Satellitome Analysis of Adalia bipunctata (Coleoptera): Revealing Centromeric Turnover and Potential Chromosome Rearrangements in a Comparative Interspecific Study
Abstract
1. Introduction
2. Results and Discussion
2.1. Adalia bipunctata Satellitome Description
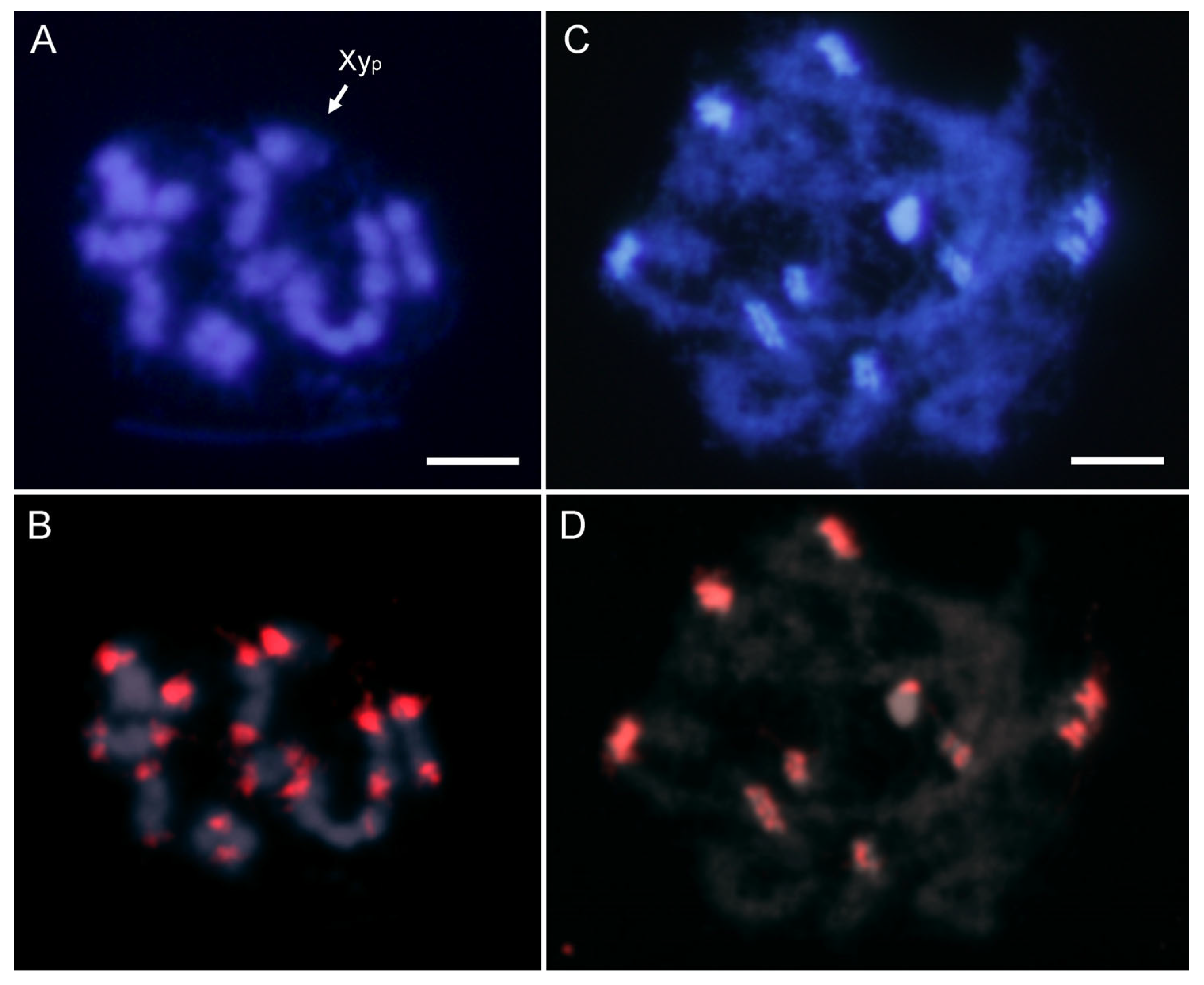
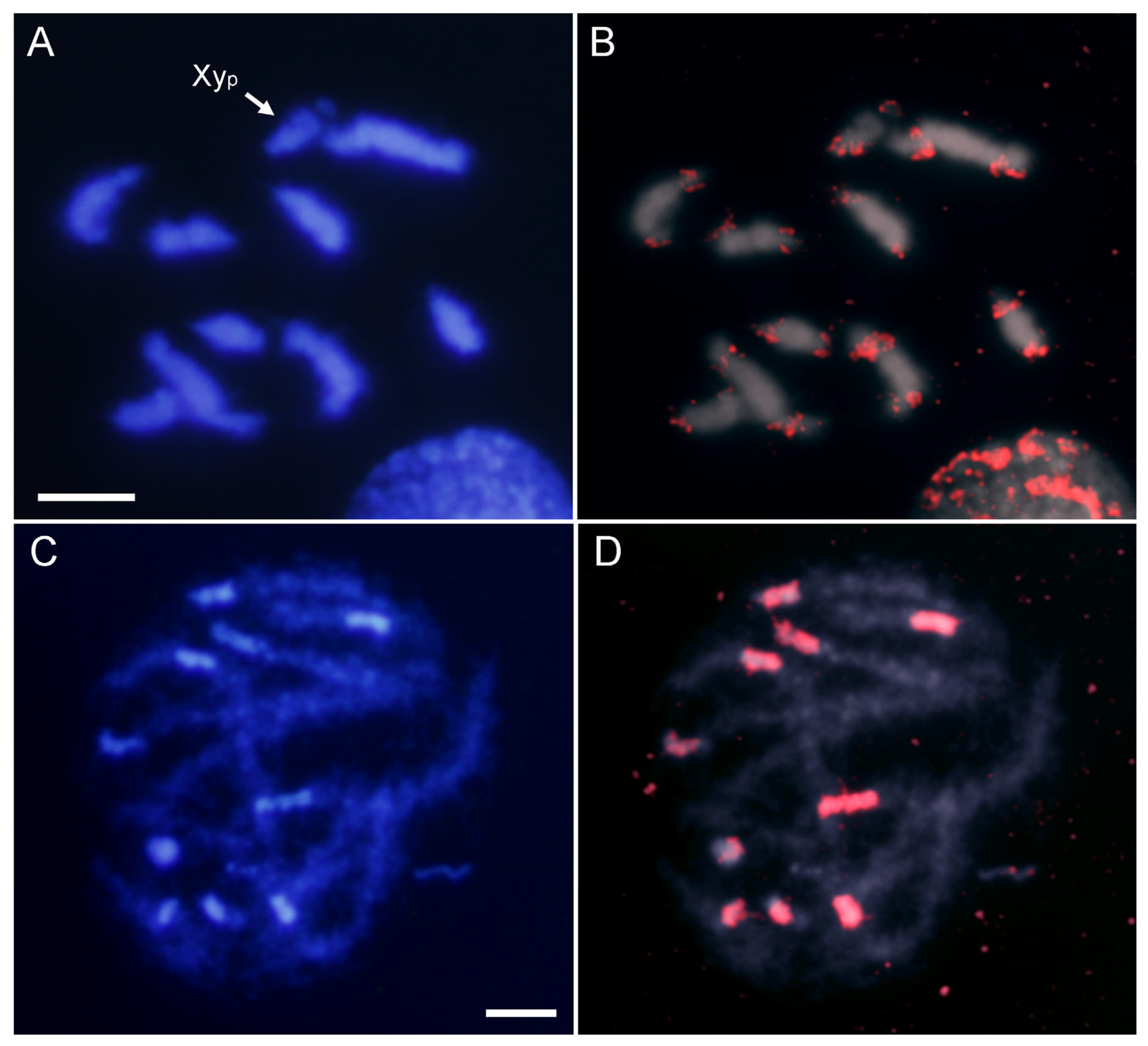
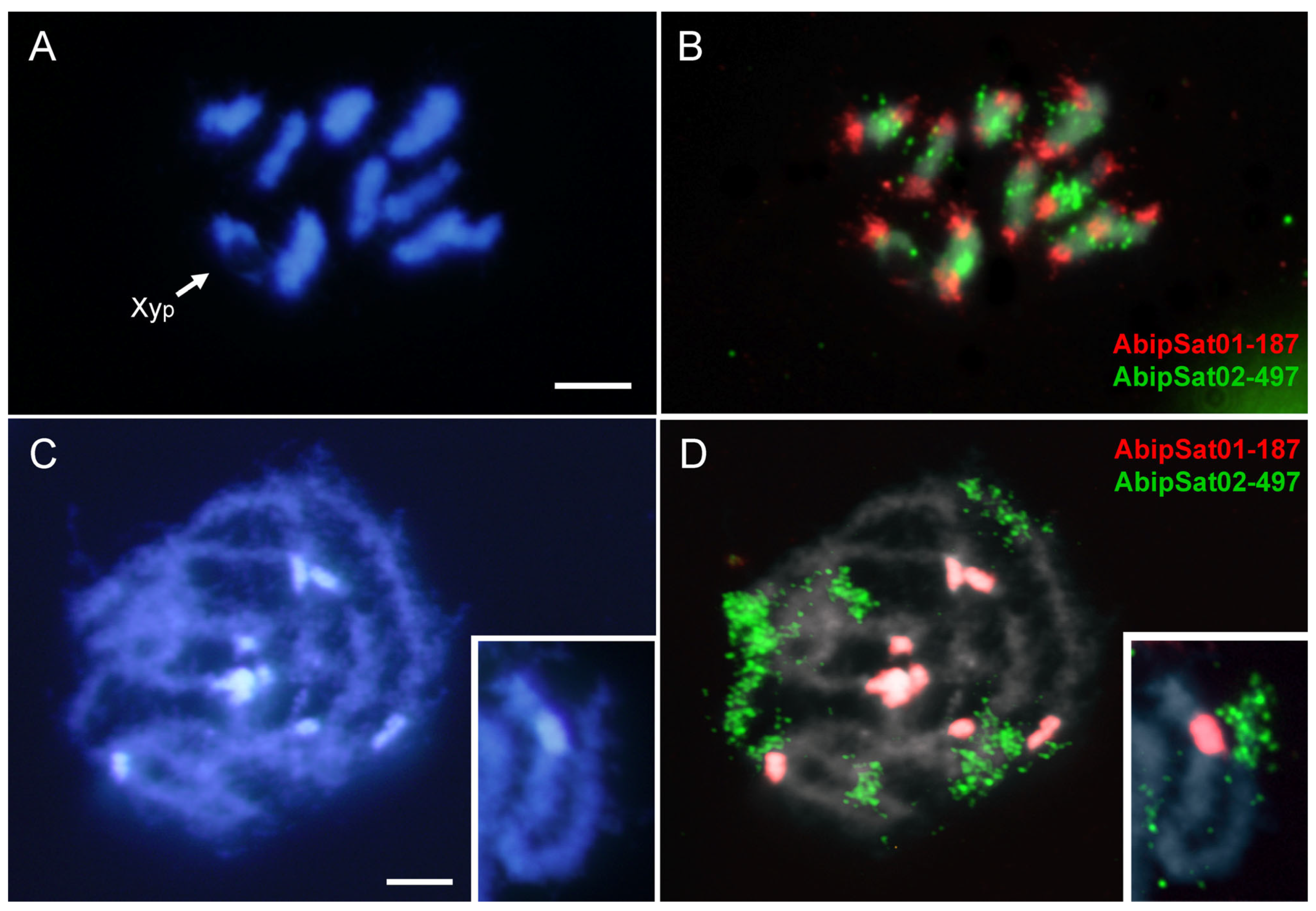
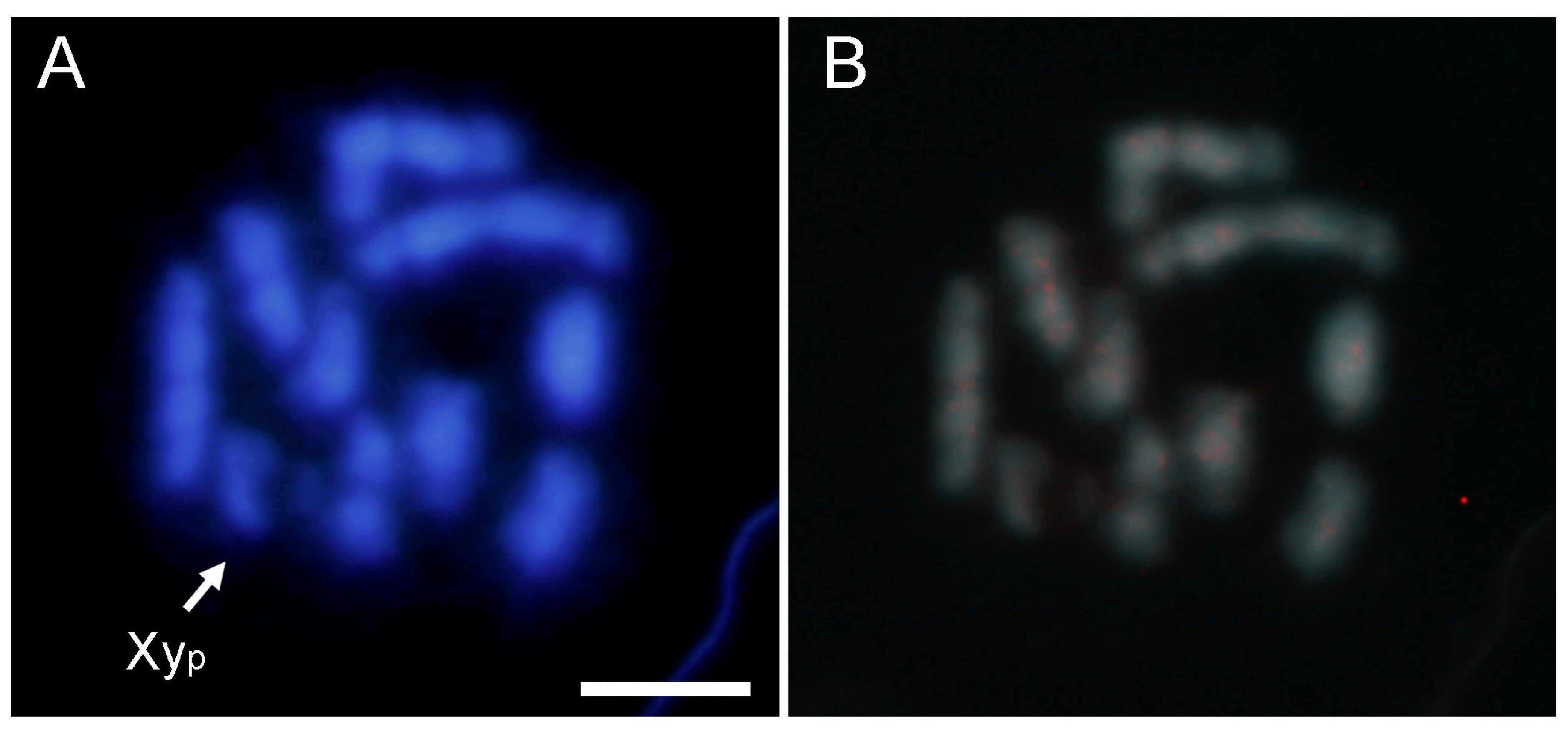
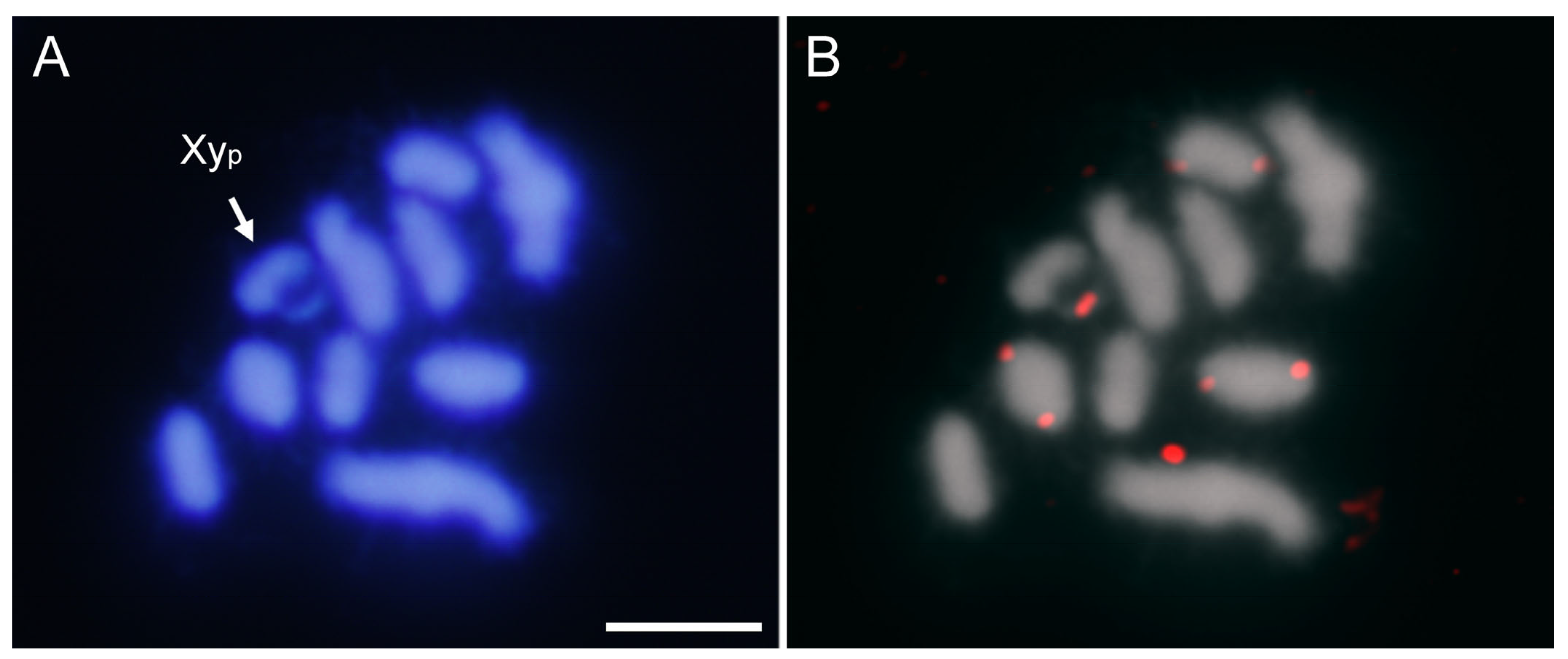
2.2. SatDNA Location in Adalia bipunctata Chromosomes by FISH and CHRISMAPP
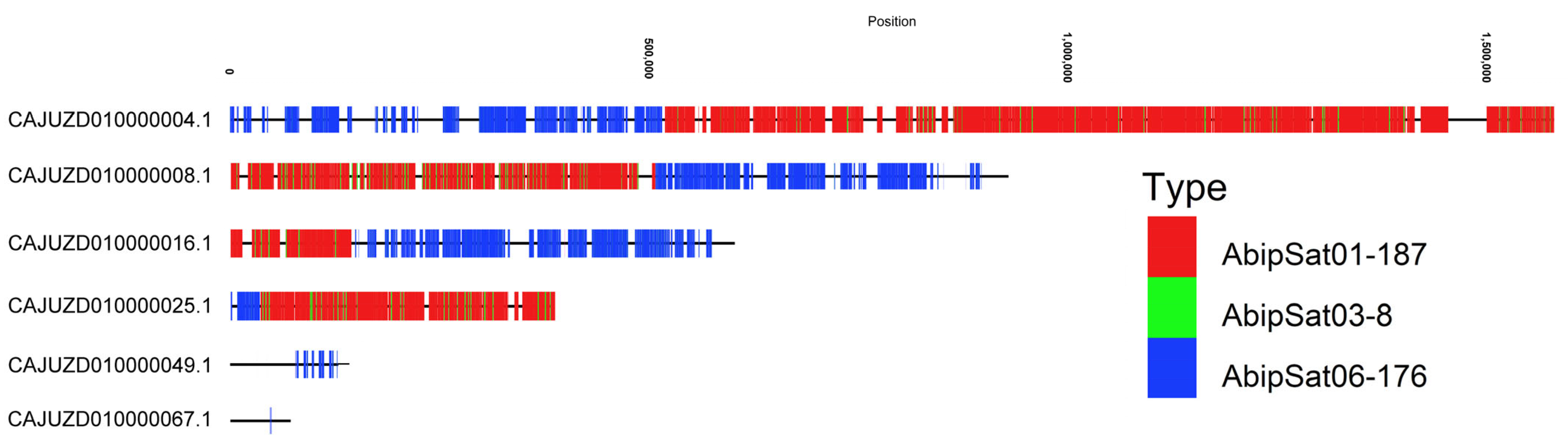
2.3. Satellitome Representation in the Adalia bipunctata Genome Assembly
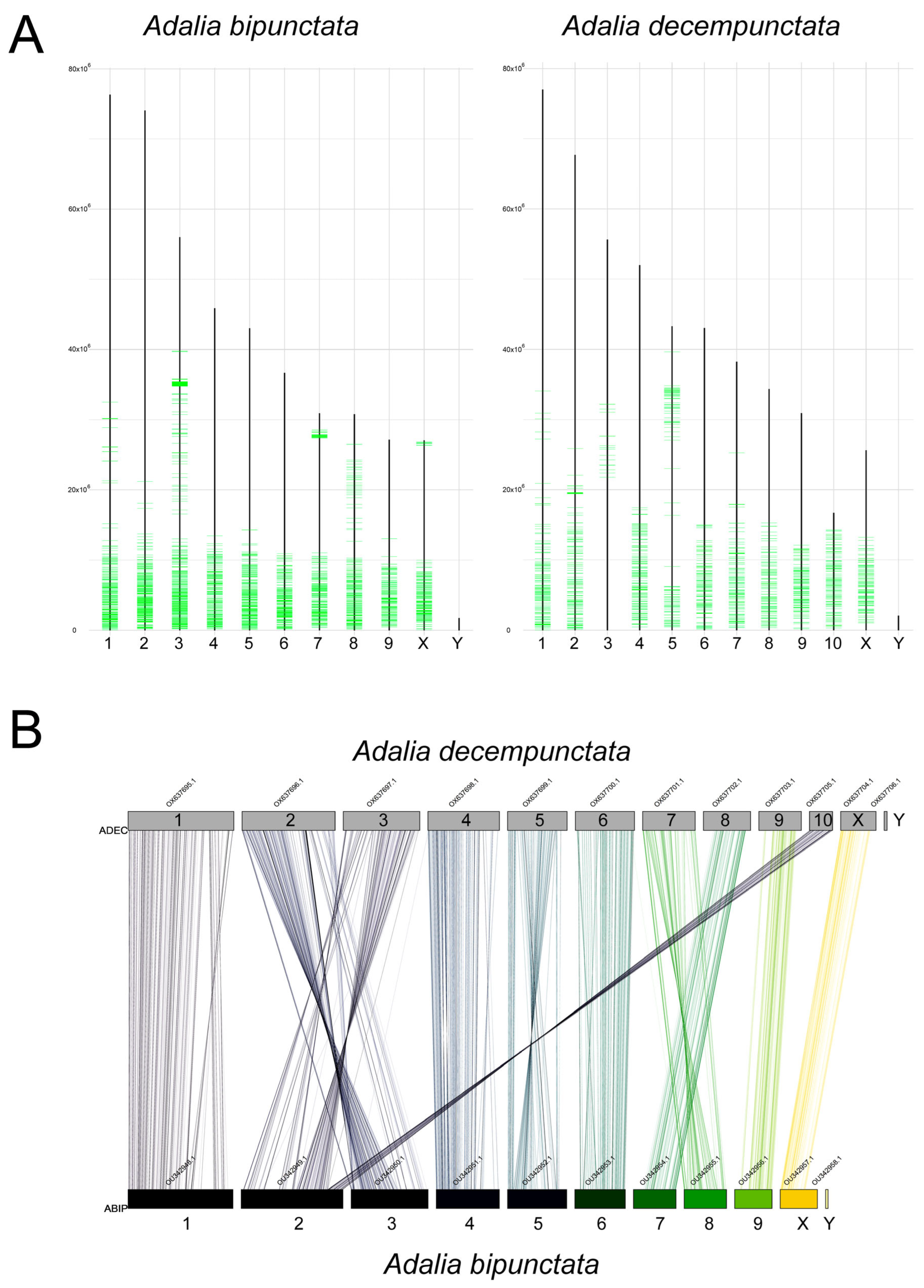
2.4. Comparative Analysis with Adalia decempunctata
3. Materials and Methods
3.1. Insects, Preparation of Chromosome Spreads, and C-Banding
3.2. Satellitome Analysis
3.3. Cytogenetic Mapping by Fluorescence In Situ Hybridization (FISH) and by Chromosome In Silico Mapping (CHRISMAPP) and Synteny Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Britten, R.J.; Kohne, D.E. Repeated sequences in DNA. Science 1968, 161, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Biscotti, M.A.; Olmo, E.; Heslop-Harrison, J.S. Repetitive DNA in eukaryotic genomes. Chromosome Res. 2015, 23, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Pons, J.; Petitpierre, E.; Juan, C. Characterization of the heterochromatin of the darkling beetle Misolampus goudoti: Cloning of two satellite DNA families and digestion of chromosomes with restriction enzymes. Hereditas 1993, 119, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Ramos, M.A. Satellite DNA: An evolving topic. Genes 2017, 8, 230. [Google Scholar] [CrossRef]
- Ugarković, Ð.; Plohl, M. Variation in satellite DNA profiles-causes and effects. EMBO J. 2002, 21, 5955–5959. [Google Scholar] [CrossRef]
- Pezer, Z.; Brajković, J.; Feliciello, I.; Ugarković, Ð. Satellite DNA-mediated effects on genome regulation. Genome Dyn. 2012, 7, 153–169. [Google Scholar] [CrossRef] [PubMed]
- Plohl, M.; Meštrović, N.; Mravinac, B. Centromere identity from the DNA point of view. Chromosoma 2014, 123, 313–325. [Google Scholar] [CrossRef]
- Rošić, S.; Köhler, F.; Erhardt, S. Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J. Cell Biol. 2014, 207, 335–349. [Google Scholar] [CrossRef]
- Perea-Resa, C.; Blower, M.D. Satellite transcripts locally promote centromere formation. Dev. Cell 2017, 42, 201–202. [Google Scholar] [CrossRef]
- Louzada, S.; Lopes, M.; Ferreira, D.; Adega, F.; Escudeiro, A.; Gama-Carvalho, M.; Chaves, R. Decoding the role of satellite DNA in genome architecture and plasticity—An evolutionary and clinical affair. Genes 2020, 11, 72. [Google Scholar] [CrossRef]
- Di Tommaso, E.; Giunta, S. Dynamic interplay between human alpha-satellite DNA structure and centromere functions. Semin. Cell Dev. Biol. 2024, 156, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Gambogi, C.W.; Dawicki-McKenna, J.M.; Logsdon, G.A.; Black, B.E. The unique kind of human artificial chromosome: Bypassing the requirement for repetitive centromere DNA. Exp. Cell Res. 2020, 1, 111978. [Google Scholar] [CrossRef]
- Ohzeki, J.I.; Otake, K.; Masumoto, H. Human artificial chromosome: Chromatin assembly mechanisms and CENP-B. Exp. Cell Res. 2020, 389, 111900. [Google Scholar] [CrossRef]
- Cáceres-Gutiérrez, R.E.; Andonegui, M.A.; Oliva-Rico, D.A.; González-Barrios, R.; Luna, F.; Arriaga-Canon, C.; López-Saavedra, A.; Prada, D.; Castro, C.; Parmentier, L.; et al. Proteasome inhibition alters mitotic progression through the upregulation of centromeric α-Satellite RNAs. FEBS J. 2022, 289, 1858–1875. [Google Scholar] [CrossRef]
- Özgür, E.; Mayer, Z.; Keskin, M.; Yörüker, E.E.; Holdenrieder, S.; Gezer, U. Satellite 2 repeat DNA in blood plasma as a candidate biomarker for the detection of cancer. Clin. Chim. Acta 2021, 514, 74–79. [Google Scholar] [CrossRef]
- Ariffen, N.A.; Ornellas, A.A.; Alves, G.; Shana’ah, A.M.; Sharma, S.; Kankel, S.; Jamali, E.; Theis, B.; Liehr, T. Amplification of different satellite-DNAs in prostate cancer. Pathol. Res. Pract. 2024, 256, 155269. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.B.; Oh, J.H.; McIver, L.J.; Rashkovetsky, E.; Michalak, K.; Garner, H.R.; Kang, L.; Nevo, E.; Korol, A.B.; Michalak, P. Divergence of Drosophila melanogaster repeatomes in response to a sharp microclimate contrast in Evolution Canyon, Israel. Proc. Natl. Acad. Sci. USA 2014, 111, 10630–10635. [Google Scholar] [CrossRef]
- Ruiz-Ruano, F.J.; López-León, M.D.; Cabrero, J.; Camacho, J.P.M. High–throughput analysis of the satellitome illuminates satellite DNA evolution. Sci. Rep. 2016, 6, 28333. [Google Scholar] [CrossRef] [PubMed]
- Novák, P.; Neumann, P.; Macas, J. Graph–based clustering and characterization of repetitive sequences in next–generation sequencing data. BMC Bioinform. 2010, 11, 378. [Google Scholar] [CrossRef]
- Novák, P.; Neumann, P.; Pech, J.; Steinhaisl, J.; Macas, J. RepeatExplorer: A Galaxy–based web server for genome–wide characterization of eukaryotic repetitive elements from next–generation sequence reads. Bioinformatics 2013, 29, 792–793. [Google Scholar] [CrossRef]
- Novák, P.; Ávila Robledillo, L.; Koblížková, A.; Vrbová, I.; Neumann, P.; Macas, J. TAREAN: A computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acids Res. 2017, 45, e111. [Google Scholar] [CrossRef] [PubMed]
- Weiss-Schneeweiss, H.; Leitch, A.R.; McCann, J.; Jang, T.S.; Macas, J. Employing next generation sequencing to explore the repeat landscape of the plant genome. In Next Generation Sequencing in Plant Systematics, Regnum Vegetabile 157; Hörandl, E., Appelhans, M., Eds.; Koeltz Scientific Books: Königstein, Germany, 2015; pp. 155–180. [Google Scholar]
- Garrido-Ramos, M.A. The genomics of plant satellite DNA. In Satellite DNAs in Physiology and Evolution. Progress in Molecular and Subcellular Biology; Ugarković, Ð., Ed.; Springer: Cham, Switzerland, 2021; Volume 60. [Google Scholar] [CrossRef]
- Kheloufi, A.; Arar, S.; Aoutti, A.; Mansouri, L.M. Coexistence of Henosepilachna elaterii (Rossi, 1794) (Coleoptera Coccinellidae) on Citrullus colocynthis Schrad. (Cucurbitaceae) around the water dam of El Outaya (Biskra, Algeria). Biodivers. J. 2018, 9, 69–72. [Google Scholar]
- Dixon, A.F.G. Insect Predator-Prey Dynamics; Ladybird Beetles and Biological Control; Cambridge University Press: Cambridge, UK, 2005. [Google Scholar]
- Khan, A.A.; Zaki, F.A.; Khan, Z.H.; Mir, R.A. Biodiversity of predacious ladybird beetles (Coleoptera: Coccinellidae) in Kashmir. J. Biol. Control 2009, 23, 43–47. [Google Scholar] [CrossRef]
- Asiri, A.; Foster, C. Temporal and climatic variation in the colour forms of Adalia bipunctata and Harmonia axyridis (Coleoptera: Coccinellidae) populations in the United Kingdom. Eur. J. Entomol. 2022, 119, 250–259. [Google Scholar] [CrossRef]
- Omkar; Pervez, A. Ecology of two-spotted ladybird, Adalia bipunctata: A review. J. Appl. Entomol. 2005, 129, 465–474. [Google Scholar] [CrossRef]
- De Clercq, P.; Bonte, M.; Van Speybroeck, K.; Bolckmans, K.; Deforce, K. Development and reproduction of Adalia bipunctata (Col., Coccinellidae) on eggs of Ephestia kuehniella (Lep., Phycitidae) and pollen. Pest Manag. Sci. 2005, 61, 1129–1132. [Google Scholar] [CrossRef]
- van Lenteren, J.C. The state of commercial augmentative biological control: Plenty of natural enemies, but a frustrating lack of uptake. BioControl 2012, 57, 1–20. [Google Scholar] [CrossRef]
- Palomeque, T.; Lorite, P. Satellite DNA in insects: A review. Heredity 2008, 100, 564–573. [Google Scholar] [CrossRef]
- Mora, P.; Vela, J.; Ruiz, A.; Palomeque, T.; Lorite, P. Characterization and transcriptional analysis of a subtelomeric satellite DNA family in the ladybird beetle Henosepilachna argus (Coleoptera: Coccinellidae). Eur. J. Entomol. 2017, 114, 481–487. [Google Scholar] [CrossRef][Green Version]
- Mora, P.; Vela, J.; Ruiz-Mena, A.; Palomeque, T.; Lorite, P. Isolation of a pericentromeric satellite DNA family in Chnootriba argus (Henosepilachna argus) with an unusual short repeat unit (TTAAAA) for beetles. Insects 2019, 10, 306. [Google Scholar] [CrossRef]
- Mora, P.; Vela, J.; Ruiz–Ruano, F.J.; Ruiz–Mena, A.; Montiel, E.E.; Palomeque, T.; Lorite, P. Satellitome analysis in the ladybird beetle Hippodamia variegata (Coleoptera, Coccinellidae). Genes 2020, 11, 783. [Google Scholar] [CrossRef]
- Rico-Porras, J.M.; Mora, P.; Palomeque, T.; Montiel, E.E.; Cabral-de-Mello, D.C.; Lorite, P. Heterochromatin is not the only place for satDNAs: The high diversity of satDNAs in the euchromatin of the beetle Chrysolina americana (Coleoptera, Chrysomelidae). Genes 2024, 15, 395. [Google Scholar] [CrossRef] [PubMed]
- Wellcome Sanger Institute Tree of Life Programme; Wellcome Sanger Institute Scientific Operations: DNA Pipelines Collective; Tree of Life Core Informatics Collective; Goate, Z.; Darwin Tree of Life Consortium. The genome sequence of the two-spot ladybird, Adalia bipunctata (Linnaeus, 1758). Wellcome Open Res. 2022, 7, 288. [Google Scholar] [CrossRef]
- Crowley, L.M.; Roy, H.E.; Brown, P.M.J.; University of Oxford and Wytham Woods Genome Acquisition Lab; Darwin Tree of Life Barcoding Collective; Wellcome Sanger Institute Tree of Life Management, Samples and Laboratory Team; Wellcome Sanger Institute Scientific Operations: Sequencing Operations; Wellcome Sanger Institute Tree of Life Core Informatics Team; Tree of Life Core Informatics Collective, Darwin Tree of Life Consortium. The genome sequence of the ten-spot ladybird, Adalia decempunctata (Linnaeus, 1758). Wellcome Open Res. 2024, 9, 106. [Google Scholar] [CrossRef]
- Kyriacou, R.G.; Mulhair, P.O.; Holland, P.W.H. GC content across insect genomes: Phylogenetic patterns, causes and consequences. J. Mol. Evol. 2024, 92, 138–152. [Google Scholar] [CrossRef] [PubMed]
- Novák, P.; Neumann, P.; Macas, J. Global analysis of repetitive DNA from unassembled sequence reads using RepeatExplorer2. Nat. Protoc. 2020, 15, 3745–3776. [Google Scholar] [CrossRef] [PubMed]
- Montiel, E.E.; Mora, P.; Rico–Porras, J.M.; Palomeque, T.; Lorite, P. Satellitome of the red palm weevil, Rhynchophorus ferrugineus (Coleoptera: Curculionidae), the most diverse among insects. Front. Ecol. Evol. 2022, 10, 826808. [Google Scholar] [CrossRef]
- Oppert, B.; Muszewska, A.; Steczkiewicz, K.; Šatović-Vukšić, E.; Plohl, M.; Fabrick, J.A.; Vinokurov, K.S.; Koloniuk, I.; Johnston, J.S.; Smith, T.P.L.; et al. The genome of Rhyzopertha dominica (Fab.) (Coleoptera: Bostrichidae): Adaptation for success. Genes 2022, 13, 446. [Google Scholar] [CrossRef]
- Oppert, B.; Dossey, A.T.; Chu, F.-C.; Šatović-Vukšić, E.; Plohl, M.; Smith, T.P.L.; Koren, S.; Olmstead, M.L.; Leierer, D.; Ragan, G.; et al. The genome of the yellow mealworm, Tenebrio molitor: It’s bigger than you think. Genes 2023, 14, 2209. [Google Scholar] [CrossRef]
- Gržan, T.; Dombi, M.; Despot-Slade, E.; Veseljak, D.; Volarić, M.; Meštrović, N.; Plohl, M.; Mravinac, B. The low-copy-number satellite DNAs of the model beetle Tribolium castaneum. Genes 2023, 14, 999. [Google Scholar] [CrossRef]
- Gregory, T.R. Animal Genome Size Database. 2024. Available online: http://www.genomesize.com (accessed on 4 April 2024).
- Sudalaimuthuasari, N.; Kundu, B.; Hazzouri, K.M.; Amiri, K.M.A. Near-chromosomal-level genome of the red palm weevil (Rhynchophorus ferrugineus), a potential resource for genome-based pest control. Sci. Data 2024, 11, 45. [Google Scholar] [CrossRef]
- Feliciello, I.; Chinali, G.; Ugarković, Ð. Structure and population dynamics of the major satellite DNA in the red flour beetle Tribolium castaneum. Genetica 2011, 139, 999–1008. [Google Scholar] [CrossRef]
- Petitpierre, E.; Gatewood, J.M.; Schmid, C.W. Satellite DNA from the beetle Tenebrio molitor. Experientia 1988, 44, 498–499. [Google Scholar] [CrossRef]
- Tørresen, O.K.; Star, B.; Mier, P.; Andrade-Navarro, M.A.; Bateman, A.; Jarnot, P.; Gruca, A.; Grynberg, M.; Kajava, A.V.; Promponas, V.J.; et al. Tandem repeats lead to sequence assembly errors and impose multi-level challenges for genome and protein databases. Nucleic Acids Res. 2019, 47, 10994–11006. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.F.; Singchat, W.; Jehangir, M.; Suntronpong, A.; Panthum, T.; Malaivijitnond, S.; Srikulnath, K. Dark matter of primate genomes: Satellite DNA repeats and their evolutionary dynamics. Cells 2020, 9, 2714. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Lorenzen, M.D.; Beeman, R.W.; Brown, S.J. Analysis of repetitive DNA distribution patterns in the Tribolium castaneum genome. Genome Biol. 2008, 9, R61. [Google Scholar] [CrossRef]
- Pavlek, M.; Gelfand, Y.; Plohl, M.; Meštrović, N. Genome-wide analysis of tandem repeats in Tribolium castaneum genome reveals abundant and highly dynamic tandem repeat families with satellite DNA features in euchromatic chromosomal arms. DNA Res. 2015, 22, 387–401. [Google Scholar] [CrossRef] [PubMed]
- Montiel, E.E.; Panzera, F.; Palomeque, T.; Lorite, P.; Pita, S. Satellitome analysis of Rhodnius prolixus, one of the main Chagas disease vector species. Int. J. Mol. Sci. 2021, 22, 6052. [Google Scholar] [CrossRef]
- Ruiz-Ruano, F.J.; Castillo-Martínez, J.; Cabrero, J.; Gómez, R.; Camacho, J.P.M.; López-León, M.D. High-throughput analysis of satellite DNA in the grasshopper Pyrgomorpha conica reveals abundance of homologous and heterologous higher-order repeats. Chromosoma 2018, 127, 323–340. [Google Scholar] [CrossRef]
- Milani, D.; Bardella, V.B.; Ferretti, A.B.S.M.; Palacios-Gimenez, O.M.; Melo, A.d.S.; Moura, R.C.; Loreto, V.; Song, H.; Cabral-de-Mello, D.C. Satellite DNAs unveil clues about the ancestry and composition of B chromosomes in three grasshopper species. Genes 2018, 9, 523. [Google Scholar] [CrossRef]
- Cabral-de-Mello, D.C.; Mora, P.; Rico-Porras, J.M.; Ferretti, A.B.S.M.; Palomeque, T.; Lorite, P. The spread of satellite DNAs in euchromatin and insights into the multiple sex chromosome evolution in Hemiptera revealed by repeatome analysis of the bug Oxycarenus hyalinipennis. Insect Mol. Biol. 2023, 32, 725–737. [Google Scholar] [CrossRef]
- Lorite, P.; Carrillo, J.A.; Tinaut, A.; Palomeque, T. Evolutionary dynamics of satellite DNA in species of the genus Formica (Hymenoptera, Formicidae). Gene 2004, 332, 159–168. [Google Scholar] [CrossRef]
- Ruiz–Ruano, F.J.; Navarro–Domínguez, B.; Camacho, J.P.M.; Garrido–Ramos, M.A. Characterization of the satellitome in lower vascular plants: The case of the endangered fern Vandenboschia speciosa. Ann. Bot. 2018, 123, 587–599. [Google Scholar] [CrossRef]
- Ruiz-Ruano, F.J.; Cabrero, J.; López-León, M.D.; Camacho, J.P.M. Satellite DNA content illuminates the ancestry of a supernumerary (B) chromosome. Chromosoma 2017, 126, 487–500. [Google Scholar] [CrossRef]
- Goes, C.A.G.; dos Santos, N.; Rodrigues, P.H.d.M.; Stornioli, J.H.F.; Silva, A.B.d.; dos Santos, R.Z.; Vidal, J.A.D.; Silva, D.M.Z.d.A.; Artoni, R.F.; Foresti, F.; et al. The satellite DNA catalogues of two Serrasalmidae (Teleostei, Characiformes): Conservation of general satDNA features over 30 million years. Genes 2023, 14, 91. [Google Scholar] [CrossRef] [PubMed]
- Stevens, N.M. Studies in Spermatogenesis with Especial Reference to the ‘Accessory Chromosome’; Carnegie Institution of Washington: Washington, DC, USA, 1905; Volume 36, pp. 1–33. [Google Scholar]
- Dutrillaux, A.M.; Dutrillaux, B. Different behaviour of C-banded peri-centromeric heterochromatin between sex chromosomes and autosomes in Polyphagan beetles. Comp. Cytogenet. 2019, 13, 179–192. [Google Scholar] [CrossRef]
- John, B.; Lewis, K. Nucleolar controlled segregation of the sex chromosomes in beetles. Heredity 1960, 15, 431–439. [Google Scholar] [CrossRef][Green Version]
- Camacho, J.P.; Ruiz-Ruano, F.J.; Martín-Blázquez, R.; López-León, M.D.; Cabrero, J.; Lorite, P.; Cabral-de-Mello, D.C.; Bakkali, M. A step to the gigantic genome of the desert locust: Chromosome sizes and repeated DNAs. Chromosoma 2015, 124, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Waring, M.; Britten, R.J. Nucleotide sequence repetition: A rapidly reassociating fraction of mouse DNA. Science 1966, 154, 791–794. [Google Scholar] [CrossRef]
- Lower, S.S.; McGurk, M.P.; Clark, A.G.; Barbash, D.A. Satellite DNA evolution: Old ideas, new approaches. Curr. Opin. Genet. Dev. 2018, 48, 70–78. [Google Scholar] [CrossRef]
- Šatović, E. Tools and databases for solving problems in detection and identification of repetitive DNA sequences. Period. Biol. 2020, 121–122, 7–14. [Google Scholar] [CrossRef]
- de Lima, L.G.; Ruiz-Ruano, F.J. In-depth satellitome analyses of 37 Drosophila species illuminate repetitive DNA evolution in the Drosophila genus. Genome Biol. Evol. 2022, 14, evac064. [Google Scholar] [CrossRef] [PubMed]
- Petitpierre, E.; Juan, C.; Pons, J.; Plohl, M.; Ugarković, Đ. Satellite DNA and constitutive heterochromatin in tenebrionid beetles. In Kew Chromosome Conference IV; Brandham, P.E., Bennet, M., Eds.; Royal Botanic Gardens: Richmond, UK, 1995; pp. 351–362. [Google Scholar]
- Mora, P.; Pita, S.; Montiel, E.E.; Rico-Porras, J.M.; Palomeque, T.; Panzera, F.; Lorite, P. Making the genome huge: The case of Triatoma delpontei, a triatominae species with more than 50% of its genome full of satellite DNA. Genes 2023, 14, 371. [Google Scholar] [CrossRef] [PubMed]
- Zakharov, I.A.; Shaikevich, Y.V. Molecular study of geographic races of ladybird beetles Adalia bipunctata and A. frigida. Russ. J. Genet. Appl. Res. 2016, 6, 138–143. [Google Scholar] [CrossRef]
- Fry, K.; Salser, W. Nucleotide sequences of HS–alpha satellite DNA from kangaroo rat Dipodomys ordii and characterization of similar sequences in other rodents. Cell 1977, 12, 1069–1084. [Google Scholar] [CrossRef]
- Mestrović, N.; Plohl, M.; Mravinac, B.; Ugarković, Ð. Evolution of satellite DNAs from the genus Palorus––experimental evidence for the “library” hypothesis. Mol. Biol. Evol. 1998, 15, 1062–1068. [Google Scholar] [CrossRef]
- Pons, J.; Bruvo, B.; Petitpierre, E.; Plohl, M.; Ugarković, Ð.; Juan, C. Complex structural features of satellite DNA sequences in the genus Pimelia (Coleoptera: Tenebrionidae): Random differential amplification from a common ‘satellite DNA library’. Hereditas 2004, 92, 418–427. [Google Scholar] [CrossRef]
- Blackmon, H.; Demuth, J.P. Coleoptera Karyotype Database. Coleopterists Bull. 2015, 69, 174–175. [Google Scholar] [CrossRef]
- Bracewell, R.; Chatla, K.; Nalley, M.J.; Bachtrog, D. Dynamic turnover of centromeres drives karyotype evolution in Drosophila. eLife 2019, 8, e49002. [Google Scholar] [CrossRef]
- Quigley, S.; Damas, J.; Larkin, D.M.; Farré, M. syntenyPlotteR: A user-friendly R package to visualize genome synteny, ideal for both experienced and novice bioinformaticians. Bioinform. Adv. 2023, 3, vbad161. [Google Scholar] [CrossRef]
- Kaiser, V.B.; Bachtrog, D. Evolution of sex chromosomes in insects. Annu. Rev. Genet. 2010, 44, 91–112. [Google Scholar] [CrossRef] [PubMed]
- Blackmon, H.; Demuth, J.P. Genomic origins of insect sex chromosomes. Curr. Opin. Insect Sci. 2015, 7, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Lorite, P.; Chica, E.; Palomeque, T. G-banding and chromosome condensation in the ant, Tapinoma nigerrimum. Chromosome Res. 1996, 4, 77–79. [Google Scholar] [CrossRef] [PubMed]
- Sumner, A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972, 75, 304–306. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones–Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Bao, W.; Kojima, K.K.; Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 2015, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI–BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Smit, A.F.A.; Hubley, R.; Green, P. RepeatMasker Open-4.0. 2013–2015. Available online: http://www.repeatmasker.org (accessed on 10 June 2024).
- RStudio Team. RStudio: Integrated Development for R; RStudio, PBC: Boston, MA, USA, 2020; Available online: http://www.rstudio.com/ (accessed on 10 June 2024).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Springer International Publishing: Cham, Switzerland, 2016; Available online: https://ggplot2.tidyverse.org (accessed on 10 June 2024).
- Negm, S.; Greenberg, A.; Larracuente, A.M.; Sproul, J.S. RepeatProfiler: A pipeline for visualization and comparative analysis of repetitive DNA profiles. Mol. Ecol. Resour. 2021, 21, 969–981. [Google Scholar] [CrossRef]
- Cabral-de-Mello, D.C.; Marec, F. Universal fluorescence in situ hybridization (FISH) protocol for mapping repetitive DNAs in insects and other arthropods. Mol. Genet. Genom. 2021, 296, 513–526. [Google Scholar] [CrossRef]
- Pinkel, D.; Straume, T.; Gray, J.M. Cytogenetic analysis using quantitative, high sensitivity fluorescence hybridization. Proc. Natl. Acad. Sci. USA 1986, 83, 2934–2938. [Google Scholar] [CrossRef] [PubMed]
- Feron, R.; Waterhouse, R.M. Assessing species coverage and assembly quality of rapidly accumulating sequenced genomes. GigaScience 2022, 11, giac006. [Google Scholar] [CrossRef] [PubMed]



| SatDNA Families | % A+T | Illumina Reads | Genome Assembly | ||
|---|---|---|---|---|---|
| Genome (%) | Divergence (Kimura-2p) | Chromosomes (%) | Chromosomes + Unplaced Scaffolds (%) | ||
| AbipSat01-187 | 70.1 | 9.9808 | 7.91 | 3.2895 | 7.0636 |
| AbipSat02-497 | 64.8 | 1.6789 | 15.99 | 1.8672 | 1.7659 |
| AbipSat03-8 | 87.5 | 0.3864 | 2.57 | 0.2758 | 0.5723 |
| AbipSat04-193 | 55.4 | 0.2678 | 6.62 | 0.2257 | 0.2134 |
| AbipSat05-258 | 65.9 | 0.2383 | 6.91 | 0.2325 | 0.2199 |
| AbipSat06-176 | 74.4 | 0.2328 | 7.01 | 0.0019 | 0.1960 |
| AbipSat07-579 | 63.4 | 0.1054 | 6.22 | 0.1396 | 0.1323 |
| AbipSat08-316 | 64.9 | 0.1017 | 8.56 | 0.1349 | 0.1276 |
| AbipSat09-473 | 68.7 | 0.0916 | 18.10 | 0.1705 | 0.1613 |
| AbipSat10-1902 | 57.1 | 0.0874 | 1.52 | 0.1012 | 0.1100 |
| AbipSat11-151 | 62.3 | 0.0816 | 9.31 | 0.1029 | 0.0976 |
| AbipSat12-233 | 62.2 | 0.0810 | 2.16 | 0.0268 | 0.0254 |
| AbipSat13-1767 | 66.3 | 0.0799 | 7.13 | 0.1924 | 0.1823 |
| AbipSat14-53 | 73.6 | 0.0751 | 3.25 | 0.0001 | 0.0008 |
| AbipSat15-179 | 65.4 | 0.0712 | 17.93 | 0.0957 | 0.0905 |
| AbipSat16-443 | 56.7 | 0.0660 | 10.05 | 0.0946 | 0.0920 |
| AbipSat17-86 | 69.8 | 0.0642 | 22.80 | 0.0826 | 0.0781 |
| AbipSat18-14 | 64.3 | 0.0636 | 6.85 | 0.0664 | 0.0641 |
| AbipSat19-1237 | 63.3 | 0.0618 | 3.28 | 0.1174 | 0.1110 |
| AbipSat20-176 | 61.9 | 0.0550 | 10.94 | 0.0540 | 0.0543 |
| AbipSat21-1899 | 65.4 | 0.0537 | 8.61 | 0.0796 | 0.0759 |
| AbipSat22-148 | 57.4 | 0.0527 | 4.00 | 0.0455 | 0.0431 |
| AbipSat23-376 | 58.8 | 0.0474 | 11.11 | 0.1068 | 0.1024 |
| AbipSat24-5-TEL | 60.0 | 0.0434 | 3.63 | 0.0068 | 0.0137 |
| AbipSat25-176 | 63.6 | 0.0416 | 18.96 | 0.0369 | 0.0349 |
| AbipSat26-2235 | 60.1 | 0.0398 | 3.47 | 0.0549 | 0.0544 |
| AbipSat27-84 | 61.9 | 0.0352 | 23.48 | 0.0007 | 0.0006 |
| AbipSat28-165 | 67.9 | 0.0330 | 16.69 | 0.0432 | 0.0408 |
| AbipSat29-174 | 64.9 | 0.0282 | 23.19 | 0.0369 | 0.0349 |
| AbipSat30-212 | 59.0 | 0.0278 | 21.05 | 0.0047 | 0.0057 |
| AbipSat31-145 | 63.4 | 0.0241 | 8.06 | 0.0353 | 0.0335 |
| AbipSat32-163 | 60.1 | 0.0237 | 8.55 | 0.0310 | 0.0321 |
| AbipSat33-284 | 62.7 | 0.0228 | 11.54 | 0.0339 | 0.0322 |
| AbipSat34-137 | 70.1 | 0.0223 | 10.47 | 0.0354 | 0.0337 |
| AbipSat35-141 | 56.0 | 0.0218 | 4.68 | 0.0167 | 0.0158 |
| AbipSat36-157 | 47.8 | 0.0194 | 4.49 | 0.0364 | 0.0345 |
| AbipSat37-297 | 65.3 | 0.0153 | 11.24 | 0.0221 | 0.0209 |
| AbipSat38-2435 | 58.2 | 0.0141 | 4.60 | 0.0210 | 0.0204 |
| AbipSat39-159 | 60.4 | 0.0132 | 6.50 | 0.0103 | 0.0097 |
| AbipSat40-152 | 61.8 | 0.0131 | 11.87 | 0.0109 | 0.0105 |
| AbipSat41-69 | 53.6 | 0.0117 | 4.52 | 0.0069 | 0.0067 |
| AbipSat42-21 | 61.9 | 0.0116 | 18.70 | 0.0330 | 0.0313 |
| AbipSat43-282 | 69.9 | 0.0111 | 7.37 | 0.0086 | 0.0092 |
| AbipSat44-148 | 56.8 | 0.0110 | 8.71 | 0.0138 | 0.0130 |
| AbipSat45-307 | 64.5 | 0.0109 | 5.95 | 0.0088 | 0.0083 |
| AbipSat46-126 | 50.8 | 0.0089 | 2.36 | 0.0061 | 0.0058 |
| AbipSat47-399 | 60.7 | 0.0080 | 5.88 | 0.0162 | 0.0153 |
| AbipSat48-18 | 50.0 | 0.0080 | 7.15 | ND | ND |
| AbipSat49-148 | 60.8 | 0.0075 | 8.48 | 0.0091 | 0.0086 |
| AbipSat50-199 | 55.8 | 0.0072 | 12.04 | 0.0042 | 0.0040 |
| AbipSat51-128 | 60.9 | 0.0071 | 4.80 | 0.0055 | 0.0052 |
| AbipSat52-177 | 66.7 | 0.0064 | 8.37 | 0.0089 | 0.0084 |
| AbipSat53-22 | 50.0 | 0.0059 | 15.97 | 0.0035 | 0.0033 |
| AbipSat54-271 | 65.3 | 0.0058 | 6.28 | 0.0002 | 0.00350 |
| AbipSat55-145 | 62.1 | 0.0054 | 4.12 | 0.0085 | 0.0082 |
| AbipSat56-30 | 46.7 | 0.0051 | 9.38 | ND | ND |
| AbipSat57-144 | 58.3 | 0.0049 | 14.89 | 0.0040 | 0.0039 |
| AbipSat58-518 | 60.0 | 0.0047 | 7.88 | 0.0060 | 0.0062 |
| AbipSat59-105 | 55.2 | 0.0039 | 1.37 | 0.0030 | 0.0026 |
| AbipSat60-160 | 60.0 | 0.0038 | 3.13 | 0.0020 | 0.0020 |
| AbipSat61-175 | 68.0 | 0.0038 | 8.32 | 0.0070 | 0.0064 |
| AbipSat62-144 | 55.6 | 0.0037 | 2.52 | 0.0020 | 0.0019 |
| AbipSat63-186 | 61.8 | 0.0031 | 4.08 | 0.0020 | 0.0017 |
| AbipSat64-201 | 62.2 | 0.0031 | 5.24 | 0.0020 | 0.0023 |
| AbipSat65-286 | 59.1 | 0.0029 | 5.91 | 0.0040 | 0.0036 |
| AbipSat66-169 | 62.1 | 0.0028 | 1.26 | ND | ND |
| AbipSat67-304 | 67.8 | 0.0027 | 2.01 | 0.0020 | 0.0019 |
| AbipSat68-237 | 54.9 | 0.0025 | 6.66 | 0.0006 | 0.0006 |
| AbipSat69-438 | 63.5 | 0.0025 | 2.03 | 0.0020 | 0.0019 |
| AbipSat70-168 | 56.0 | 0.0024 | 3.23 | 0.0009 | 0.0008 |
| AbipSat71-438 | 61.9 | 0.0023 | 1.93 | 0.0012 | 0.0011 |
| AbipSat72-426 | 64.1 | 0.0022 | 2.45 | 0.0019 | 0.0018 |
| AbipSat73-396 | 71.0 | 0.0020 | 7.27 | 0.0012 | 0.0011 |
| AbipSat74-344 | 61.0 | 0.0019 | 7.28 | 0.0023 | 0.0022 |
| AbipSat75-471 | 61.6 | 0.0019 | 7.51 | 0.0011 | 0.0011 |
| AbipSat76-140 | 76.4 | 0.0018 | 4.76 | 0.0006 | 0.0005 |
| AbipSat77-155 | 54.8 | 0.0018 | 4.58 | 0.0024 | 0.0022 |
| AbipSat78-399 | 63.9 | 0.0017 | 4.18 | 0.0021 | 0.0020 |
| AbipSat79-309 | 67.0 | 0.0017 | 1.39 | 0.0034 | 0.0032 |
| AbipSat80-337 | 57.3 | 0.0016 | 3.51 | 0.0019 | 0.0018 |
| AbipSat81-258 | 66.7 | 0.0016 | 3.95 | 0.0006 | 0.0005 |
| AbipSat82-291 | 59.8 | 0.0014 | 2.87 | 0.0009 | 0.0008 |
| AbipSat83-191 | 64.9 | 0.0012 | 0.93 | 0.0009 | 0.0008 |
| AbipSat84-169 | 52.7 | 0.0011 | 0.57 | 0.0005 | 0.0004 |
| AbipSat85-284 | 65.1 | 0.0011 | 5.45 | 0.0010 | 0.0009 |
| AbipSat86-294 | 61.9 | 0.0009 | 3.25 | 0.0016 | 0.0015 |
| Satellite proportion | 14.6677 | 8.1351 | 12.1966 | ||
| SatDNA Family | Oligonucleotide Name | Sequence |
|---|---|---|
| AbipSat01-187 | Abip-CL1-F | ACTGCTGTGAATAATCGG |
| Abip-CL1-R | GACTTTTTGATCTGAGGG | |
| AbipSat02-497 | Abip-CL4-F | AATATGGTGTTTTTGGGG |
| Abip-CL4-R | TTTCACCTGTCAAATGGC | |
| AbipSat03-8 | AbipCL-13 | GTCAAATGTCATCTGTCAAA |
| AbipSat04-193 | Abip-CL46-F | CAAATTTTTTCTCTTCGC |
| Abip-CL46-R | ACCATGCATGCTGTATTG | |
| AbipSat05-258 | Abip-CL42-F | TTTCGAAAAATTATATTCC |
| Abip-CL42-R | CTTTGTAGCTCAATTGTTC | |
| AbipSat06-176 | Abip-CL46-RE2-F | AACGAATTCCACCAAGGG |
| Abip-CL46-RE2-R | TCGTTCAAGTAGCCGAGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mora, P.; Rico-Porras, J.M.; Palomeque, T.; Montiel, E.E.; Pita, S.; Cabral-de-Mello, D.C.; Lorite, P. Satellitome Analysis of Adalia bipunctata (Coleoptera): Revealing Centromeric Turnover and Potential Chromosome Rearrangements in a Comparative Interspecific Study. Int. J. Mol. Sci. 2024, 25, 9214. https://doi.org/10.3390/ijms25179214
Mora P, Rico-Porras JM, Palomeque T, Montiel EE, Pita S, Cabral-de-Mello DC, Lorite P. Satellitome Analysis of Adalia bipunctata (Coleoptera): Revealing Centromeric Turnover and Potential Chromosome Rearrangements in a Comparative Interspecific Study. International Journal of Molecular Sciences. 2024; 25(17):9214. https://doi.org/10.3390/ijms25179214
Chicago/Turabian StyleMora, Pablo, José M. Rico-Porras, Teresa Palomeque, Eugenia E. Montiel, Sebastián Pita, Diogo C. Cabral-de-Mello, and Pedro Lorite. 2024. "Satellitome Analysis of Adalia bipunctata (Coleoptera): Revealing Centromeric Turnover and Potential Chromosome Rearrangements in a Comparative Interspecific Study" International Journal of Molecular Sciences 25, no. 17: 9214. https://doi.org/10.3390/ijms25179214
APA StyleMora, P., Rico-Porras, J. M., Palomeque, T., Montiel, E. E., Pita, S., Cabral-de-Mello, D. C., & Lorite, P. (2024). Satellitome Analysis of Adalia bipunctata (Coleoptera): Revealing Centromeric Turnover and Potential Chromosome Rearrangements in a Comparative Interspecific Study. International Journal of Molecular Sciences, 25(17), 9214. https://doi.org/10.3390/ijms25179214








