Staphylococcus aureus and induces a T helper type 1 response in atopic dermatitis. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2005, 35, 1088–1095. [Google Scholar] [CrossRef]
- Queck, S.Y.; Jameson-Lee, M.; Villaruz, A.E.; Bach, T.H.; Khan, B.A.; Sturdevant, D.E.; Ricklefs, S.M.; Li, M.; Otto, M. RNAIII-independent target gene control by the agr quorum-sensing system: Insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 2008, 32, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Matsumoto, M.; Katayama, Y.; Oguma, R.; Wakabayashi, S.; Nygaard, T.; Saijo, S.; Inohara, N.; Otto, M.; Matsue, H.; et al. Staphylococcus aureus Virulent PSMalpha Peptides Induce Keratinocyte Alarmin Release to Orchestrate IL-17-Dependent Skin Inflammation. Cell Host Microbe 2017, 22, 667–677.e5. [Google Scholar] [CrossRef]
- Liu, H.; Archer, N.K.; Dillen, C.A.; Wang, Y.; Ashbaugh, A.G.; Ortines, R.V.; Kao, T.; Lee, S.K.; Cai, S.S.; Miller, R.J.; et al. Staphylococcus aureus Epicutaneous Exposure Drives Skin Inflammation via IL-36-Mediated T Cell Responses. Cell Host Microbe 2017, 22, 653–666.e5. [Google Scholar] [CrossRef]
- Syed, A.K.; Reed, T.J.; Clark, K.L.; Boles, B.R.; Kahlenberg, J.M. Staphlyococcus aureus phenol-soluble modulins stimulate the release of proinflammatory cytokines from keratinocytes and are required for induction of skin inflammation. Infect. Immun. 2015, 83, 3428–3437. [Google Scholar] [CrossRef]
- Nakamura, Y.; Takahashi, H.; Takaya, A.; Inoue, Y.; Katayama, Y.; Kusuya, Y.; Shoji, T.; Takada, S.; Nakagawa, S.; Oguma, R.; et al. Staphylococcus Agr virulence is critical for epidermal colonization and associates with atopic dermatitis development. Sci. Transl. Med. 2020, 12, eaay4068. [Google Scholar] [CrossRef]
- Burian, M.; Rautenberg, M.; Kohler, T.; Fritz, M.; Krismer, B.; Unger, C.; Hoffmann, W.H.; Peschel, A.; Wolz, C.; Goerke, C. Temporal expression of adhesion factors and activity of global regulators during establishment of Staphylococcus aureus nasal colonization. J. Infect. Dis. 2010, 201, 1414–1421. [Google Scholar] [CrossRef] [PubMed]
- Burian, M.; Wolz, C.; Goerke, C. Regulatory adaptation of Staphylococcus aureus during nasal colonization of humans. PLoS ONE 2010, 5, e10040. [Google Scholar] [CrossRef] [PubMed]
- Burian, M.; Wolz, C.; Yazdi, A.S. Transcriptional adaptation of staphylococci during colonization of the authentic human environment: An overview of transcriptomic changes and their relationship to physiological conditions. Front. Cell Infect. Microbiol. 2022, 12, 1062329. [Google Scholar] [CrossRef]
- Kong, H.H.; Andersson, B.; Clavel, T.; Common, J.E.; Jackson, S.A.; Olson, N.D.; Segre, J.A.; Traidl-Hoffmann, C. Performing Skin Microbiome Research: A Method to the Madness. J. Investig. Dermatol. 2017, 137, 561–568. [Google Scholar] [CrossRef]
- Moran, M.C.; Cahill, M.P.; Brewer, M.G.; Yoshida, T.; Knowlden, S.; Perez-Nazario, N.; Schlievert, P.M.; Beck, L.A. Staphylococcal Virulence Factors on the Skin of Atopic Dermatitis Patients. mSphere 2019, 4, e00616-19. [Google Scholar] [CrossRef]
- Poh, S.E.; Koh, W.L.C.; Lim, S.Y.D.; Wang, E.C.E.; Yew, Y.W.; Common, J.E.A.; Oon, H.H.; Li, H. Expression of Staphylococcus aureus Virulence Factors in Atopic Dermatitis. JID Innov. 2022, 2, 100130. [Google Scholar] [CrossRef] [PubMed]
- Levin, J.; Friedlander, S.F.; Del Rosso, J.Q. Atopic dermatitis and the stratum corneum: Part 1: The role of filaggrin in the stratum corneum barrier and atopic skin. J. Clin. Aesthet. Dermatol. 2013, 6, 16–22. [Google Scholar]
- Clausen, M.L.; Edslev, S.M.; Andersen, P.S.; Clemmensen, K.; Krogfelt, K.A.; Agner, T. Staphylococcus aureus colonization in atopic eczema and its association with filaggrin gene mutations. Br. J. Dermatol. 2017, 177, 1394–1400. [Google Scholar] [CrossRef] [PubMed]
- Alsterholm, M.; Strombeck, L.; Ljung, A.; Karami, N.; Widjestam, J.; Gillstedt, M.; Ahren, C.; Faergemann, J. Variation in Staphylococcus aureus Colonization in Relation to Disease Severity in Adults with Atopic Dermatitis during a Five-month Follow-up. Acta Derm. Venereol. 2017, 97, 802–807. [Google Scholar] [CrossRef][Green Version]
- Yamazaki, Y.; Ito, T.; Tamai, M.; Nakagawa, S.; Nakamura, Y. The role of Staphylococcus aureus quorum sensing in cutaneous and systemic infections. Inflamm. Regen. 2024, 44, 9. [Google Scholar] [CrossRef]
- Kwon, S.; Choi, J.Y.; Shin, J.W.; Huh, C.H.; Park, K.C.; Du, M.H.; Yoon, S.; Na, J.I. Changes in Lesional and Non-lesional Skin Microbiome During Treatment of Atopic Dermatitis. Acta Derm. Venereol. 2019, 99, 284–290. [Google Scholar] [CrossRef]
- Piewngam, P.; Otto, M. Staphylococcus aureus colonisation and strategies for decolonisation. Lancet Microbe 2024, 5, e606–e618. [Google Scholar] [CrossRef] [PubMed]
- Gehrke, A.E.; Giai, C.; Gómez, M.I. Staphylococcus aureus Adaptation to the Skin in Health and Persistent/Recurrent Infections. Antibiotics 2023, 12, 1520. [Google Scholar] [CrossRef]
- Burian, M.; Plange, J.; Schmitt, L.; Kaschke, A.; Marquardt, Y.; Huth, L.; Baron, J.M.; Hornef, M.W.; Wolz, C.; Yazdi, A.S. Adaptation of Staphylococcus aureus to the Human Skin Environment Identified Using an ex vivo Tissue Model. Front. Microbiol. 2021, 12, 728989. [Google Scholar] [CrossRef] [PubMed]
- Cruz, A.R.; van Strijp, J.A.G.; Bagnoli, F.; Manetti, A.G.O. Virulence Gene Expression of Staphylococcus aureus in Human Skin. Front. Microbiol. 2021, 12, 692023. [Google Scholar] [CrossRef] [PubMed]
- Wright, J.S., 3rd; Jin, R.; Novick, R.P. Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc. Natl. Acad. Sci. USA 2005, 102, 1691–1696. [Google Scholar] [CrossRef]
- Weidinger, S.; Beck, L.A.; Bieber, T.; Kabashima, K.; Irvine, A.D. Atopic dermatitis. Nat. Rev. Dis. Primers 2018, 4, 1. [Google Scholar] [CrossRef]
- Tamai, M.; Yamazaki, Y.; Ito, T.; Nakagawa, S.; Nakamura, Y. Pathogenic role of the staphylococcal accessory gene regulator quorum sensing system in atopic dermatitis. Front. Cell Infect. Microbiol. 2023, 13, 1178650. [Google Scholar] [CrossRef]
- Soong, G.; Paulino, F.; Wachtel, S.; Parker, D.; Wickersham, M.; Zhang, D.; Brown, A.; Lauren, C.; Dowd, M.; West, E.; et al. Methicillin-resistant Staphylococcus aureus adaptation to human keratinocytes. mBio 2015, 6, e00289-15. [Google Scholar] [CrossRef]
- Foster, T.J.; Geoghegan, J.A.; Ganesh, V.K.; Hook, M. Adhesion, invasion and evasion: The many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 2014, 12, 49–62. [Google Scholar] [CrossRef]
- Foster, T.J. Surface Proteins of Staphylococcus aureus. Microbiol. Spectr. 2019, 7, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Schwermann, N.; Winstel, V. Functional diversity of staphylococcal surface proteins at the host-microbe interface. Front. Microbiol. 2023, 14, 1196957. [Google Scholar] [CrossRef]
- Weidenmaier, C.; Peschel, A. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat. Rev. Microbiol. 2008, 6, 276–287. [Google Scholar] [CrossRef]
- Hrestak, D.; Matijasic, M.; Cipcic Paljetak, H.; Ledic Drvar, D.; Ljubojevic Hadzavdic, S.; Peric, M. Skin Microbiota in Atopic Dermatitis. Int. J. Mol. Sci. 2022, 23, 3503. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.H.; Strickland, I.; Boguniewicz, M.; Leung, D.Y. Fibronectin and fibrinogen contribute to the enhanced binding of Staphylococcus aureus to atopic skin. J. Allergy Clin. Immunol. 2001, 108, 269–274. [Google Scholar] [CrossRef]
- Wertheim, H.F.L.; Walsh, E.; Choudhurry, R.; Melles, D.C.; Boelens, H.A.M.; Miajlovic, H.; Verbrugh, H.A.; Foster, T.; van Belkum, A. Key role for clumping factor B in Staphylococcus aureus nasal colonization of humans. PLoS Med. 2008, 5, 104–112. [Google Scholar] [CrossRef]
- Weidenmaier, C.; Kokai-Kun, J.F.; Kristian, S.A.; Chanturiya, T.; Kalbacher, H.; Gross, M.; Nicholson, G.; Neumeister, B.; Mond, J.J.; Peschel, A. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat. Med. 2004, 10, 243–245. [Google Scholar] [CrossRef]
- Wolz, C.; Goerke, C.; Landmann, R.; Zimmerli, W.; Fluckiger, U. Transcription of clumping factor A in attached and unattached Staphylococcus aureus in vitro and during device-related infection. Infect. Immun. 2002, 70, 2758–2762. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Buist, G.; van Dijl, J.M. Staphylococcus aureus cell wall maintenance—The multifaceted roles of peptidoglycan hydrolases in bacterial growth, fitness, and virulence. Fems Microbiol. Rev. 2022, 46, fuac025. [Google Scholar] [CrossRef]
- Stapleton, M.R.; Horsburgh, M.J.; Hayhurst, E.J.; Wright, L.; Jonsson, I.M.; Tarkowski, A.; Kokai-Kun, J.F.; Mond, J.J.; Foster, S.J. Characterization of IsaA and SceD, two putative lytic transglycosylases of Staphylococcus aureus. J. Bacteriol. 2007, 189, 7316–7325. [Google Scholar] [CrossRef]
- Peschel, A. How do bacteria resist human antimicrobial peptides? Trends Microbiol. 2002, 10, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Bera, A.; Biswas, R.; Herbert, S.; Götz, F. The presence of peptidoglycan O-acetyltransferase in various staphylococcal species correlates with lysozyme resistance and pathogenicity. Infect. Immun. 2006, 74, 4598–4604. [Google Scholar] [CrossRef]
- Thammavongsa, V.; Kim, H.K.; Missiakas, D.; Schneewind, O. Staphylococcal manipulation of host immune responses. Nat. Rev. Microbiol. 2015, 13, 529–543. [Google Scholar] [CrossRef]
- Howden, B.P.; Giulieri, S.G.; Wong Fok Lung, T.; Baines, S.L.; Sharkey, L.K.; Lee, J.Y.H.; Hachani, A.; Monk, I.R.; Stinear, T.P. Staphylococcus aureus host interactions and adaptation. Nat. Rev. Microbiol. 2023, 21, 380–395. [Google Scholar] [CrossRef] [PubMed]
- de Jong, N.W.M.; van Kessel, K.P.M.; van Strijp, J.A.G. Immune Evasion by Staphylococcus aureus. Microbiol. Spectr. 2019, 7, 1–27. [Google Scholar] [CrossRef]
- Cruz, A.R.; Boer, M.A.D.; Strasser, J.; Zwarthoff, S.A.; Beurskens, F.J.; de Haas, C.J.C.; Aerts, P.C.; Wang, G.; de Jong, R.N.; Bagnoli, F.; et al. Staphylococcal protein A inhibits complement activation by interfering with IgG hexamer formation. Proc. Natl. Acad. Sci. USA 2021, 118, e2016772118. [Google Scholar] [CrossRef] [PubMed]
- Falugi, F.; Kim, H.K.; Missiakas, D.M.; Schneewind, O. Role of Protein A in the Evasion of Host Adaptive Immune Responses by Staphylococcus aureus. mBio 2013, 4, e00575-13. [Google Scholar] [CrossRef]
- Sieprawska-Lupa, M.; Mydel, P.; Krawczyk, K.; Wojcik, K.; Puklo, M.; Lupa, B.; Suder, P.; Silberring, J.; Reed, M.; Pohl, J.; et al. Degradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinases. Antimicrob. Agents Chemother. 2004, 48, 4673–4679. [Google Scholar] [CrossRef]
- Pietrocola, G.; Nobile, G.; Rindi, S.; Speziale, P. Staphylococcus aureus Manipulates Innate Immunity through Own and Host-Expressed Proteases. Front. Cell. Infect. Microbiol. 2017, 7, 166. [Google Scholar] [CrossRef]
- Hirasawa, Y.; Takai, T.; Nakamura, T.; Mitsuishi, K.; Gunawan, H.; Suto, H.; Ogawa, T.; Wang, X.L.; Ikeda, S.; Okumura, K.; et al. Staphylococcus aureus extracellular protease causes epidermal barrier dysfunction. J. Investig. Dermatol. 2010, 130, 614–617. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Chen, T.H.; Two, A.M.; Chun, K.A.; Narala, S.; Geha, R.S.; Hata, T.R.; Gallo, R.L. Staphylococcus aureus Exploits Epidermal Barrier Defects in Atopic Dermatitis to Trigger Cytokine Expression. J. Investig. Dermatol. 2016, 136, 2192–2200. [Google Scholar] [CrossRef]
- Kim, J.E.; Kim, B.J.; Jeong, M.S.; Seo, S.J.; Kim, M.N.; Hong, C.K.; Ro, B.I. Expression and modulation of LL-37 in normal human keratinocytes, HaCaT cells, and inflammatory skin diseases. J. Korean Med. Sci. 2005, 20, 649–654. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.H.; Fang, S.Y. The expression of human antimicrobial peptide LL-37 in the human nasal mucosa. Am. J. Rhinol. 2004, 18, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Laarman, A.J.; Ruyken, M.; Malone, C.L.; van Strijp, J.A.; Horswill, A.R.; Rooijakkers, S.H. Staphylococcus aureus metalloprotease aureolysin cleaves complement C3 to mediate immune evasion. J. Immunol. 2011, 186, 6445–6453. [Google Scholar] [CrossRef]
- Potempa, J.; Dubin, A.; Korzus, G.; Travis, J. Degradation of elastin by a cysteine proteinase from Staphylococcus aureus. J. Biol. Chem. 1988, 263, 2664–2667. [Google Scholar] [CrossRef] [PubMed]
- Potempa, J.; Watorek, W.; Travis, J. The inactivation of human plasma alpha 1-proteinase inhibitor by proteinases from Staphylococcus aureus. J. Biol. Chem. 1986, 261, 14330–14334. [Google Scholar] [CrossRef]
- Ohbayashi, T.; Irie, A.; Murakami, Y.; Nowak, M.; Potempa, J.; Nishimura, Y.; Shinohara, M.; Imamura, T. Degradation of fibrinogen and collagen by staphopains, cysteine proteases released from Staphylococcus aureus. Microbiology 2011, 157, 786–792. [Google Scholar] [CrossRef]
- Sebulsky, M.T.; Heinrichs, D.E. Identification and characterization of fhuD1 and fhuD2, two genes involved in iron-hydroxamate uptake in Staphylococcus aureus. J. Bacteriol. 2001, 183, 4994–5000. [Google Scholar] [CrossRef]
- Morrissey, J.A.; Cockayne, A.; Brummell, K.; Williams, P. The staphylococcal ferritins are differentially regulated in response to iron and manganese and via PerR and Fur. Infect. Immun. 2004, 72, 972–979. [Google Scholar] [CrossRef]
- Teichmann, P.; Both, A.; Wolz, C.; Hornef, M.W.; Rohde, H.; Yazdi, A.S.; Burian, M. The Staphylococcus epidermidis Transcriptional Profile During Carriage. Front. Microbiol. 2022, 13, 896311. [Google Scholar] [CrossRef]
- Goerke, C.; Fluckiger, U.; Steinhuber, A.; Bisanzio, V.; Ulrich, M.; Bischoff, M.; Patti, J.M.; Wolz, C. Role of Staphylococcus aureus global regulators sae and sigmaB in virulence gene expression during device-related infection. Infect. Immun. 2005, 73, 3415–3421. [Google Scholar] [CrossRef] [PubMed]
- Harmsen, D.; Claus, H.; Witte, W.; Rothganger, J.; Claus, H.; Turnwald, D.; Vogel, U. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 2003, 41, 5442–5448. [Google Scholar] [CrossRef] [PubMed]

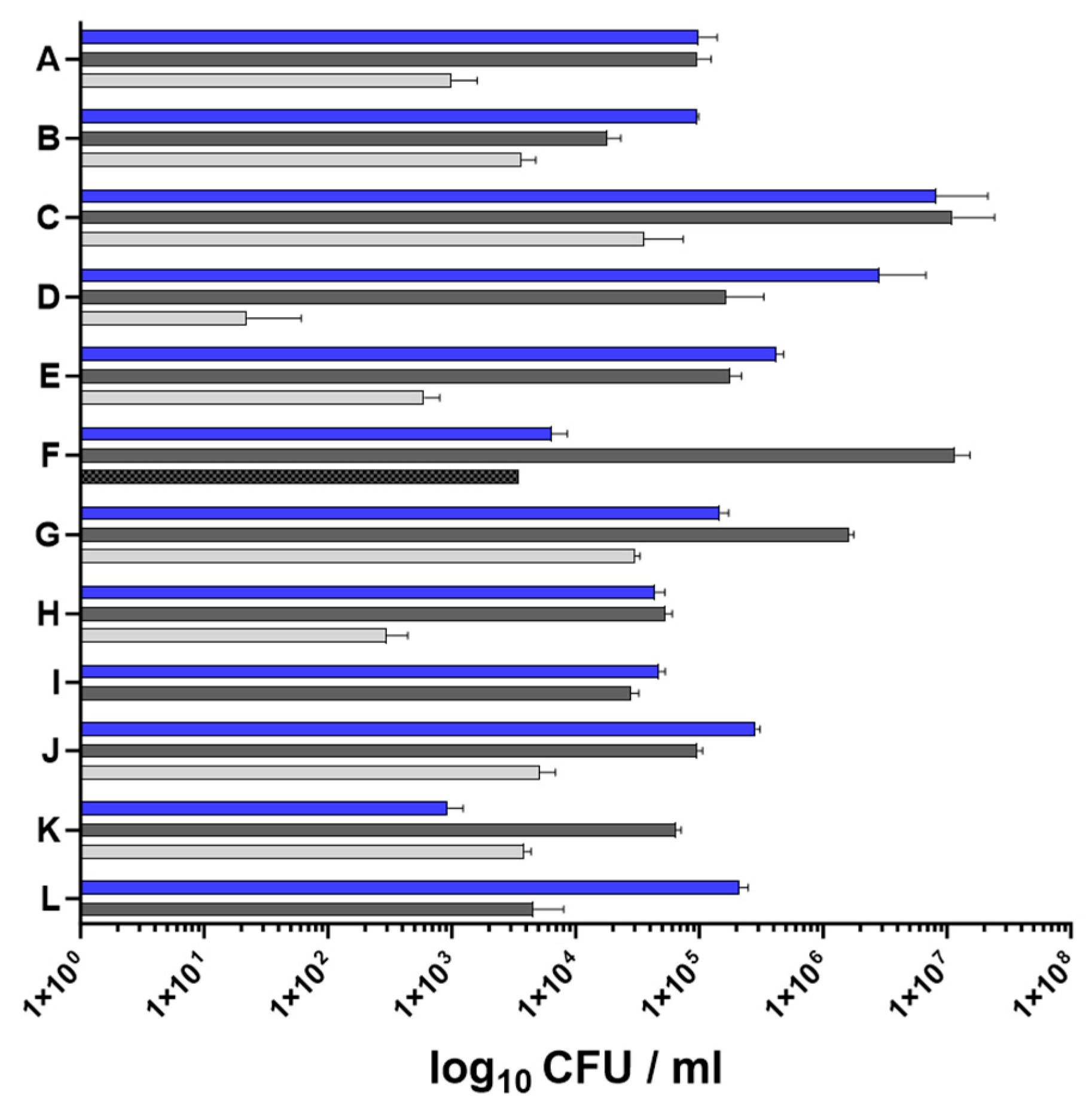


| Patient | Age | Sex | SCORAD | Comorbidities | S. aureus Isolate | Spa Typing ID | Deduced MLST CC | Accessory Gene Content | Phenotype |
|---|---|---|---|---|---|---|---|---|---|
| A | 19 | female | 77 | - | nose lesional skin non-lesional skin | t304 | - | sak, scn | hemolytic hemolytic hemolytic |
| B | 61 | female | 72.6 | Asthma bronchiale Milk product allergy | nose lesional skin non-lesional skin | t1451 | - | chp, scn | hemolytic hemolytic hemolytic |
| C | 39 | male | 80.4 | Asthma bronchiale Rhinoconjunctivitis | nose lesional skin non-lesional skin | t008 | CC8 | - | hemolytic hemolytic hemolytic |
| D | 36 | male | 34.4 | Asthma bronchiale Rhinoconjunctivitis | nose lesional skin | t674 t1943 | - | sak, scn | hemolytic hemolytic |
| E | 54 | male | 10.5 | Rhinoconjunctivitis | nose lesional skin non-lesional skin | t094 | - | chp, scn | hemolytic hemolytic hemolytic |
| F | 38 | female | 47 | - | nose lesional skin 1 lesional skin 2 | t091 t091 t3697 | - | sak, scn | hemolytic hemolytic non-hemolytic |
| G | 48 | male | 48 | Asthma bronchiale Rhinoconjunctivitis | nose lesional skin non-lesional skin | t190 | CC8 | sak, chp, scn | hemolytic hemolytic hemolytic |
| H | 41 | male | 41.3 | Asthma bronchiale Contact allergy (Amerchol) | nose lesional skin non-lesional skin | t002 | CC5 | sak, chp, scn | hemolytic hemolytic hemolytic |
| I | 53 | male | 48.6 | Contact allergy (Isoeugenol) | nose lesional skin | t026 | CC45 | sak, chp, scn | hemolytic hemolytic |
| J | 19 | male | 45.5 | Asthma bronchiale Rhinoconjunctivitis | nose lesional skin non-lesional skin | t091 | - | sak, scn | hemolytic hemolytic hemolytic |
| K | 38 | male | 38.5 | Asthma bronchiale Rhinoconjunctivitis Contact allergy | nose lesional skin non-lesional skin | t2953 | - | sak, chp, scn | hemolytic hemolytic hemolytic |
| L | 35 | female | 29 | Asthma bronchiale Rhinoconjunctivitis | nose lesional skin | t091 t1268 | - | sak, scn sak, chp, scn | hemolytic hemolytic |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, P.; Schulte, J.; Wurpts, G.; Hornef, M.W.; Wolz, C.; Yazdi, A.S.; Burian, M. Transcriptional Profiling of Staphylococcus aureus during the Transition from Asymptomatic Nasal Colonization to Skin Colonization/Infection in Patients with Atopic Dermatitis. Int. J. Mol. Sci. 2024, 25, 9165. https://doi.org/10.3390/ijms25179165
Li P, Schulte J, Wurpts G, Hornef MW, Wolz C, Yazdi AS, Burian M. Transcriptional Profiling of Staphylococcus aureus during the Transition from Asymptomatic Nasal Colonization to Skin Colonization/Infection in Patients with Atopic Dermatitis. International Journal of Molecular Sciences. 2024; 25(17):9165. https://doi.org/10.3390/ijms25179165
Chicago/Turabian StyleLi, Peijuan, Julia Schulte, Gerda Wurpts, Mathias W. Hornef, Christiane Wolz, Amir S. Yazdi, and Marc Burian. 2024. "Transcriptional Profiling of Staphylococcus aureus during the Transition from Asymptomatic Nasal Colonization to Skin Colonization/Infection in Patients with Atopic Dermatitis" International Journal of Molecular Sciences 25, no. 17: 9165. https://doi.org/10.3390/ijms25179165
APA StyleLi, P., Schulte, J., Wurpts, G., Hornef, M. W., Wolz, C., Yazdi, A. S., & Burian, M. (2024). Transcriptional Profiling of Staphylococcus aureus during the Transition from Asymptomatic Nasal Colonization to Skin Colonization/Infection in Patients with Atopic Dermatitis. International Journal of Molecular Sciences, 25(17), 9165. https://doi.org/10.3390/ijms25179165







