The Role of Furin and Its Therapeutic Potential in Cardiovascular Disease Risk
Abstract
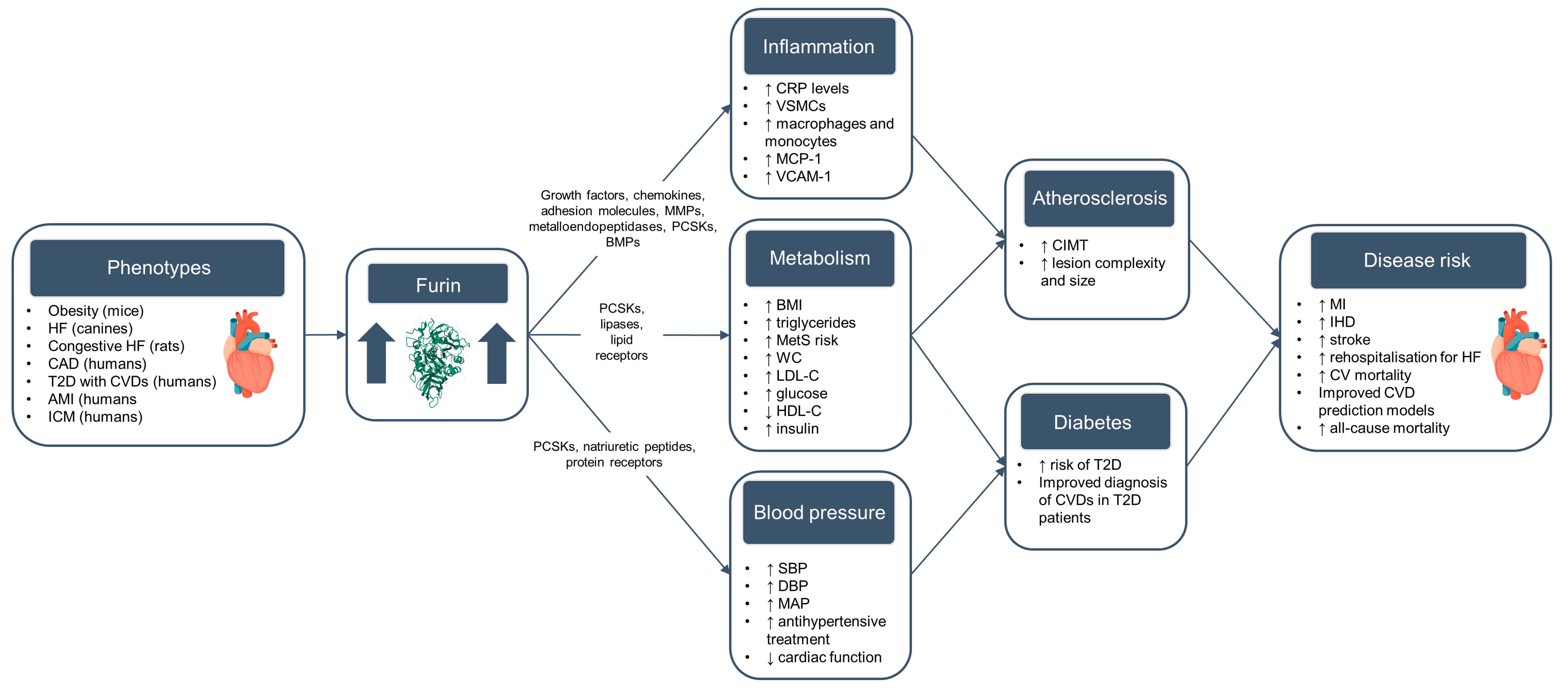
:1. Introduction
2. Risk Factors for Cardiovascular Diseases
2.1. Blood Pressure Traits
2.2. Blood Lipids
| Study Design | Population | Primary Endpoints | Protein Measurement | Key Findings | Reference |
|---|---|---|---|---|---|
| Blood pressure traits: | |||||
| Cross-sectional and prospective (Suzhou, China) | Middle-aged and elderly residents in the community (N = 2312) | Hypertension, SBP, DBP, MAP | Furin serum levels measured using ELISA | Cross-sectional: lowest quartile of furin concentration compared to the highest had higher SBP, DBP, and MAP Prospective: those in the lowest quartile of furin compared to the highest had a higher risk of hypertension | [21] |
| Prospective (Malmö, Sweden) | Residents aged 45 to 69 years (N = 4678) | Diabetes, CAD, all-cause mortality, cause-specific mortality | Furin serum levels measured using PEA (Olink) | Baseline furin concentration strongly associated with SBP, DBP, and antihypertensive treatment | [22] |
| Blood lipids: | |||||
| Prospective (Malmö, Sweden) | Residents aged 45 to 69 years (N = 4678) | Diabetes, CAD, all-cause mortality, cause-specific mortality | Furin serum levels measured using PEA (Olink) | Baseline furin concentration strongly associated LDL and HDL | [22] |
| Nested case-control (Västerbotten, Sweden) | Residents aged > 40 years (N = 276) | MetS (based on score developed using BMI, triglyceride levels, total-C levels, mid-blood pressure, and fasting glucose levels) and CRC | Furin serum levels measured using PEA (Olink) | Furin levels positively associated with MetS score, BMI, and triglyceride levels (Bonferroni-adjusted p < 0.5) | [51] |
| Cross-sectional (Suita, Japan) | CAD patients with HeFH (N = 138) | Intravascular ultrasound measures (PAV, vessel volume, imaged length, and lumen volume) | Total, mature, and furin-cleaved PCSK9 (fc-PCSK9) measured using ELISA | No significant association between fc-PCSK9 and PAV, vessel volume, or lumen volume Fc-PCSK9 was not a significant predictor of PAV | [52] |
| Prospective, (Suita, Japan) | City residents aged 30–79 years (N = 1436) | CAD (including AMI, sudden cardiac death, and stable CAD) | Total, mature, and fc-PCSK9 measured using ELISA | Fc-PCSK9 was positively associated with SBP, BMI, high-sensitivity CRP levels, and DBP at baseline | [53] |
| Randomised clinical trial (Iwate, Japan) | STEMI patients (N = 36) | Serum mature and fc-PCSK9 and serum Lp(a) | Mature and fc-PCSK9 measured using ELISA | Fc-PCSK9 levels were significantly reduced with administration of evolocumab | [54] |
| Diabetes and adiposity: | |||||
| Prospective (Malmö, Sweden) | Residents aged 45 to 69 years (N = 4678) | Diabetes, CAD, all-cause mortality, cause-specific mortality | Furin serum levels measured using PEA (Olink) | Baseline furin concentration strongly associated with BMI, glucose, insulin After adjusting for conventional risk factors, increase in furin concentration is associated with higher risk of diabetes Higher furin concentration associated with lower survival rate | [22] |
| Prospective (China) | Middle-aged and elderly residents in the community in Suzhou (N = 892) | Abdominal obesity (WC ≥ 85 cm for males and 80 cm for females) | Furin serum levels measured using ELISA | Lower furin serum levels were associated with higher BMI, WC, and prevalent diabetes at baseline Individuals in the lowest tertile of serum furin had a higher risk of developing abdominal obesity compared to those with highest tertile | [55] |
| Case-control (Gaza Strip, Palestine) | Hospital patients referred to diabetic clinic, hospital cardio care unit and healthy subjects from routine check-ups (N = 75) | T2D with and without cardiovascular comorbidities | Furin serum levels measured using ELISA | Significantly higher furin concentration in T2D patients with CVDs than those without and healthy controls Furin had a high sensitivity (80%) and specificity (96%) for diagnosing CVDs in T2D patients | [56] |
| Cross-sectional (Suzhou, China) | Middle-aged and elderly residents in the community (N = 2172) | Diabetes (>7.0 mmol/L) and prediabetes (5.6–6.9 mmol/L) | Furin serum levels measured using ELISA | Lower log-furin levels associated with higher levels of fasting plasma glucose Lower log-furin levels borderline associated with increased risk of diabetes and prediabetes | [57] |
2.3. Diabetes and Adiposity
3. Cardiovascular Diseases
3.1. Coronary Artery Disease (CAD)
3.2. Stroke
3.3. Other Cardiovascular Diseases
4. Summary and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| α-1-PDX | α1-antitrypsin Portland |
| ADHF | Acute decompensated heart failure |
| AMI | Acute myocardial infarction |
| BBJ | Biobank Japan |
| bi-shRNA | Bifunctional small hairpin RNA |
| BMI | Body mass index |
| BNP | Brain natriuretic peptide/B-type natriuretic peptide |
| CAD | Coronary artery disease |
| CC4D | CARDIoGRAMplusC4D |
| CHF | Congestive heart failure |
| CIMT | Carotid intima media thickness |
| CKB | China Kadoorie Biobank |
| CRP | C-reactive protein |
| CVD | Cardiovascular disease |
| DALYs | Disability adjusted life years |
| DBP | Diastolic blood pressure |
| DCM | Dilated cardiomyopathy |
| ENaC | Epithelial sodium channel |
| eQTL | Expression quantitative trait loci |
| Fc-PCSK9 | Furin-cleaved PCSK9 |
| FDR | False discovery rate |
| GBD | Global Burden of Disease study |
| GEO | Gene Expression Omnibus |
| GTEx | Genotype Tissue Expression database |
| GWAS | Genome-wide association studies |
| HDL-C | High-density lipoprotein cholesterol |
| HeFH | Heterozygous familial hypercholesterolemia |
| HF | Heart failure |
| HR | Hazard ratio |
| ICBP | International consortium for blood pressure |
| ICM | Ischaemic cardiomyopathy |
| ICU | Intensive care units |
| IHD | Ischaemic heart disease |
| IL1-β | Interleukin-1 beta |
| LAS | Large-artery atherosclerotic stroke |
| LDL-C | Low-density lipoprotein cholesterol |
| LDLR | Low-density lipoprotein lipase receptors |
| MACE | Major adverse coronary events |
| MAP | Mean arterial pressure |
| MCAO | Middle cerebral artery occlusion |
| MCP-1 | Monocyte chemotactic protein-1 |
| MetS | Metabolic syndrome |
| MeCP2 | Methyl-CpG binding protein 2 |
| MI | Myocardial infarction |
| MiRNA | MicroRNA |
| mRNA | Messenger RNA |
| MMPs | Matrix metalloproteinases |
| MR | Mendelian randomisation |
| NSTEMI | Non-ST segment elevation myocardial infarction |
| OR | Odds ratio |
| PCSK6 | Proprotein convertase subtilisin/kexin type 6 |
| PCSK9 | Proprotein convertase subtilisin/kexin type 9 |
| PCR | Polymerase chain reaction |
| PheWAS | Phenome-wide association studies |
| PLTP | Phospholipid transfer protein |
| pQTL | Protein quantitative trait loci |
| PRR | Pro-renin receptor |
| SBP | Systolic blood pressure |
| SD | Standard deviation |
| shRNA | Short-hairpin RNA |
| SNP | Single nucleotide polymorphism |
| STEMI | ST-elevation myocardial infarction |
| T2D | Type 2 diabetes |
| TAA | Thoracic aortic aneurysms |
| TGF-β1 | Transforming growth factor beta 1 |
| TIA | Transient ischaemic attack |
| TNF-α | Tumour necrosis factor alpha |
| UKB | UK Biobank |
| VCAM-1 | Vascular cell adhesion molecule-1 |
References
- Mensah, G.A.; Fuster, V.; Murray, C.J.L.; Roth, G.A.; Global Burden of Cardiovascular, D.; Risks, C. Global Burden of Cardiovascular Diseases and Risks, 1990–2022. J. Am. Coll. Cardiol. 2023, 82, 2350–2473. [Google Scholar] [CrossRef]
- Palstrom, N.B.; Matthiesen, R.; Rasmussen, L.M.; Beck, H.C. Recent Developments in Clinical Plasma Proteomics-Applied to Cardiovascular Research. Biomedicines 2022, 10, 162. [Google Scholar] [CrossRef]
- Ferkingstad, E.; Sulem, P.; Atlason, B.A.; Sveinbjornsson, G.; Magnusson, M.I.; Styrmisdottir, E.L.; Gunnarsdottir, K.; Helgason, A.; Oddsson, A.; Halldorsson, B.V.; et al. Large-scale integration of the plasma proteome with genetics and disease. Nat. Genet. 2021, 53, 1712–1721. [Google Scholar] [CrossRef]
- Mazidi, M.; Wright, N.; Yao, P.; Kartsonaki, C.; Millwood, I.Y.; Fry, H.; Said, S.; Pozarickij, A.; Pei, P.; Chen, Y.; et al. Plasma Proteomics to Identify Drug Targets for Ischemic Heart Disease. J. Am. Coll. Cardiol. 2023, 82, 1906–1920. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Thomas, G. Furin at the cutting edge: From protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 2002, 3, 753–766. [Google Scholar] [CrossRef]
- Seidah, N.G.; Prat, A. The biology and therapeutic targeting of the proprotein convertases. Nat. Rev. Drug Discov. 2012, 11, 367–383. [Google Scholar] [CrossRef]
- Osman, E.E.A.; Rehemtulla, A.; Neamati, N. Why All the Fury over Furin? J. Med. Chem. 2022, 65, 2747–2784. [Google Scholar] [CrossRef]
- Seidah, N.G.; Sadr, M.S.; Chretien, M.; Mbikay, M. The multifaceted proprotein convertases: Their unique, redundant, complementary, and opposite functions. J. Biol. Chem. 2013, 288, 21473–21481. [Google Scholar] [CrossRef] [PubMed]
- Peacock, T.P.; Goldhill, D.H.; Zhou, J.; Baillon, L.; Frise, R.; Swann, O.C.; Kugathasan, R.; Penn, R.; Brown, J.C.; Sanchez-David, R.Y.; et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021, 6, 899–909. [Google Scholar] [CrossRef]
- Coto, E.; Albaiceta, G.M.; Amado-Rodriguez, L.; Garcia-Clemente, M.; Cuesta-Llavona, E.; Vazquez-Coto, D.; Alonso, B.; Iglesias, S.; Melon, S.; Alvarez-Arguelles, M.E.; et al. FURIN gene variants (rs6224/rs4702) as potential markers of death and cardiovascular traits in severe COVID-19. J. Med. Virol. 2022, 94, 3589–3595. [Google Scholar] [CrossRef] [PubMed]
- Ming, Y.; Qiang, L. Involvement of Spike Protein, Furin, and ACE2 in SARS-CoV-2-Related Cardiovascular Complications. SN Compr. Clin. Med. 2020, 2, 1103–1108. [Google Scholar] [CrossRef] [PubMed]
- Langnau, C.; Rohlfing, A.K.; Gekeler, S.; Gunter, M.; Poschel, S.; Petersen-Uribe, A.; Jaeger, P.; Avdiu, A.; Harm, T.; Kreisselmeier, K.P.; et al. Platelet Activation and Plasma Levels of Furin Are Associated with Prognosis of Patients with Coronary Artery Disease and COVID-19. Arter. Thromb. Vasc. Biol. 2021, 41, 2080–2096. [Google Scholar] [CrossRef] [PubMed]
- Roebroek, A.J.; Umans, L.; Pauli, I.G.; Robertson, E.J.; van Leuven, F.; Van de Ven, W.J.; Constam, D.B. Failure of ventral closure and axial rotation in embryos lacking the proprotein convertase Furin. Development 1998, 125, 4863–4876. [Google Scholar] [CrossRef]
- Wichaiyo, S.; Koonyosying, P.; Morales, N.P. Functional Roles of Furin in Cardio-Cerebrovascular Diseases. ACS Pharmacol. Transl. Sci. 2024, 7, 570–585. [Google Scholar] [CrossRef]
- Chen, Z.; Lu, S.; Xu, M.; Liu, P.; Ren, R.; Ma, W. Role of miR-24, Furin, and Transforming Growth Factor-beta1 Signal Pathway in Fibrosis After Cardiac Infarction. Med. Sci. Monit. 2017, 23, 65–70. [Google Scholar] [CrossRef]
- Semenov, A.G.; Tamm, N.N.; Seferian, K.R.; Postnikov, A.B.; Karpova, N.S.; Serebryanaya, D.V.; Koshkina, E.V.; Krasnoselsky, M.I.; Katrukha, A.G. Processing of pro-B-type natriuretic peptide: Furin and corin as candidate convertases. Clin. Chem. 2010, 56, 1166–1176. [Google Scholar] [CrossRef] [PubMed]
- Ren, K.; Jiang, T.; Zheng, X.L.; Zhao, G.J. Proprotein convertase furin/PCSK3 and atherosclerosis: New insights and potential therapeutic targets. Atherosclerosis 2017, 262, 163–170. [Google Scholar] [CrossRef]
- Li, N.; Luo, W.; Juhong, Z.; Yang, J.; Wang, H.; Zhou, L.; Chang, J. Associations between genetic variations in the FURIN gene and hypertension. BMC Med. Genet. 2010, 11, 124. [Google Scholar] [CrossRef]
- Turpeinen, H.; Seppala, I.; Lyytikainen, L.P.; Raitoharju, E.; Hutri-Kahonen, N.; Levula, M.; Oksala, N.; Waldenberger, M.; Klopp, N.; Illig, T.; et al. A genome-wide expression quantitative trait loci analysis of proprotein convertase subtilisin/kexin enzymes identifies a novel regulatory gene variant for FURIN expression and blood pressure. Hum. Genet. 2015, 134, 627–636. [Google Scholar] [CrossRef]
- He, Y.; Ren, L.; Zhang, Q.; Zhang, M.; Shi, J.; Hu, W.; Peng, H.; Zhang, Y. Serum furin as a biomarker of high blood pressure: Findings from a longitudinal study in Chinese adults. Hypertens. Res. 2019, 42, 1808–1815. [Google Scholar] [CrossRef]
- Fernandez, C.; Rysa, J.; Almgren, P.; Nilsson, J.; Engstrom, G.; Orho-Melander, M.; Ruskoaho, H.; Melander, O. Plasma levels of the proprotein convertase furin and incidence of diabetes and mortality. J. Intern. Med. 2018, 284, 377–387. [Google Scholar] [CrossRef]
- CARDIoGRAMplusC4D Consortium; Deloukas, P.; Kanoni, S.; Willenborg, C.; Farrall, M.; Assimes, T.L.; Thompson, J.R.; Ingelsson, E.; Saleheen, D.; Erdmann, J.; et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 2013, 45, 25–33. [Google Scholar] [CrossRef]
- Stawowy, P.; Marcinkiewicz, J.; Graf, K.; Seidah, N.; Chretien, M.; Fleck, E.; Marcinkiewicz, M. Selective expression of the proprotein convertases furin, pc5, and pc7 in proliferating vascular smooth muscle cells of the rat aorta in vitro. J. Histochem. Cytochem. 2001, 49, 323–332. [Google Scholar] [CrossRef]
- Senzer, N.; Barve, M.; Kuhn, J.; Melnyk, A.; Beitsch, P.; Lazar, M.; Lifshitz, S.; Magee, M.; Oh, J.; Mill, S.W.; et al. Phase I trial of “bi-shRNAi(furin)/GMCSF DNA/autologous tumor cell” vaccine (FANG) in advanced cancer. Mol. Ther. 2012, 20, 679–686. [Google Scholar] [CrossRef]
- Oh, J.; Barve, M.; Matthews, C.M.; Koon, E.C.; Heffernan, T.P.; Fine, B.; Grosen, E.; Bergman, M.K.; Fleming, E.L.; DeMars, L.R.; et al. Phase II study of Vigil(R) DNA engineered immunotherapy as maintenance in advanced stage ovarian cancer. Gynecol Oncol. 2016, 143, 504–510. [Google Scholar] [CrossRef]
- Vaduganathan, M.; Mensah, G.A.; Turco, J.V.; Fuster, V.; Roth, G.A. The Global Burden of Cardiovascular Diseases and Risk: A Compass for Future Health. J. Am. Coll. Cardiol. 2022, 80, 2361–2371. [Google Scholar] [CrossRef]
- Cilhoroz, B.T.; Schifano, E.D.; Panza, G.A.; Ash, G.I.; Corso, L.; Chen, M.H.; Deshpande, V.; Zaleski, A.; Farinatti, P.; Santos, L.P.; et al. FURIN variant associations with postexercise hypotension are intensity and race dependent. Physiol. Rep. 2019, 7, 13952. [Google Scholar] [CrossRef]
- Ganesh, S.K.; Tragante, V.; Guo, W.; Guo, Y.; Lanktree, M.B.; Smith, E.N.; Johnson, T.; Castillo, B.A.; Barnard, J.; Baumert, J.; et al. Loci influencing blood pressure identified using a cardiovascular gene-centric array. Hum. Mol. Genet. 2013, 22, 1663–1678. [Google Scholar] [CrossRef]
- International Consortium for Blood Pressure Genome-Wide Association Studies; Ehret, G.B.; Munroe, P.B.; Rice, K.M.; Bochud, M.; Johnson, A.D.; Chasman, D.I.; Smith, A.V.; Tobin, M.D.; Verwoert, G.C.; et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 2011, 478, 103–109. [Google Scholar] [CrossRef]
- Zhang, H.; Mo, X.B.; Xu, T.; Bu, X.Q.; Lei, S.F.; Zhang, Y.H. Novel Genes Affecting Blood Pressure Detected Via Gene-Based Association Analysis. G3 2015, 5, 1035–1042. [Google Scholar] [CrossRef]
- Feitosa, M.F.; Kraja, A.T.; Chasman, D.I.; Sung, Y.J.; Winkler, T.W.; Ntalla, I.; Guo, X.; Franceschini, N.; Cheng, C.Y.; Sim, X.; et al. Novel genetic associations for blood pressure identified via gene-alcohol interaction in up to 570K individuals across multiple ancestries. PLoS ONE 2018, 13, 0198166. [Google Scholar] [CrossRef]
- Sakaue, S.; Kanai, M.; Tanigawa, Y.; Karjalainen, J.; Kurki, M.; Koshiba, S.; Narita, A.; Konuma, T.; Yamamoto, K.; Akiyama, M.; et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet. 2021, 53, 1415–1424. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, T.J.; Ehret, G.B.; Nandakumar, P.; Ranatunga, D.; Schaefer, C.; Kwok, P.Y.; Iribarren, C.; Chakravarti, A.; Risch, N. Genome-wide association analyses using electronic health records identify new loci influencing blood pressure variation. Nat. Genet. 2017, 49, 54–64. [Google Scholar] [CrossRef]
- Plotnikov, D.; Huang, Y.; Khawaja, A.P.; Foster, P.J.; Zhu, Z.; Guggenheim, J.A.; He, M. High Blood Pressure and Intraocular Pressure: A Mendelian Randomization Study. Investig. Ophthalmol. Vis. Sci. 2022, 63, 29. [Google Scholar] [CrossRef] [PubMed]
- Shi, G. Genome-wide variance quantitative trait locus analysis suggests small interaction effects in blood pressure traits. Sci. Rep. 2022, 12, 12649. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Richard, M.; Musani, S.K.; Sung, Y.J.; Winkler, T.W.; Schwander, K.; Chai, J.F.; Guo, X.; Kilpelainen, T.O.; Vojinovic, D.; et al. Multi-Ancestry Genome-wide Association Study Accounting for Gene-Psychosocial Factor Interactions Identifies Novel Loci for Blood Pressure Traits. HGG Adv. 2021, 2, 100013. [Google Scholar] [CrossRef]
- Guindo-Martinez, M.; Amela, R.; Bonas-Guarch, S.; Puiggros, M.; Salvoro, C.; Miguel-Escalada, I.; Carey, C.E.; Cole, J.B.; Rueger, S.; Atkinson, E.; et al. The impact of non-additive genetic associations on age-related complex diseases. Nat. Commun. 2021, 12, 2436. [Google Scholar] [CrossRef]
- Liu, C.; Kraja, A.T.; Smith, J.A.; Brody, J.A.; Franceschini, N.; Bis, J.C.; Rice, K.; Morrison, A.C.; Lu, Y.; Weiss, S.; et al. Meta-analysis identifies common and rare variants influencing blood pressure and overlapping with metabolic trait loci. Nat. Genet. 2016, 48, 1162–1170. [Google Scholar] [CrossRef]
- Kanai, M.; Akiyama, M.; Takahashi, A.; Matoba, N.; Momozawa, Y.; Ikeda, M.; Iwata, N.; Ikegawa, S.; Hirata, M.; Matsuda, K.; et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat. Genet. 2018, 50, 390–400. [Google Scholar] [CrossRef]
- Takeuchi, F.; Akiyama, M.; Matoba, N.; Katsuya, T.; Nakatochi, M.; Tabara, Y.; Narita, A.; Saw, W.Y.; Moon, S.; Spracklen, C.N.; et al. Interethnic analyses of blood pressure loci in populations of East Asian and European descent. Nat. Commun. 2018, 9, 5052. [Google Scholar] [CrossRef] [PubMed]
- Kiiskinen, T.; Helkkula, P.; Krebs, K.; Karjalainen, J.; Saarentaus, E.; Mars, N.; Lehisto, A.; Zhou, W.; Cordioli, M.; Jukarainen, S.; et al. Genetic predictors of lifelong medication-use patterns in cardiometabolic diseases. Nat. Med. 2023, 29, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Tyrmi, J.S.; Kaartokallio, T.; Lokki, A.I.; Jaaskelainen, T.; Kortelainen, E.; Ruotsalainen, S.; Karjalainen, J.; Ripatti, S.; Kivioja, A.; Laisk, T.; et al. Genetic Risk Factors Associated with Preeclampsia and Hypertensive Disorders of Pregnancy. JAMA Cardiol. 2023, 8, 674–683. [Google Scholar] [CrossRef]
- Honigberg, M.C.; Truong, B.; Khan, R.R.; Xiao, B.; Bhatta, L.; Vy, H.M.T.; Guerrero, R.F.; Schuermans, A.; Selvaraj, M.S.; Patel, A.P.; et al. Polygenic prediction of preeclampsia and gestational hypertension. Nat. Med. 2023, 29, 1540–1549. [Google Scholar] [CrossRef] [PubMed]
- Abe, S.; Tokoro, F.; Matsuoka, R.; Arai, M.; Noda, T.; Watanabe, S.; Horibe, H.; Fujimaki, T.; Oguri, M.; Kato, K.; et al. Association of genetic variants with dyslipidemia. Mol. Med. Rep. 2015, 12, 5429–5436. [Google Scholar] [CrossRef]
- Ueyama, C.; Horibe, H.; Yamase, Y.; Fujimaki, T.; Oguri, M.; Kato, K.; Arai, M.; Watanabe, S.; Murohara, T.; Yamada, Y. Association of FURIN and ZPR1 polymorphisms with metabolic syndrome. Biomed. Rep. 2015, 3, 641–647. [Google Scholar] [CrossRef]
- Li, Z.; Chyr, J.; Jia, Z.; Wang, L.; Hu, X.; Wu, X.; Song, C. Identification of Hub Genes Associated with Hypertension and Their Interaction with miRNA Based on Weighted Gene Coexpression Network Analysis (WGCNA) Analysis. Med. Sci. Monit. 2020, 26, 923514. [Google Scholar] [CrossRef]
- Jin, W.; Wang, X.; Millar, J.S.; Quertermous, T.; Rothblat, G.H.; Glick, J.M.; Rader, D.J. Hepatic proprotein convertases modulate HDL metabolism. Cell Metab. 2007, 6, 129–136. [Google Scholar] [CrossRef]
- Jin, W.; Fuki, I.V.; Seidah, N.G.; Benjannet, S.; Glick, J.M.; Rader, D.J. Proprotein convertases [corrected] are responsible for proteolysis and inactivation of endothelial lipase. J. Biol. Chem. 2005, 280, 36551–36559. [Google Scholar] [CrossRef]
- Wang, Y.K.; Tang, J.N.; Han, L.; Liu, X.D.; Shen, Y.L.; Zhang, C.Y.; Liu, X.B. Elevated FURIN levels in predicting mortality and cardiovascular events in patients with acute myocardial infarction. Metabolism 2020, 111, 154323. [Google Scholar] [CrossRef] [PubMed]
- Harlid, S.; Myte, R.; Van Guelpen, B. The Metabolic Syndrome, Inflammation, and Colorectal Cancer Risk: An Evaluation of Large Panels of Plasma Protein Markers Using Repeated, Prediagnostic Samples. Mediators Inflamm. 2017, 2017, 4803156. [Google Scholar] [CrossRef]
- Kataoka, Y.; Harada-Shiba, M.; Nakao, K.; Nakashima, T.; Kawakami, S.; Fujino, M.; Kanaya, T.; Nagai, T.; Tahara, Y.; Asaumi, Y.; et al. Mature proprotein convertase subtilisin/kexin type 9, coronary atheroma burden, and vessel remodeling in heterozygous familial hypercholesterolemia. J. Clin. Lipidol. 2017, 11, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, Y.; Harada-Shiba, M.; Hori, M.; Watanabe, M.; Kokubo, Y.; Noguchi, T.; Yasuda, S.; Miyamoto, Y. Circulating Furin-Cleaved Proprotein Convertase Subtilisin/Kexin Type 9 Concentration Predicts Future Coronary Events in Japanese Subjects. JACC Asia 2021, 1, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Kanazawa, M.; Kagaya, Y.; Kondo, M.; Sato, K.; Endo, H.; Nozaki, E. Plasma kinetics of mature PCSK9, furin-cleaved PCSK9, and Lp(a) with or without administration of PCSK9 inhibitors in acute myocardial infarction. J. Cardiol. 2020, 76, 395–401. [Google Scholar] [CrossRef]
- He, Y.; Ren, L.; Zhang, Q.; Zhang, M.; Shi, J.; Hu, W.; Peng, H. Deficient serum furin predicts risk of abdominal obesity: Findings from a prospective cohort of Chinese adults. Postgrad. Med. J. 2021, 97, 234–238. [Google Scholar] [CrossRef]
- Fathy, S.A.; Abdel Hamid, F.F.; Zabut, B.M.; Jamee, A.F.; Ali, M.A.; Abu Mustafa, A.M. Diagnostic utility of BNP, corin and furin as biomarkers for cardiovascular complications in type 2 diabetes mellitus patients. Biomarkers 2015, 20, 460–469. [Google Scholar] [CrossRef]
- He, Y.; Zhu, H.; Zhang, M.; Li, J.; Ma, S.; Lu, Y.; Chen, L.; Zhang, M.; Peng, H. Association Between Serum Furin and Fasting Glucose: A Cross-Sectional Study in Chinese Adults. Front. Endocrinol. 2021, 12, 781890. [Google Scholar] [CrossRef]
- Zhao, G.; Yang, W.; Wu, J.; Chen, B.; Yang, X.; Chen, J.; McVey, D.G.; Andreadi, C.; Gong, P.; Webb, T.R.; et al. Influence of a Coronary Artery Disease-Associated Genetic Variant on FURIN Expression and Effect of Furin on Macrophage Behavior. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 1837–1844. [Google Scholar] [CrossRef]
- Yang, X.; Yang, W.; McVey, D.G.; Zhao, G.; Hu, J.; Poston, R.N.; Ren, M.; Willeit, K.; Coassin, S.; Willeit, J.; et al. FURIN Expression in Vascular Endothelial Cells Is Modulated by a Coronary Artery Disease-Associated Genetic Variant and Influences Monocyte Transendothelial Migration. J. Am. Heart Assoc. 2020, 9, e014333. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, G.; Sakurada, M.; Kitagawa, K.; Kondo, T.; Takahashi, M.; Ueno, Y. Effect of FURIN SNP rs17514846 on coronary atherosclerosis in human cardiac specimens: An autopsy study of 106 cases. Leg. Med. 2022, 55, 102006. [Google Scholar] [CrossRef]
- Zorkoltseva, I.; Shadrina, A.; Belonogova, N.; Kirichenko, A.; Tsepilov, Y.; Axenovich, T. In silico genome-wide gene-based association analysis reveals new genes predisposing to coronary artery disease. Clin. Genet. 2022, 101, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, K.N.; Fisher, E.A.; Breslow, J.L. Overexpression of PCSK9 accelerates the degradation of the LDLR in a post-endoplasmic reticulum compartment. Proc. Natl. Acad. Sci. USA 2005, 102, 2069–2074. [Google Scholar] [CrossRef] [PubMed]
- Rashid, S.; Curtis, D.E.; Garuti, R.; Anderson, N.N.; Bashmakov, Y.; Ho, Y.K.; Hammer, R.E.; Moon, Y.A.; Horton, J.D. Decreased plasma cholesterol and hypersensitivity to statins in mice lacking Pcsk9. Proc. Natl. Acad. Sci. USA 2005, 102, 5374–5379. [Google Scholar] [CrossRef] [PubMed]
- Oleaga, C.; Hay, J.; Gurcan, E.; David, L.L.; Mueller, P.A.; Tavori, H.; Shapiro, M.D.; Pamir, N.; Fazio, S. Insights into the kinetics and dynamics of the furin-cleaved form of PCSK9. J. Lipid. Res. 2021, 62, 100003. [Google Scholar] [CrossRef]
- Sawaguchi, J.; Saeki, Y.; Oda, M.; Takamura, T.A.; Fujibayashi, K.; Wakasa, M.; Akao, H.; Kitayama, M.; Kawai, Y.; Kajinami, K. The circulating furin-cleaved/mature PCSK9 ratio has a potential prognostic significance in statin-naive patients with acute ST elevation myocardial infarction. Atheroscler. Plus 2022, 50, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Basu, D.; Li, Z.; Zhang, M.; Rudic, R.D.; Jiang, X.C.; Jin, W. Hepatic overexpression of the prodomain of furin lessens progression of atherosclerosis and reduces vascular remodeling in response to injury. Atherosclerosis 2014, 236, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Lei, X.; Jiang, H.; Li, Z.; Creemers, J.W.M.; Zhang, M.; Qin, S.; Jin, W.; Jiang, X.C. Prodomain of Furin Promotes Phospholipid Transfer Protein Proteasomal Degradation in Hepatocytes. J. Am. Heart Assoc. 2018, 7. [Google Scholar] [CrossRef]
- Zhong, M.; Munzer, J.S.; Basak, A.; Benjannet, S.; Mowla, S.J.; Decroly, E.; Chretien, M.; Seidah, N.G. The prosegments of furin and PC7 as potent inhibitors of proprotein convertases. In vitro and ex vivo assessment of their efficacy and selectivity. J. Biol. Chem. 1999, 274, 33913–33920. [Google Scholar] [CrossRef]
- Yang, W.; Cao, J.; McVey, D.G.; Ye, S. Allele-Specific Epigenetic Regulation of FURIN Expression at a Coronary Artery Disease Susceptibility Locus. Cells 2023, 12, 1681. [Google Scholar] [CrossRef]
- Kappert, K.; Meyborg, H.; Fritzsche, J.; Urban, D.; Kruger, J.; Wellnhofer, E.; Kintscher, U.; Fleck, E.; Stawowy, P. Proprotein convertase subtilisin/kexin type 3 promotes adipose tissue-driven macrophage chemotaxis and is increased in obesity. PLoS ONE 2013, 8, 70542. [Google Scholar] [CrossRef]
- Stawowy, P.; Meyborg, H.; Stibenz, D.; Borges Pereira Stawowy, N.; Roser, M.; Thanabalasingam, U.; Veinot, J.P.; Chretien, M.; Seidah, N.G.; Fleck, E.; et al. Furin-like proprotein convertases are central regulators of the membrane type matrix metalloproteinase-pro-matrix metalloproteinase-2 proteolytic cascade in atherosclerosis. Circulation 2005, 111, 2820–2827. [Google Scholar] [CrossRef]
- Turpeinen, H.; Raitoharju, E.; Oksanen, A.; Oksala, N.; Levula, M.; Lyytikainen, L.P.; Jarvinen, O.; Creemers, J.W.; Kahonen, M.; Laaksonen, R.; et al. Proprotein convertases in human atherosclerotic plaques: The overexpression of FURIN and its substrate cytokines BAFF and APRIL. Atherosclerosis 2011, 219, 799–806. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, L.; Mi, S.; Wang, H.; Wang, C.; Jia, W.; Gong, L.; Dong, H.; Xu, B.; Jing, Y.; et al. FURIN suppresses the progression of atherosclerosis by promoting macrophage autophagy. FASEB J. 2023, 37, 22933. [Google Scholar] [CrossRef] [PubMed]
- Nikpay, M.; Goel, A.; Won, H.H.; Hall, L.M.; Willenborg, C.; Kanoni, S.; Saleheen, D.; Kyriakou, T.; Nelson, C.P.; Hopewell, J.C.; et al. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015, 47, 1121–1130. [Google Scholar] [CrossRef] [PubMed]
- Temprano-Sagrera, G.; Sitlani, C.M.; Bone, W.P.; Martin-Bornez, M.; Voight, B.F.; Morrison, A.C.; Damrauer, S.M.; de Vries, P.S.; Smith, N.L.; Sabater-Lleal, M. Multi-phenotype analyses of hemostatic traits with cardiovascular events reveal novel genetic associations. J. Thromb. Haemost. 2022, 20, 1331–1349. [Google Scholar] [CrossRef]
- Matsunaga, H.; Ito, K.; Akiyama, M.; Takahashi, A.; Koyama, S.; Nomura, S.; Ieki, H.; Ozaki, K.; Onouchi, Y.; Sakaue, S.; et al. Transethnic Meta-Analysis of Genome-Wide Association Studies Identifies Three New Loci and Characterizes Population-Specific Differences for Coronary Artery Disease. Circ. Genom. Precis. Med. 2020, 13, e002670. [Google Scholar] [CrossRef] [PubMed]
- van der Harst, P.; Verweij, N. Identification of 64 Novel Genetic Loci Provides an Expanded View on the Genetic Architecture of Coronary Artery Disease. Circ. Res. 2018, 122, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Vivar, J.; Nelson, C.P.; Willenborg, C.; Segre, A.V.; Makinen, V.P.; Nikpay, M.; Erdmann, J.; Blankenberg, S.; O’Donnell, C.; et al. Systems Genetics Analysis of Genome-Wide Association Study Reveals Novel Associations Between Key Biological Processes and Coronary Artery Disease. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 1712–1722. [Google Scholar] [CrossRef]
- Sun, Q.X.; Zhou, H.M.; Du, Q.W. Association of Rs2071410 on Furin with Transient Ischemic Attack Susceptibility and Prognosis in a Chinese Population. Med. Sci. Monit. 2016, 22, 3828–3834. [Google Scholar] [CrossRef]
- Malik, R.; Chauhan, G.; Traylor, M.; Sargurupremraj, M.; Okada, Y.; Mishra, A.; Rutten-Jacobs, L.; Giese, A.K.; van der Laan, S.W.; Gretarsdottir, S.; et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat. Genet. 2018, 50, 524–537. [Google Scholar] [CrossRef]
- Mishra, A.; Malik, R.; Hachiya, T.; Jurgenson, T.; Namba, S.; Posner, D.C.; Kamanu, F.K.; Koido, M.; Le Grand, Q.; Shi, M.; et al. Stroke genetics informs drug discovery and risk prediction across ancestries. Nature 2022, 611, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.W.; Ma, Q.; Liu, J.; Li, J.W.; Chen, Y.D. The association between plasma furin and cardiovascular events after acute myocardial infarction. BMC Cardiovasc. Disord. 2021, 21, 468. [Google Scholar] [CrossRef] [PubMed]
- Vodovar, N.; Seronde, M.F.; Laribi, S.; Gayat, E.; Lassus, J.; Boukef, R.; Nouira, S.; Manivet, P.; Samuel, J.L.; Logeart, D.; et al. Post-translational modifications enhance NT-proBNP and BNP production in acute decompensated heart failure. Eur. Heart J. 2014, 35, 3434–3441. [Google Scholar] [CrossRef]
- Tarazon, E.; Rosello-Lleti, E.; Rivera, M.; Ortega, A.; Molina-Navarro, M.M.; Trivino, J.C.; Lago, F.; Gonzalez-Juanatey, J.R.; Orosa, P.; Montero, J.A.; et al. RNA sequencing analysis and atrial natriuretic peptide production in patients with dilated and ischemic cardiomyopathy. PLoS ONE 2014, 9, 90157. [Google Scholar] [CrossRef]
- Yakala, G.K.; Cabrera-Fuentes, H.A.; Crespo-Avilan, G.E.; Rattanasopa, C.; Burlacu, A.; George, B.L.; Anand, K.; Mayan, D.C.; Corliano, M.; Hernandez-Resendiz, S.; et al. FURIN Inhibition Reduces Vascular Remodeling and Atherosclerotic Lesion Progression in Mice. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 387–401. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, Q.; Liao, Z. Microarray Data Analysis of Molecular Mechanism Associated with Stroke Progression. J. Mol. Neurosci. 2019, 67, 424–433. [Google Scholar] [CrossRef]
- Akerman, A.W.; Collins, E.N.; Peterson, A.R.; Collins, L.B.; Harrison, J.K.; DeVaughn, A.; Townsend, J.M.; Vanbuskirk, R.L.; Riopedre-Maqueira, J.; Reyes, A.; et al. miR-133a Replacement Attenuates Thoracic Aortic Aneurysm in Mice. J. Am. Heart Assoc. 2021, 10, e019862. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Liu, X.; Zhao, L.; Yan, S.; Song, Q.; Zou, C.; Li, X. MiR-22-3p in exosomes increases the risk of heart failure after down-regulation of FURIN. Chem. Biol. Drug Des. 2023, 101, 550–567. [Google Scholar] [CrossRef]
- Khoury, E.E.; Knaney, Y.; Fokra, A.; Kinaneh, S.; Azzam, Z.; Heyman, S.N.; Abassi, Z. Pulmonary, cardiac and renal distribution of ACE2, furin, TMPRSS2 and ADAM17 in rats with heart failure: Potential implication for COVID-19 disease. J. Cell Mol. Med. 2021, 25, 3840–3855. [Google Scholar] [CrossRef]
- Ichiki, T.; Boerrigter, G.; Huntley, B.K.; Sangaralingham, S.J.; McKie, P.M.; Harty, G.J.; Harders, G.E.; Burnett, J.C., Jr. Differential expression of the pro-natriuretic peptide convertases corin and furin in experimental heart failure and atrial fibrosis. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013, 304, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Stawowy, P.; Fleck, E. Proprotein convertases furin and PC5: Targeting atherosclerosis and restenosis at multiple levels. J. Mol. Med. 2005, 83, 865–875. [Google Scholar] [CrossRef] [PubMed]
- Klein-Szanto, A.J.; Bassi, D.E. Proprotein convertase inhibition: Paralyzing the cell’s master switches. Biochem. Pharmacol. 2017, 140, 8–15. [Google Scholar] [CrossRef]
- Seidah, N.G.; Abifadel, M.; Prost, S.; Boileau, C.; Prat, A. The Proprotein Convertases in Hypercholesterolemia and Cardiovascular Diseases: Emphasis on Proprotein Convertase Subtilisin/Kexin 9. Pharmacol. Rev. 2017, 69, 33–52. [Google Scholar] [CrossRef] [PubMed]
- Suur, B.E.; Chemaly, M.; Lindquist Liljeqvist, M.; Djordjevic, D.; Stenemo, M.; Bergman, O.; Karlof, E.; Lengquist, M.; Odeberg, J.; Hurt-Camejo, E.; et al. Therapeutic potential of the Proprotein Convertase Subtilisin/Kexin family in vascular disease. Front. Pharmacol. 2022, 13, 988561. [Google Scholar] [CrossRef]
- Scamuffa, N.; Calvo, F.; Chretien, M.; Seidah, N.G.; Khatib, A.M. Proprotein convertases: Lessons from knockouts. FASEB J. 2006, 20, 1954–1963. [Google Scholar] [CrossRef]
- Zaragoza-Huesca, D.; Martinez-Cortes, C.; Banegas-Luna, A.J.; Perez-Garrido, A.; Vegara-Meseguer, J.M.; Penas-Martinez, J.; Rodenas, M.C.; Espin, S.; Perez-Sanchez, H.; Martinez-Martinez, I. Identification of Kukoamine A, Zeaxanthin, and Clexane as New Furin Inhibitors. Int. J. Mol. Sci. 2022, 23, 2796. [Google Scholar] [CrossRef]
- Thomas, G.; Couture, F.; Kwiatkowska, A. The Path to Therapeutic Furin Inhibitors: From Yeast Pheromones to SARS-CoV-2. Int. J. Mol. Sci. 2022, 23, 3435. [Google Scholar] [CrossRef]
- Devi, K.P.; Pourkarim, M.R.; Thijssen, M.; Sureda, A.; Khayatkashani, M.; Cismaru, C.A.; Neagoe, I.B.; Habtemariam, S.; Razmjouei, S.; Khayat Kashani, H.R. A perspective on the applications of furin inhibitors for the treatment of SARS-CoV-2. Pharmacol. Rep. 2022, 74, 425–430. [Google Scholar] [CrossRef]
- Fitzgerald, K. Furin Protease: From SARS CoV-2 to Anthrax, Diabetes, and Hypertension. Perm. J. 2020, 24, 4. [Google Scholar] [CrossRef]
- Rocconi, R.P.; Grosen, E.A.; Ghamande, S.A.; Chan, J.K.; Barve, M.A.; Oh, J.; Tewari, D.; Morris, P.C.; Stevens, E.E.; Bottsford-Miiler, J.N.; et al. Gemogenovatucel-T (Vigil) immunotherapy as maintenance in frontline stage III/IV ovarian cancer (VITAL): A randomised, double-blind, placebo-controlled, phase 2b trial. Lancet Oncol. 2020, 21, 1661–1672. [Google Scholar] [CrossRef] [PubMed]
- Ghisoli, M.; Barve, M.; Mennel, R.; Lenarsky, C.; Horvath, S.; Wallraven, G.; Pappen, B.O.; Whiting, S.; Rao, D.; Senzer, N.; et al. Three-year Follow up of GMCSF/bi-shRNA. DNA-transfected Autologous Tumor Immunotherapy (Vigil) in Metastatic Advanced Ewing’s Sarcoma. Mol. Ther. 2016, 24, 1478–1483. [Google Scholar] [CrossRef]
- Wouters, O.J.; McKee, M.; Luyten, J. Estimated Research and Development Investment Needed to Bring a New Medicine to Market, 2009–2018. JAMA 2020, 323, 844–853. [Google Scholar] [CrossRef] [PubMed]
- Hughes, J.P.; Rees, S.; Kalindjian, S.B.; Philpott, K.L. Principles of early drug discovery. Br. J. Pharmacol. 2011, 162, 1239–1249. [Google Scholar] [CrossRef] [PubMed]
- Minikel, E.V.; Painter, J.L.; Dong, C.C.; Nelson, M.R. Refining the impact of genetic evidence on clinical success. Nature 2024, 629, 624–629. [Google Scholar] [CrossRef]
- Trajanoska, K.; Bherer, C.; Taliun, D.; Zhou, S.; Richards, J.B.; Mooser, V. From target discovery to clinical drug development with human genetics. Nature 2023, 620, 737–745. [Google Scholar] [CrossRef]
- Sun, B.B.; Chiou, J.; Traylor, M.; Benner, C.; Hsu, Y.H.; Richardson, T.G.; Surendran, P.; Mahajan, A.; Robins, C.; Vasquez-Grinnell, S.G.; et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature 2023, 622, 329–338. [Google Scholar] [CrossRef]


| Study Design | Study Population | SNP | Chr | Mapped Genes | Minor Allele | Associated Trait(s) | Reference |
|---|---|---|---|---|---|---|---|
| Blood pressure and hypertension: | |||||||
| Case-control | Turkic N = 934 | rs2071410 | 15 | FURIN | A/G/T | Hypertension (G allele = ↑ risk) | [19] |
| Cross-sectional | European N = 1428 | rs4702 | 15 | FURIN | A/C | Protein expression in blood (A allele = ↓ levels), SBP and DBP (A allele = ↑ levels) | [20] |
| Intervention | Multiple ancestries N = 23 | rs12917264 | 15 | FURIN | T | Post-exercise hypotension ** | [28] |
| rs75493298 | 15 | FURIN | A/T | Post-exercise hypotension ** | |||
| rs74037507 | 15 | FURIN | A/C | Post-exercise hypotension ** | |||
| rs1573644 | 15 | FURIN | C | Post-exercise hypotension ** | |||
| Cross-sectional | European N = 61,619 | rs2071410 | 15 | FURIN | A/G/T | DBP ** | [29] |
| rs6227 | 15 | FURIN | T | SBP, MAP ** | |||
| Case-control, cross-sectional | Multiple ancestries N = 276,527 | rs2521501 | 15 | FES | C/T | SBP, DBP (T allele = ↑ levels) | [30] |
| Case-control, cross-sectional | European N = 69,395 | rs8032315 | 15 | FURIN | A/C | SBP | [31] |
| Cross-sectional | Multiple ancestries N = 130,828 | rs6227 | 15 | FURIN | T | SBP ** | [32] * |
| rs1573643 | 15 | FURIN | C/G | DBP, MAP ** | |||
| rs8027450 | 15 | FURIN | A/T | PP ** | |||
| rs8029440 | 15 | RN7SL363P, FURIN | A | DBP ** | |||
| Cross-sectional | Multiple ancestries N = 178,726 | rs7166599 | 15 | RN7SL363P, FURIN | C/G | SBP, MAP (G allele = ↑ levels) | [33] * |
| rs12906125 | 15 | FES—FURIN | A | SBP (A allele = ↑ levels) | |||
| rs1573643 | 15 | FURIN | C/G | MAP (C allele = ↑ levels) | |||
| Longitudinal | Multiple ancestries N = 99,785 | rs4932371 | 15 | RN7SL363P, FURIN | C | SBP, DBP, PP (T allele = ↓ levels) | [34] * |
| Cross-sectional | Europeans N = 226,997 | rs8032315 | 15 | FURIN | A/C | SBP (A allele = ↑ levels) | [35] * |
| Cross-sectional | Europeans N = 396,077 | rs8032315 | 15 | FURIN | A/C | SBP (A allele = ↑ levels) | [36] * |
| Cross-sectional | Europeans N = 68,450 | rs2071410 | 15 | FURIN | A/G/T | DBP/depression ** | [37] * |
| Case-control | Europeans N = 56,637 | rs57515981 | 15 | FURIN | AAAGGAAG, AAAGGCAG, AAGGCAG | Hypertension (AAAGGCAG alleles = ↑ risk) | [38] * |
| Cross-sectional, case-control | Multiple ancestries N = 146,562 | rs17514846 | 15 | FURIN | A/G/T | SBP, DBP, MAP (C allele = ↓ levels) | [39] * |
| Cross-sectional | East Asian N = 162,255 | rs17514846 | 15 | FURIN | A/G/T | SBP ** | [40] * |
| Cross-sectional | East Asian N = 130,777 | rs17514846 | 15 | FURIN | A/G/T | SBP and MAP (A allele = ↑ levels) and hypertension (A allele = ↑ risk) | [41] * |
| rs8039305 | 15 | FURIN | C | Hypertension (T allele = ↓ risk) | |||
| Case-control | Europeans N = 218,792 | rs8032315 | 15 | FURIN | A/C | Antihypertensive use ** | [42] * |
| Case control | European N = 15,200 | rs6224 | 15 | FURIN | C/T | Preeclampsia or other hypertensive disorders during pregnancy (T allele = ↑ risk) | [43] * |
| Case-control | Multiple ancestry N = 468,391 | rs6224 | 15 | FURIN | C/T | Preeclampsia and gestational hypertension (T allele = ↑ risk) | [44] |
| Blood lipids: | |||||||
| Cross-sectional | Multiple ancestries N = 178,726 | rs8039305 | 15 | FURIN | C | Taking lipid-lowering medication—HMG-CoA reductase inhibitors (C10AA; C allele = ↑ use) | [33] * |
| Case-control | East Asian N = 5460 | rs17514846 | 15 | FURIN | A/G/T | Serum triglycerides (A allele = ↓ levels) | [45] |
| Case-control | East Asian N = 2918 | rs17514846 | 15 | FURIN | A/G/T | MetS (A allele = ↓ risk), serum triglycerides (A allele = ↓ levels), and serum HDL-C (A allele = ↑ levels) | [46] |
| Study Design | Study Population | SNP | Chr | Mapped Genes | Minor Allele | Associated Trait(s) | Reference |
|---|---|---|---|---|---|---|---|
| Coronary artery disease: | |||||||
| Two sample MR | Multiple ancestries N = >20,000 | rs6227 | 15 | FURIN | C/T | IHD ** | [4] |
| rs1029420 | 15 | FURIN | C | IHD ** | |||
| Case-control | Multiple ancestries N = 194,427 | rs17514846 | 15 | FURIN | A/G/T | CAD (A allele = ↑ risk) | [23] |
| Case-control, cross-sectional | European N = 69,395 | rs8032315 | 15 | FURIN | A/C | CAD ** | [31] |
| Cross-sectional | Multiple ancestries N = 178,726 | rs57515981 | 15 | FURIN | AAAGGAAG, AAAGGCAG, AAGGCAG | UAP (AAAGGCAG alleles = ↑ risk) and WBC (AAAGGCAG alleles = ↑ levels) | [33] * |
| Longitudinal | Multiple ancestries N = 99,785 | rs4932371 | 15 | RN7SL363P, FURIN | C | CAD (C allele = ↑ risk) | [34] * |
| Cross-sectional, case-control | Multiple ancestries N = 146,562 | rs17514846 | 15 | FURIN | A/G/T | CAD and MI (C allele = ↓ risk) | [39] * |
| Case-control | Europeans N = 218,792 | rs8032315 | 15 | FURIN | A/C | CAD | [42] * |
| Cross-sectional | European N = 826 | rs17514846 | 15 | FURIN | A/G/T | Circulating MCP-1 (A allele = ↑ levels), and CIMT (A allele = ↑ thickness) | [59] |
| Case-control | European N = 408,458 | rs6227 | 15 | FURIN | T | IHD ** | [61] |
| rs17514846 | 15 | FURIN | A/G/T | IHD ** | |||
| Case-control | Multiple ancestries N = 184,305 | rs17514846 | 15 | FURIN | A/G/T | CAD (A allele = ↑ risk) | [74] * |
| Case-control | Multiple ancestries N = 738,986 | rs8027450 | 15 | FURIN | A/T | CAD with haemostatic traits ** | [75] * |
| rs8032315 | 15 | FURIN | A/C | CAD, PAI-1, tPA ** | |||
| Case-control | Multiple ancestries N = 392,241 | rs4932371 | 15 | RN7SL363P, FURIN | C | CAD (C allele = ↑ risk) | [76] * |
| Case-control | Multiple ancestries N = 296,525 | rs17514846 | 15 | FURIN | A/G/T | CAD (A allele = ↑ risk) | [77] * |
| rs4932373 | 15 | FES | C/T | CAD (A allele = ↓ risk) | |||
| Case-control | European N = 20,978 | rs17514846 | 15 | FURIN | A/G/T | Identified as critical component in several different CAD pathways | [78] |
| Stroke: | |||||||
| Case-control | East Asian N = 753 | rs2071410 | 15 | FURIN | A/G/T | TIA (G allele = ↑ risk), 90-day prognosis (G allele = ↓ prognosis) | [79] |
| Case-control | Multiple ancestries N = 521,612 | rs4932370 | 15 | RN7SL363P, FURIN | A/C/T | AIS ** | [80] * |
| Case-control | Multiple ancestries N = 1,614,080 | rs1573644 | 15 | FURIN | C | LAS, AS and AIS (C allele = ↑ risk) | [81] |
| Study Design | Population | Primary Endpoints | Protein Measurement | Key Findings | Reference |
|---|---|---|---|---|---|
| Coronary artery disease | |||||
| Case-subcohort (China) | Incident CAD cases and subcohort from CKB prospective cohort study (N = 3977) | CAD | Plasma furin levels measured using Olink Explore 1500 assay | Higher risk of CAD associated with higher plasma furin levels | [4] |
| Case control (Shanghai, China) | STEMI and non-STEMI patients admitted to Tongji Hospital and Shanghai East Hospital, Tongji University (N = 1312) | MACE (including all-cause mortality, hospitalisation for HF and recurrent MI) | Furin serum levels measured using ELISA | Risk of MACE and all-cause mortality significantly higher for those in the highest furin concentration Risk of recurrent MI, cardiovascular death, and rehospitalisation for HF were also significantly higher for those with the highest furin concentrations Furin significantly improved prediction models | [50] |
| Prospective, (Suita, Japan) | City residents aged 30–79 years (N = 1436) | CAD (including AMI, sudden cardiac death, and stable CAD) | Total, mature, and fc-PCSK9 measured using ELISA | Highest tertile of fc-PCSK9 was associated with a higher risk of coronary and composite events compared to the lowest tertile Fc-PCSK9 predicted future coronary events whereas the mature subtype did not demonstrate any association with CV outcomes | [53] |
| Prospective, (Kanazawa, Japan) | Hospitalised STEMI patients (N = 126) | MACE (composite of all-cause mortality, non-fatal MI and stroke, and new angina pectoris) | Mature and fc-PCSK9 measured using ELISA | Patients in the highest tertile of fc/m-PCSK9 serum levels were at significantly higher risk of MACE during the follow-up period | [65] |
| Prospective (Beijing, China) | Hospitalised AMI patients admitted to the People’s Liberation Army General Hospital (N = 1100) | MACE (composite of CV death, non-fatal MI, non-fatal stroke) | Furin serum levels measured using ELISA | Significant association between elevated furin levels at baseline and recurrent non-fatal MI, but not with MACE or CV death | [82] |
| Other cardiovascular diseases | |||||
| Cross-sectional (Monastir, Tunisia; Paris, France; Finland) | Emergency room patients presenting with shortness of breath (“Biomarcoeurs” cohort) and AHF patients from 14 hospitals (FINN-AKVA cohort) (N = 683) | ADHF and CHF | Furin serum levels measured using ELISA, furin activity measured using fluorescence | No significant difference in furin plasma concentrations of between groups Furin activity was significantly higher in ADHF cases No correlation between furin concentration and plasma levels of proBNP, NT-proBNP, and BNP | [83] |
| Case-control (Valencia, Spain) | Patients at “La Fe” University Hospital (N = 73) | DCM and ICM | Furin tissue protein levels of measured by gel electrophoresis and western blot | No significant difference in furin protein levels between HF patients and controls Furin levels were significantly higher in ICM patients compared to DCM patients | [84] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fry, H.; Mazidi, M.; Kartsonaki, C.; Clarke, R.; Walters, R.G.; Chen, Z.; Millwood, I.Y. The Role of Furin and Its Therapeutic Potential in Cardiovascular Disease Risk. Int. J. Mol. Sci. 2024, 25, 9237. https://doi.org/10.3390/ijms25179237
Fry H, Mazidi M, Kartsonaki C, Clarke R, Walters RG, Chen Z, Millwood IY. The Role of Furin and Its Therapeutic Potential in Cardiovascular Disease Risk. International Journal of Molecular Sciences. 2024; 25(17):9237. https://doi.org/10.3390/ijms25179237
Chicago/Turabian StyleFry, Hannah, Mohsen Mazidi, Christiana Kartsonaki, Robert Clarke, Robin G. Walters, Zhengming Chen, and Iona Y. Millwood. 2024. "The Role of Furin and Its Therapeutic Potential in Cardiovascular Disease Risk" International Journal of Molecular Sciences 25, no. 17: 9237. https://doi.org/10.3390/ijms25179237






