Genomics, Proteomics, and Antifungal Activity of Chitinase from the Antarctic Marine Bacterium Curtobacterium sp. CBMAI 2942
Abstract
1. Introduction
2. Results
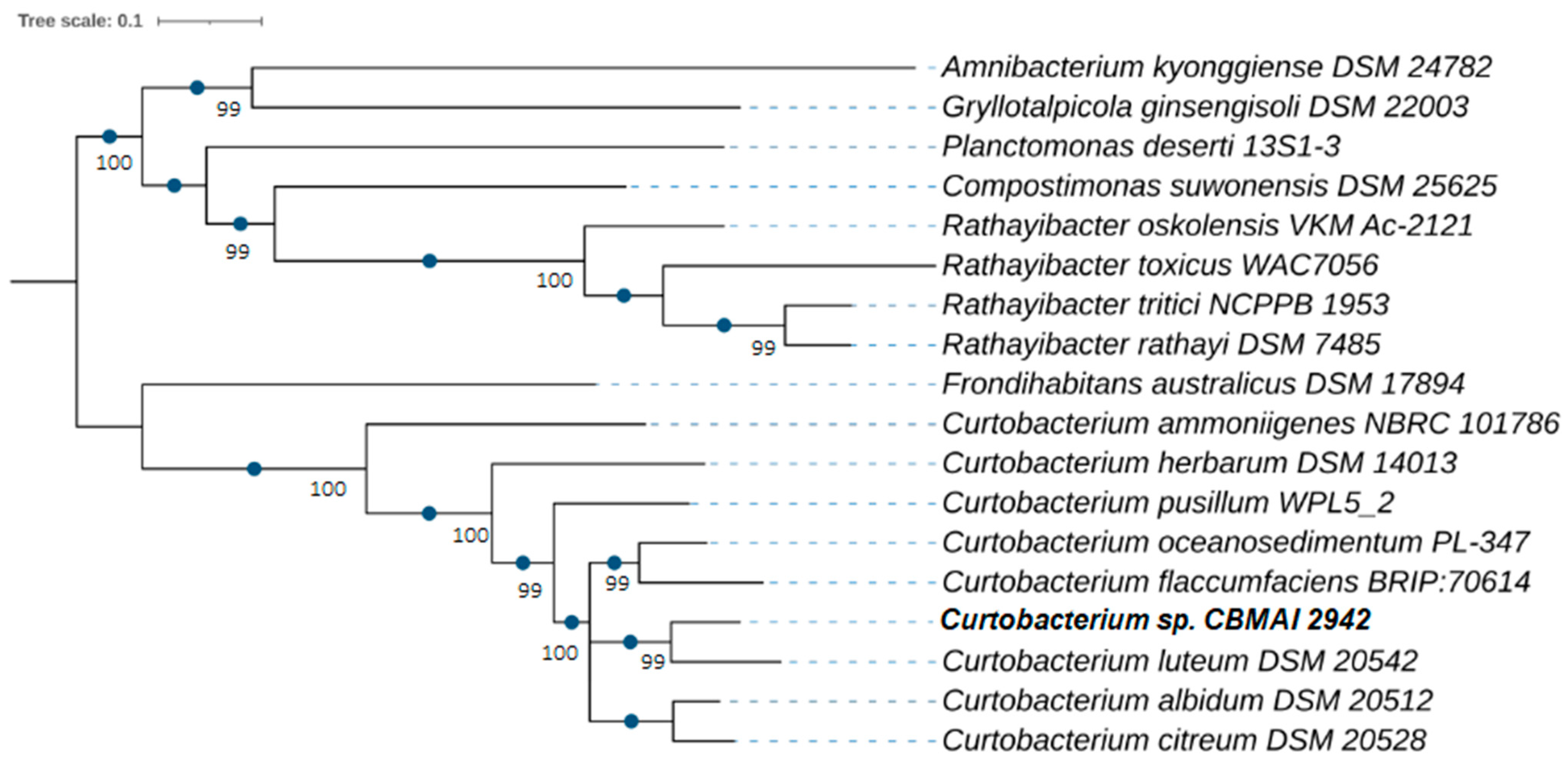
2.1. Sequencing, Assembly, and Taxonomic Affiliation of Marine Curtobacterium sp. CBMAI 2942 Genome
2.2. Genomic Potential for Chitin Degradation
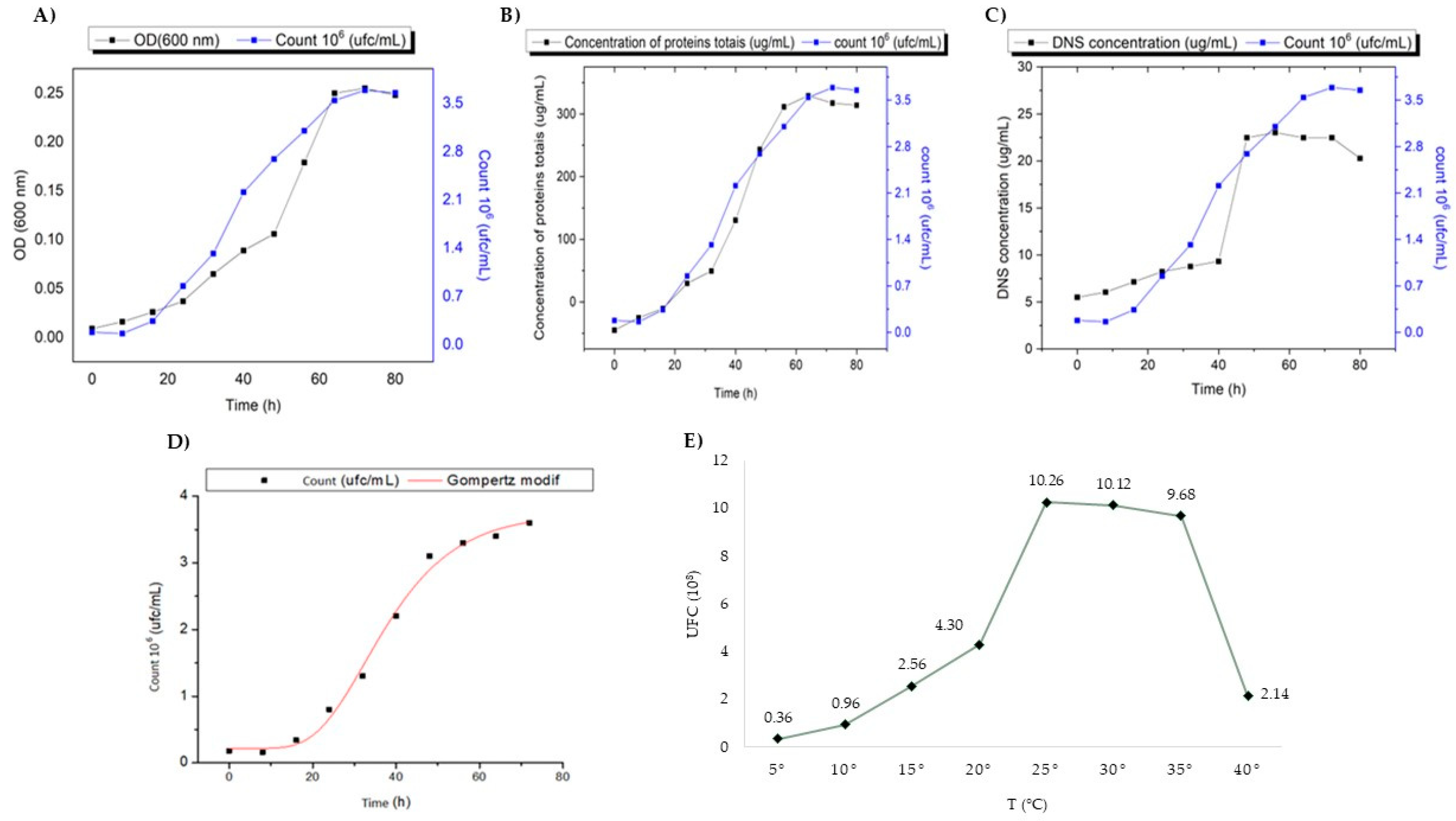
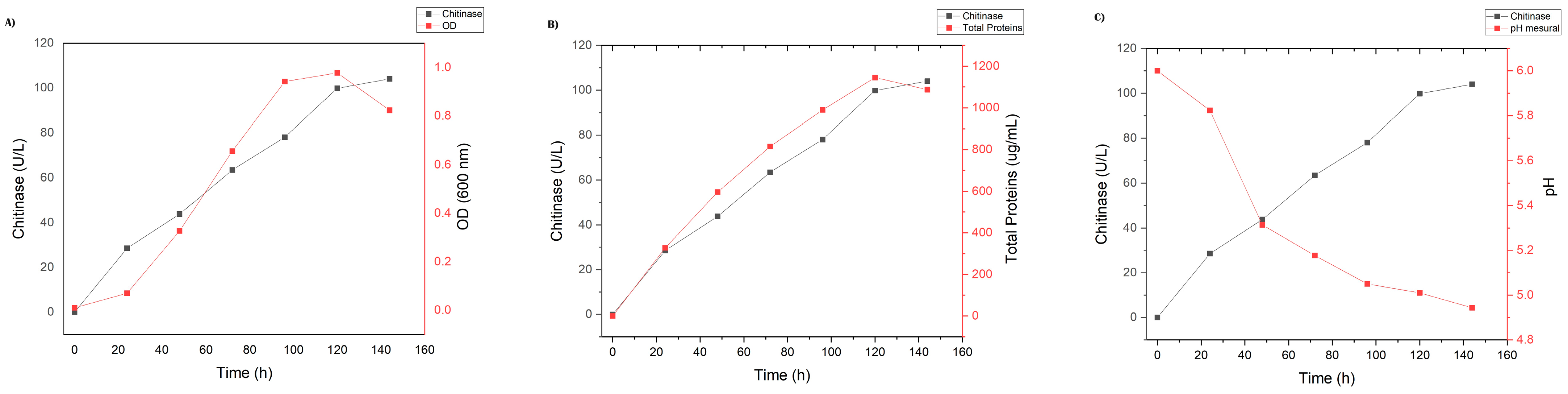
2.3. Production of Chitinase in Liquid Medium
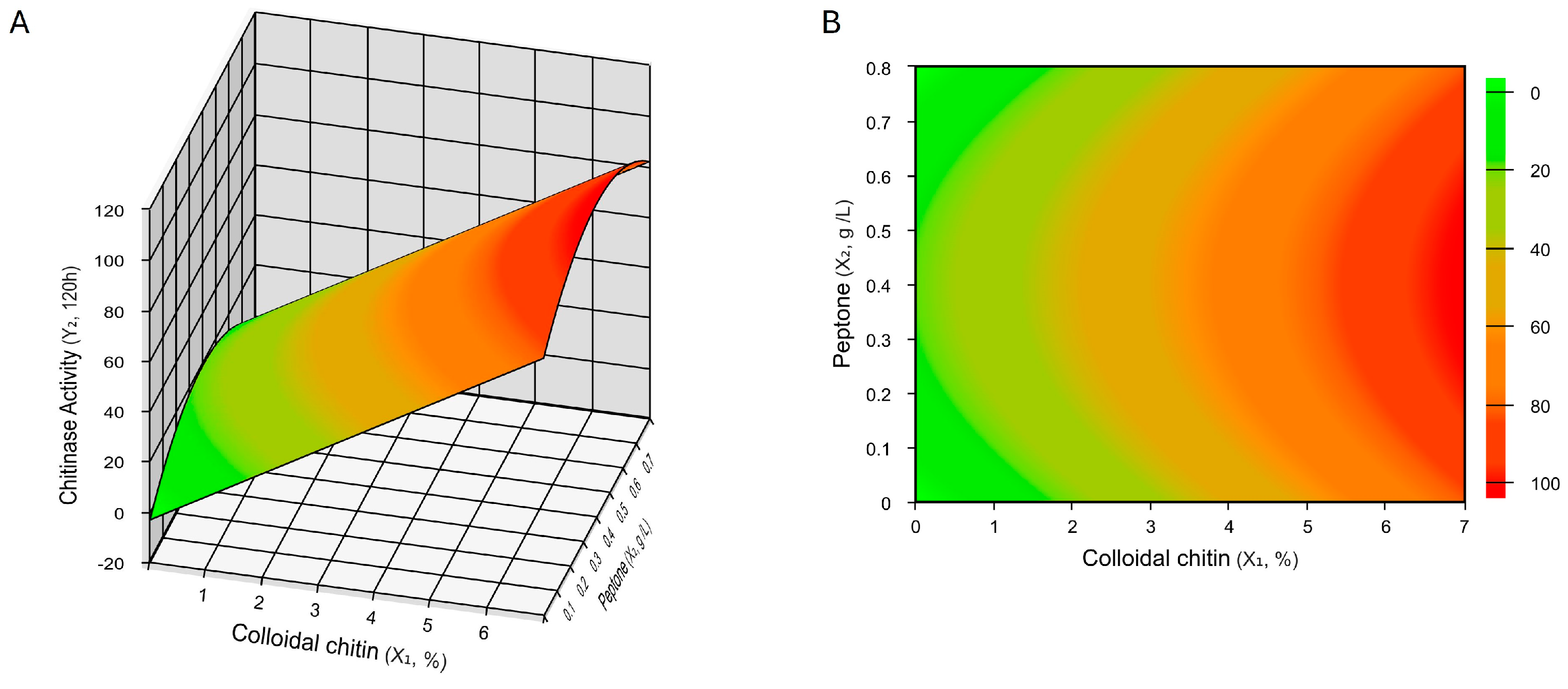
2.4. Optimization of Chitinase Production
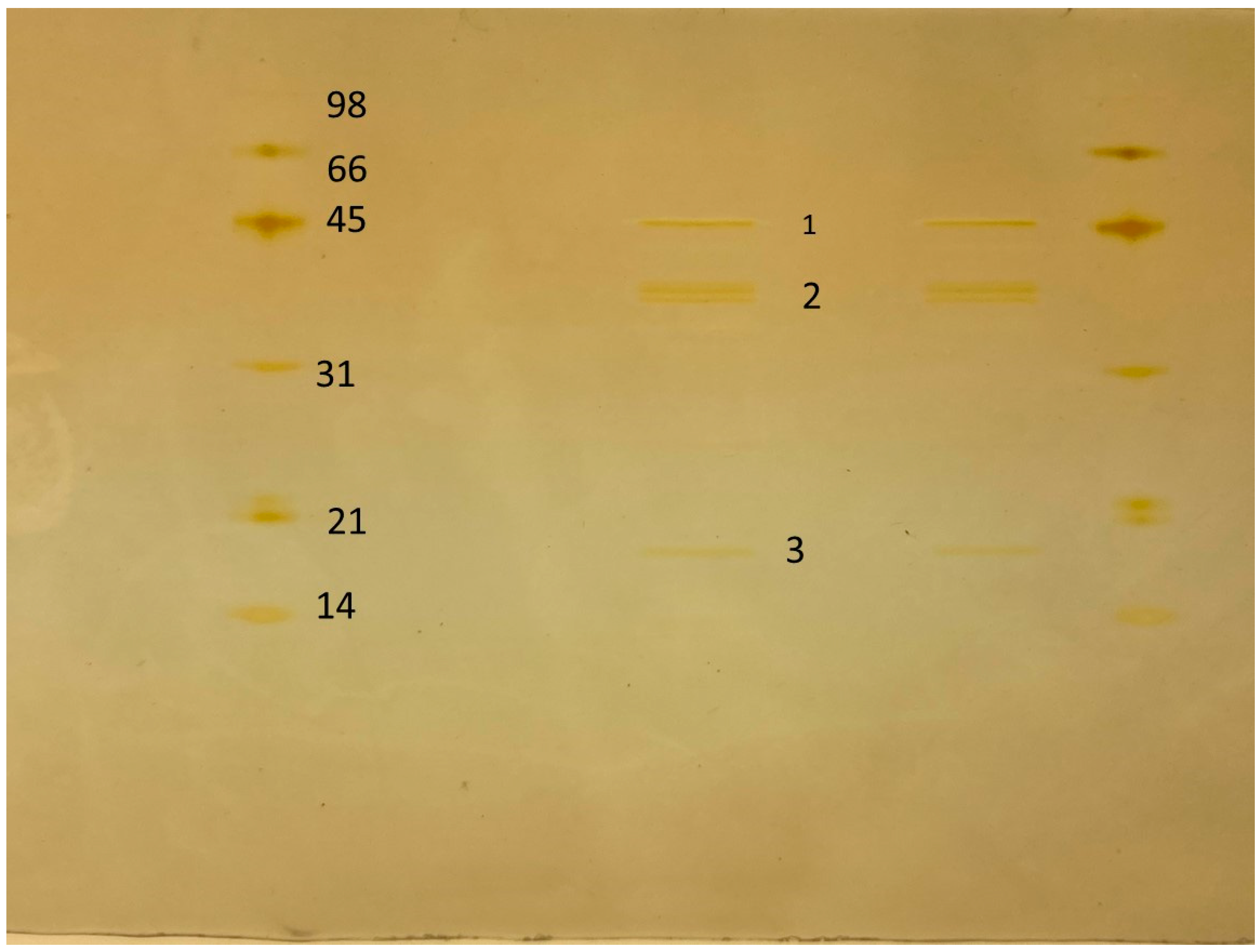
2.5. Isolation and Proteomic Analysis of Chitinase
2.6. Antifungal Activity of Chitinase Produced by Curtobacterium sp. CBMAI 2942
3. Discussion
4. Materials and Methods
4.1. Bacterial Strain
4.2. Sequencing, Assembly, and Annotation of the Curtobacterium sp. CBMAI 2942 Genome
4.2.1. Genomic DNA Extraction and Sequencing
4.2.2. Assembly and Annotation
4.2.3. Phylogenomic Identification
4.3. Production of Chitinase in Liquid Medium
4.3.1. Cultivation Conditions
4.3.2. Determination of Biomass
4.3.3. Quantification of Total Proteins
4.3.4. Chitinase Enzyme Activity
- CGlc = concentration of N-acetylglucosamine (GlcNAc) determined by the standard curve.
- Vt = total volume in the enzymatic reaction (2 mL).
- PM = N-acetylglucosamine (GlcNAc) molecular mass (221.208 mg/mmol).
- Ve = volume of enzyme preparation used (1 mL).
- T = reaction time (6 h).
- Dil = dilution applied to the enzyme preparation.
4.3.5. Evaluation of the Optimal Growth Temperature
4.4. Statistical Optimization Design for Chitinase Production
4.4.1. Screening of Variables by Plackett–Burman Design
4.4.2. Central Composite Design
4.5. Separation by SDS-PAGE and Identification of Chitinases via Mass Spectrometry
4.6. Antifungal Activity of Chitinases
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Le, B.; Yang, S.H. Microbial chitinases: Properties, current state and biotechnological applications. World J. Microbiol. Biotechnol. 2019, 35, 1–12. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Beygmoradi, A.; Homaei, A.; Hemmati, R.; Santos-Moriano, P.; Hormigo, D.; Fernández-Lucas, J. Marine chitinolytic enzymes, a biotechnological treasure hidden in the ocean? Appl. Microbiol. Biotechnol. 2018, 102, 9937–9948. [Google Scholar] [CrossRef]
- Loc, N.H.; Huy, N.D.; Quang, H.T.; Lan, T.T.; Thu Ha, T.T. Characterisation and antifungal activity of extracellular chitinase from a biocontrol fungus, Trichoderma asperellum PQ34. Mycology 2020, 11, 38–48. [Google Scholar] [CrossRef]
- Hassan, A.A.; Ismail, S.A. Production of antifungal N-acetyl-β-glucosaminidase chitinolytic enzyme using shrimp byproducts. Biocatal. Agric. Biotechnol. 2021, 34, 102027. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, X.; Yuan, F.; Deng, B.; Yu, X. Biocatalysis of heterogenously-expressed chitosanase for the preparation of desirable chitosan oligosaccharides applied against phytopathogenic fungi. ACS Sustain. Chem. Eng. 2020, 8, 4781–4791. [Google Scholar] [CrossRef]
- Hao, W.; Li, K.; Ma, Y.; Li, R.; Xing, R.; Yu, H.; Li, P. Preparation and antioxidant activity of chitosan dimers with different sequences. Mar. Drugs 2021, 19, 366. [Google Scholar] [CrossRef]
- Taokaew, S.; Kriangkrai, W. Chitinase-assisted bioconversion of chitinous waste for development of value-added chito-oligosaccharides products. Biology 2023, 12, 87. [Google Scholar] [CrossRef]
- Govindaraj, V.; Subramani, A.K.; Gopalakrishnan, R.; Kim, S.K.; Raval, R.; Raval, K. Bioethanol: A New Synergy between Marine Chitinases from Bacillus haynesii and ethanol production by Mucor circinelloides. Fermentation 2023, 9, 40. [Google Scholar] [CrossRef]
- Liu, K.; Ding, H.; Yu, Y.; Chen, B. A cold-adapted chitinase-producing bacterium from Antarctica and its potential in biocontrol of plant pathogenic fungi. Mar. Drugs 2019, 17, 695. [Google Scholar] [CrossRef]
- Ramli, A.N.; Mahadi, N.M.; Rabu, A.; Murad, A.M.A.; Bakar, F.D.B.; Illias, R.M. Molecular cloning, expression and biochemical characterization of a cold-adapted novel recombinat chitinase from Glaciozyma antarctica PI12. Microb. Cell Fact. 2011, 10, 1–13. [Google Scholar] [CrossRef]
- Aliabadi, N.; Aminzadeh, S.; Karkhane, A.A.; Haghbeen, K. Thermostable chitinase from Cohnella sp. A01: Isolation and product optimization. Braz. J. Microbiol. 2016, 47, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Meriem, G.; Mahmoud, K. Optimization of chitinase production by a new Streptomyces griseorubens C9 isolate using response surface methodology. Ann. Microbiol. 2017, 67, 175–183. [Google Scholar] [CrossRef]
- Kumar, M.; Brar, A.; Vivekanand, V.; Pareek, N. Production of chitinase from thermophilic Humicola grisea and its application in production of bioactive chitooligosaccharides. Int. J. Biol. Macromol. 2017, 104, 1641–1647. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Singh, M.P.; Verma, V.; Singh, P.; Srivastava, R.; Singh, A.K. Characteristics of cold adapted enzyme and its comparison with mesophilic and thermophilic counterpart. Cell Mol. Biol. 2016, 62, 144. [Google Scholar] [CrossRef]
- Da Silva, M.K.; Da Silva, A.V.; Fernandez, P.M.; Montone, R.C.; Alves, R.P.; De Queiroz, A.C.; De Oliveira, V.M.; Dos Santos, V.P.; Putzke, J.; Rosa, L.H.; et al. Extracelular hydrolytic enzymes produced by yeasts from Antarctic lichens. An. Acad. Bras. Cienc. 2022, 94, e20210540. [Google Scholar] [CrossRef]
- Vasquez YM, S.C.; Gomes, M.B.; ESilva, T.R.; Duarte AW, F.; Rosa, L.H.; De Oliveira, V.M. Cold-adapted chitinases from Antarctic bacteria: Taxonomic assessment and enzyme production optimization. Biocatal. Agric. Biotechnol. 2021, 34, 102029. [Google Scholar] [CrossRef]
- Da Silva, A.V.; de Oliveira, A.J.; Tanabe IS, B.; Silva, J.V.; da Silva Barros, T.W.; da Silva, M.K.; França PH, B.; Leite, J.; Putzke, J.; Montone, R.; et al. Antarctic lichens as a source of phosphate-solubilizing bacteria. Extremophiles 2021, 25, 181–191. [Google Scholar] [CrossRef]
- Derringer, G.; Suichi, R.J. Simultaneous optimization of several response variables. J. Qual. Technol. 1980, 12, 214–219. [Google Scholar] [CrossRef]
- Bulgari, D.; Minio, A.; Casati, P.; Quaglino, F.; Delledonne, M.; Bianco, P.A. Curtobacterium sp. genome sequencing underlines plant growth promotion-related traits. Genome Announc. 2014, 2, e00592-14. [Google Scholar] [CrossRef]
- Zaychikov, V.A.; Potekhina, N.V.; Dmitrenok, A.S.; Fan, D.; Tul’skaya, E.M.; Dorofeeva, L.V.; Evtushenko, L.I. Cell Wall Rhamnan in Actinobacteria of the Genus Curtobacterium. Microbiology 2021, 90, 343–348. [Google Scholar] [CrossRef]
- Garrido, L.M.; Alves JM, P.; Oliveira, L.S.; Gruber, A.; Padilla, G.; Araújo, W.L. Draft genome sequence of Curtobacterium sp. strain ER1/6, an endophytic strain isolated from Citrus sinensis with potential to be used as a biocontrol agent. Genome Announc. 2016, 4, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Kamei, I.; Yoshida, T.; Enami, D.; Meguro, S. Coexisting Curtobacterium bacterium promotes growth of white-rot fungus Stereum sp. Curr. Microbiol. 2012, 64, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Evseev, P.; Lukianova, A.; Tarakanov, R.; Tokmakova, A.; Shneider, M.; Ignatov, A.; Miroshnikov, K. Curtobacterium spp. and Curtobacterium flaccumfaciens: Phylogeny, genomics-based taxonomy, pathogenicity, and diagnostics. Curr. Issues Mol. Biol. 2022, 44, 889–927. [Google Scholar] [CrossRef] [PubMed]
- Araújo FD, D.S.; Santos, D.S.; Pagotto, C.C.; De Araújo, W.L.; Eberlin, M.N. Mass spectrometry characterization of endophytic bacterium Curtobacterium sp. strain ER1/6 isolated from Citrus sinensis. J. Mass. Spectrom. 2018, 53, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Hung SH, W.; Chiu, M.C.; Huang, C.C.; Kuo, C.H. Complete Genome Sequence of Curtobacterium sp. C1, a Beneficial Endophyte with the Potential for In-Plant Salinity Stress Alleviation. Mol. Plant-Microbe Interact. MPMI 2022, 35, 731–735. [Google Scholar] [CrossRef]
- Hazarika, S.N.; Saikia, K.; Thakur, D. Characterization and selection of endophytic actinobacteria for growth and disease management of Tea (Camellia sinensis L.). Front. Plant Sci. 2022, 13, 989794. [Google Scholar] [CrossRef]
- Chase, A.B.; Arevalo, P.; Polz, M.F.; Berlemont, R.; Martiny, J.B. Evidence for ecological flexibility in the cosmopolitan genus Curtobacterium. Front. Microbiol. 2016, 7, 1874. [Google Scholar] [CrossRef]
- Evtushenko, L.I.; Takeuchi, M. The Family Microbacteriaceae. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006. [Google Scholar] [CrossRef]
- Clarke, L.J.; Suter, L.; King, R.; Bissett, A.; Deagle, B.E. Antarctic Krill Are Reservoirs for Distinct Southern Ocean Microbial Communities. Front. Microbiol. 2019, 9, 3226. [Google Scholar] [CrossRef]
- Yu, Y.; Liu, X.; Miao, J.; Leng, K. Chitin from Antarctic krill shell: Eco-preparation, detection, and characterization. Int. J. Biol. Macromol. 2020, 164, 4125–4137. [Google Scholar] [CrossRef]
- Kaya, M.; Baublys, V.; Šatkauskienė, I.; Akyuz, B.; Bulut, E.; Tubelytė, V. First chitin extraction from Plumatella repens (Bryozoa) with comparison to chitins of insect and fungal origin. Int. J. Biol. Macromol. 2015, 79, 126–132. [Google Scholar] [CrossRef]
- Dimkić, I.; Bhardwaj, V.; Carpentieri-Pipolo, V.; Kuzmanović, N.; Degrassi, G. The chitinolytic activity of the Curtobacterium sp. isolated from field-grown soybean and analysis of its genome sequence. PLoS ONE 2021, 16, e0259465. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, Y.; Tan, H.; Chi, N.; Zhang, Q.; Du, Y.; Yin, H. Characterisation of a chitinase from Pseudoalteromonas sp. DL-6, a marine psychrophilic bacterium. Int. J. Biol. Macromol. 2014, 70, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Tan, X. Comprehensive analysis of the chitinase family genes in tomato (Solanum lycopersicum). Plants 2019, 8, 52. [Google Scholar] [CrossRef]
- Yeh, S.; Moffatt, B.A.; Griffith, M.; Xiong, F.; Yang, D.S.; Wiseman, S.B.; Sarhan, F.; Danyluk, J.; Xue, Y.Q.; Hew, C.L.; et al. Chitinase genes responsive to cold encode antifreeze proteins in winter cereals. Plant Physiol. 2020, 124, 1251–1264. [Google Scholar] [CrossRef] [PubMed]
- Tronelli, D.; Maugini, E.; Bossa, F.; Pascarella, S. Structural adaptation to low temperatures− analysis of the subunit interface of oligomeric psychrophilic enzymes. FEBS J. 2007, 274, 4595–4608. [Google Scholar] [CrossRef] [PubMed]
- Lonhienne, T.; Mavromatis, K.; Vorgias, C.E.; Buchon, L.; Gerday, C.; Bouriotis, V. Cloning, sequences, and characterization of two chitinase genes from the Antarctic Arthrobacter sp. strain TAD20: Isolation and partial characterization of the enzymes. J. Bacteriol. 2001, 183, 1773–1779. [Google Scholar] [CrossRef]
- Orikoshi, H.; Baba, N.; Nakayama, S.; Kashu, H.; Miyamoto, K.; Yasuda, M.; Inamori, Y.; Tsujibo, H. Molecular analysis of the gene encoding a novel cold-adapted chitinase (ChiB) from a marine bacterium, Alteromonas sp. strain O-7. J. Bacteriol. 2003, 185, 1153–1160. [Google Scholar] [CrossRef][Green Version]
- Okay, S.; Alshehri, W.A. Overexpression of chitinase a gene from Serratia marcescens in Bacillus subtilis and characterization of enhanced chitinolytic activity. Braz. Arch. Biol. Technol. 2020, 63, e20200061. [Google Scholar] [CrossRef]
- Tasharrofi, N.; Adrangi, S.; Fazeli, M.; Rastegar, H.; Khoshayand, M.R.; Faramarzi, M.A. Optimization of chitinase production by Bacillus pumilus using Plackett-Burman design and response surface methodology. Iran. J. Pharm. Res. 2011, 10, 759. [Google Scholar]
- Tjørve, K.M.; Tjørve, E. The use of Gompertz models in growth analyses, and new Gompertz-model approach: An addition to the Unified-Richards family. PLoS ONE 2017, 12, e0178691. [Google Scholar] [CrossRef]
- Chitra, M.; Sutha, S.; Pappa, N. Application of deep neural techniques in predictive modelling for the estimation of Escherichia coli growth rate. J. Appl. Microbiol. 2021, 130, 1645–1655. [Google Scholar] [CrossRef]
- Harish, B.S.; Uppuluri, K.B. Modeling of growth kinetics for an isolated marine bacterium, Oceanimonas sp. BPMS22 during the production of a trypsin inhibitor. Prep. Biochem. Biotech. 2018, 48, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Rolfe, M.D.; Rice, C.J.; Lucchini, S.; Pin, C.; Thompson, A.; Cameron, A.D.; Alston, M.; Stringer, M.F.; Betts, R.P.; Baranyi, J.; et al. Lag phase is a distinct growth phase that prepares bacteria for exponential growth and involves transient metal accumulation. J. Bacteriol. 2012, 194, 686–701. [Google Scholar] [CrossRef]
- Siddiqui, K.S.; Cavicchioli, R. Cold-adapted enzymes. Annu. Rev. Biochem. 2006, 75, 403–433. [Google Scholar] [CrossRef]
- Margesin, R. Effect of temperature on growth parameters of psychrophilic bacteria and yeasts. Extremophiles 2009, 13, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Pesciaroli, C.; Cupini, F.; Selbmann, L.; Barghini, P.; Fenice, M. Temperature preferences of bacteria isolated from seawater collected in Kandalaksha Bay, White Sea, Russia. Polar Biol. 2012, 35, 435–445. [Google Scholar] [CrossRef]
- Kuddus, M.; Ramteke, P.W. A cold-active extracellular metalloprotease from Curtobacterium luteum (MTCC 7529): Enzyme production and characterization. J. Gen. Appl. Microbiol. 2008, 54, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Kuddus, M. Statistical optimization of cold-active chitinase production by mutagenized cells of multi-enzyme producing Bacillus cereus GA6. Rend Lincei. 2015, 26, 271–280. [Google Scholar] [CrossRef]
- Jha, S.; Modi, H.A.; Jha, C.K. Characterization of extracellular chitinase produced from Streptomyces rubiginosus isolated from rhizosphere of Gossypium sp. Cogent Food Agric. 2016, 2, 1198225. [Google Scholar] [CrossRef]
- Reihani, S.F.S.; Khosravi-Darani, K. Influencing factors on single-cell protein production by submerged fermentation: A review. Electron. J. Biotechnol. 2019, 37, 34–40. [Google Scholar] [CrossRef]
- Wierzchowska, K.; Zieniuk, B.; Nowak, D.; Fabiszewska, A. Phosphorus and nitrogen limitation as a part of the strategy to stimulate microbial lipid biosynthesis. Appl. Sci. 2021, 11, 11819. [Google Scholar] [CrossRef]
- Aounallah, M.A.; Slimene-Debez, I.B.; Djebali, K.; Gharbi, D.; Hammami, M.; Azaiez, S.; Limam, F.; Tabbene, O. Enhancement of exochitinase production by Bacillus licheniformis AT6 strain and improvement of N-acetylglucosamine production. Appl. Biochem. Biotechnol. 2017, 181, 650–666. [Google Scholar] [CrossRef] [PubMed]
- Han, S.J.; Park, H.; Lee, S.G.; Lee, H.K.; Yim, J.H. Optimization of cold-active chitinase production from the Antarctic bacterium Sanguibacter antarcticus KOPRI 21702. Appl. Microbiol. Biotechnol. 2011, 89, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.S.; Seo, D.J.; Jung, W.J. Identification, purification, and expression patterns of chitinase from psychrotolerant Pedobacter sp. PR-M6 and antifungal activity in vitro. Microb. Pathog. 2017, 107, 62–68. [Google Scholar] [CrossRef]
- Wegulo, S.N.; Zwingman, M.V.; Breathnach, J.A.; Baenziger, P.S. Economic returns from fungicide application to control foliar fungal diseases in winter wheat. Crop Prot. 2011, 30, 685–692. [Google Scholar] [CrossRef]
- Karthik, N.; Binod, P.; Pandey, A. Chitinases. In Current Developments in Biotechnology and Bioengineering; Ashok, P., Sanfeeta, N., Carlose, R.S., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 335–368. [Google Scholar] [CrossRef]
- Sharma, S.; Kumar, S.; Khajuria, A.; Ohri, P.; Kaur, R.; Kaur, R. Biocontrol potential of chitinases produced by newly isolated Chitinophaga sp. S167. World J. Microbiol. Biotechnol. 2020, 36, 1–15. [Google Scholar] [CrossRef]
- Philip, N.V.; Koteshwara, A.; Kiran, G.A.; Raja, S.; Subrahmanyam, V.M.; Chandrashekar, H.R. Statistical optimization for coproduction of chitinase and beta 1, 4-endoglucanase by chitinolytic Paenibacillus elgii PB1 having antifungal activity. Appl. Biochem. Biotechnol. 2020, 191, 135–150. [Google Scholar] [CrossRef]
- Singh, G.; Arya, S.K. Antifungal and insecticidal potential of chitinases: A credible choice for the eco-friendly farming. Biocatal. Agric. Biotechnol. 2019, 20, 101289. [Google Scholar] [CrossRef]
- Silva, T.R.; Duarte, A.W.; Passarini, M.R.; Ruiz, A.L.T.; Franco, C.H.; Moraes, C.B.; de Melo, I.S.; Rodrigues, R.A.; Fantinatti-Garboggini, F.; Oliveira, V.M. Bacteria from Antarctic environments: Diversity and detection of antimicrobial, antiproliferative, and antiparasitic activities. Polar Biol. 2018, 41, 1505–1519. [Google Scholar] [CrossRef]
- Andrews, S. Fastqc: A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics: Cambridge, UK, 2010. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bushmanova, E.; Antipov, D.; Lapidus, A.; Prjibelski, A.D. rnaSPAdes: A de novo transcriptome assembler and its application to RNA-Seq. data. GigaScience 2019, 8, giz100. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 1–11. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, I.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Buchfink, B.; Reuter, K.; Drost, H.G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Asnicar, F.; Thomas Am Beghini, F.; Mengoni, C.; Manara, S.; Manghi, P.; Zhu, Q.; Bolzan, M.; Cumbo, F.; May, U.; Sanders Jg Zolfo, M.; et al. Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nat. Commun. 2020, 11, 2500. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Ulhoa, C.J.; Peberdy, J.F. Purification and some properties of the extracellular chitinase produced by Trichoderma harzianum. Enzyme Microb. Technol. 1992, 14, 236–240. [Google Scholar] [CrossRef]

| Sequence Data | Curtobacterium sp. CBMAI 2942 |
|---|---|
| Coverage (X) | 350 |
| # Contigs | 1 |
| Total compression | 3,685,083 |
| Largest contig | 3,679,616 |
| Smallest contig | 5467 |
| N50 | 3,679,616 |
| % CG | 71.48 |
| % Completeness | 99.44 |
| % Contamination | 0.76 |
| Marine Bacterial Strain | Related Bacteria | ANIb (%) | dDDH (%) | Difference in G + C Content (%) |
|---|---|---|---|---|
| Curtobacterium sp. CBMAI 2942 | Curtobacterium luteum DSM 20542T | 87.71 | 33.60 | 0.22 |
| Curtobacterium oceanosedimentum NS359 | 82.66 | 26.10 | 0.26 | |
| Curtobacterium citreum DSM 20528T | 82.79 | 26.60 | 0.46 | |
| Curtobacterium pusillum ATCC 19096T | 83.52 | 27.00 | 0.63 |
| Bacterium | Gene ID | Gene Name | Start | Final | Size (pb) | COG ID | Function COG |
|---|---|---|---|---|---|---|---|
| Curtobacterium sp. CBMAI 2942 | >NODE_1_length_3679616_cov_83.786059_76 | Chitin binding | 91,936 | 93,246 | 1310 | COG3979 | ChBD of chitin binding (Chi C) |
| >NODE_1_length_3679616_cov_83.786059_3152 | Chitin binding | 3,317,000 | 3,318,553 | 1553 | COG3979 | ChBD of chitin binding (Chi C) |
| Assay | Colloidal Chitin (%) | Yeast Extract (g/L) | Peptone (g/L) | K2HPO4 (g/L) | KH2PO4 (g/L) | MgSO4·7H2O (g/L) | NH4NO3 (g/L) | NaCl (g/L) | pH | U/L (96 h) | U/L (120 h) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 (3) | −1 (0.1) | −1 (0.1) | −1(0.2) | 1 (0.5) | −1 (0.1) | −1 (0.2) | 1 (1.8) | 1(8) | 28.18 | 28.96 |
| 2 | 1 (3) | 1 (0.5) | −1 (0.1) | −1(0.2) | −1 (0.1) | 1 (0.9) | −1 (0.2) | −1 (0.2) | 1(8) | 31.89 | 38.54 |
| 3 | 1 (3) | 1 (0.5) | 1 (0.5) | −1(0.2) | −1 (0.1) | −1 (0.1) | 1 (3.8) | −1 (0.2) | −1 (6) | 30.72 | 32.67 |
| 4 | 1 (3) | 1 (0.5) | 1 (0.5) | 1 (1.2) | −1 (0.1) | −1 (0.1) | −1 (0.2) | 1 (1.8) | −1 (6) | 28.18 | 29.15 |
| 5 | −1 (1) | 1 (0.5) | 1 (0.5) | 1 (1.2) | 1 (0.5) | −1 (0.1) | −1 (0.2) | −1 (0.2) | 1(8) | 28.96 | 30.52 |
| 6 | 1 (3) | −1 (0.1) | 1 (0.5) | 1 (1.2 | 1 (0.5) | 1 (0.9) | −1 (0.2) | −1 (0.2) | −1 (6) | 39.33 | 39.72 |
| 7 | −1 (1) | 1 (0.5) | −1 (0.1) | 1 (1.2) | 1 (0.5) | 1 (0.9) | 1 (3.8) | −1 (0.2) | −1 (6) | 30.52 | 32.09 |
| 8 | 1 (3) | −1 (0.1) | 1 (0.5) | −1(0.2) | 1 (0.5) | 1 (0.9) | 1 (3.8) | 1 (1.8) | −1 (6) | 44.22 | 46.76 |
| 9 | 1 (3) | 1 (0.5) | −1 (0.1) | 1 (1.2) | −1 (0.1) | 1 (0.9) | 1 (3.8) | 1 (1.8) | 1(8) | 27.98 | 28.18 |
| 10 | −1 (1) | 1 (0.5) | 1 (0.5) | −1(0.2) | 1 (0.5) | −1 (0.1) | 1 (3.8) | 1 (1.8) | 1(8) | 30.72 | 32.09 |
| 11 | −1 (1) | −1 (0.1) | 1 (0.5) | 1 (1.2) | −1 (0.1) | 1 (0.9) | −1 (0.2) | 1 (1.8) | 1(8) | 30.52 | 32.28 |
| 12 | 1 (3) | −1 (0.1) | −1 (0.1) | 1 (1.2) | 1 (0.5) | −1 (0.1) | 1 (3.8) | −1 (0.2) | 1(8) | 28.18 | 30.33 |
| 13 | −1 (1) | 1 (0.5) | −1 (0.1) | −1(0.2) | 1 (0.5) | 1 (0.9) | −1 (0.2) | 1 (1.8) | −1 (6) | 25.83 | 26.22 |
| 14 | −1 (1) | −1 (0.1) | 1 (0.5) | −1(0.2) | −1 (0.1) | 1 (0.9) | 1 (3.8) | −1 (0.2) | 1(8) | 26.81 | 32.09 |
| 15 | −1 (1) | −1 (0.1) | −1 (0.1) | 1 (1.2) | −1 (0.1) | −1 (0.1) | 1 (3.8) | 1 (1.8) | −1 (6) | 25.24 | 26.22 |
| 16 | −1 (1) | −1 (0.1) | −1 (0.1) | −1(0.2) | −1 (0.1) | −1 (0.1) | −1 (0.2) | −1 (0.2) | −1 (6) | 26.02 | 28.57 |
| 17 | 0 (2) | 0 (0.3) | 0 (0.3) | 0 (0.7) | 0 (0.3) | 0 (0.5) | 0 (2.0) | 0 (1.0) | 0 (7) | 27.20 | 27.98 |
| 18 | 0 (2) | 0 (0.3) | 0 (0.3) | 0 (0.7) | 0 (0.3) | 0 (0.5) | 0 (2.0) | 0 (1.0) | 0 (7) | 27.78 | 28.18 |
| 19 | 0 (2) | 0 (0.3) | 0 (0.3) | 0 (0.7) | 0 (0.3) | 0 (0.5) | 0 (2.0) | 0 (1.0) | 0 (7) | 27.00 | 28.96 |
| Assay | Colloidal Chitin (%) | Peptone (g/L) | KH2PO4 (g/L) | U/L (96 h) | U/L (120 h) |
|---|---|---|---|---|---|
| 1 | (1.50) | (0.20) | (0.20) | 32.09 | 35.41 |
| 2 | (5.00) | (0.20) | (0.20) | 58.50 | 71.80 |
| 3 | (1.50) | (0.60) | (0.20) | 30.33 | 32.28 |
| 4 | (5.00) | (0.60) | (0.20) | 60.26 | 80.01 |
| 5 | (1.50) | (0.20) | (0.50) | 31.70 | 34.83 |
| 6 | (5.00) | (0.20) | (0.50) | 54.00 | 71.21 |
| 7 | (1.50) | (0.60) | (0.50) | 30.13 | 31.50 |
| 8 | (5.00) | (0.60) | (0.50) | 61.82 | 75.91 |
| 9 | (0.31) | (0.40) | (0.35) | 23.28 | 24.46 |
| 10 | (6.19) | (0.40) | (0.35) | 86.27 | 99.19 |
| 11 | (3.25) | (0.06) | (0.35) | 36.78 | 42.26 |
| 12 | (3.25) | (0.74) | (0.35) | 38.74 | 39.72 |
| 13 | (3.25) | (0.40) | (0.10) | 39.52 | 41.09 |
| 14 | (3.25) | (0.40) | (0.60) | 58.69 | 59.28 |
| 15 | (3.25) | (0.40) | (0.35) | 57.32 | 58.69 |
| 16 | (3.25) | (0.40) | (0.35) | 57.13 | 61.04 |
| 17 | (3.25) | (0.40) | (0.35) | 56.54 | 58.89 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vasquez, Y.M.S.-C.; Cueva-Yesquen, L.G.; Duarte, A.W.F.; Rosa, L.H.; Valladão, R.; Lopes, A.R.; Costa Bonugli-Santos, R.; Oliveira, V.M.d. Genomics, Proteomics, and Antifungal Activity of Chitinase from the Antarctic Marine Bacterium Curtobacterium sp. CBMAI 2942. Int. J. Mol. Sci. 2024, 25, 9250. https://doi.org/10.3390/ijms25179250
Vasquez YMS-C, Cueva-Yesquen LG, Duarte AWF, Rosa LH, Valladão R, Lopes AR, Costa Bonugli-Santos R, Oliveira VMd. Genomics, Proteomics, and Antifungal Activity of Chitinase from the Antarctic Marine Bacterium Curtobacterium sp. CBMAI 2942. International Journal of Molecular Sciences. 2024; 25(17):9250. https://doi.org/10.3390/ijms25179250
Chicago/Turabian StyleVasquez, Yesenia Melissa Santa-Cruz, Luis Gabriel Cueva-Yesquen, Alysson Wagner Fernandes Duarte, Luiz Henrique Rosa, Rodrigo Valladão, Adriana Rios Lopes, Rafaella Costa Bonugli-Santos, and Valéria Maia de Oliveira. 2024. "Genomics, Proteomics, and Antifungal Activity of Chitinase from the Antarctic Marine Bacterium Curtobacterium sp. CBMAI 2942" International Journal of Molecular Sciences 25, no. 17: 9250. https://doi.org/10.3390/ijms25179250
APA StyleVasquez, Y. M. S.-C., Cueva-Yesquen, L. G., Duarte, A. W. F., Rosa, L. H., Valladão, R., Lopes, A. R., Costa Bonugli-Santos, R., & Oliveira, V. M. d. (2024). Genomics, Proteomics, and Antifungal Activity of Chitinase from the Antarctic Marine Bacterium Curtobacterium sp. CBMAI 2942. International Journal of Molecular Sciences, 25(17), 9250. https://doi.org/10.3390/ijms25179250






