Transcriptome-Based Screening of Candidate Low-Temperature-Associated Genes and Analysis of the BocARR-B Transcription Factor Gene Family in Kohlrabi (Brassica oleracea L. var. caulorapa L.)
Abstract
1. Introduction
2. Results
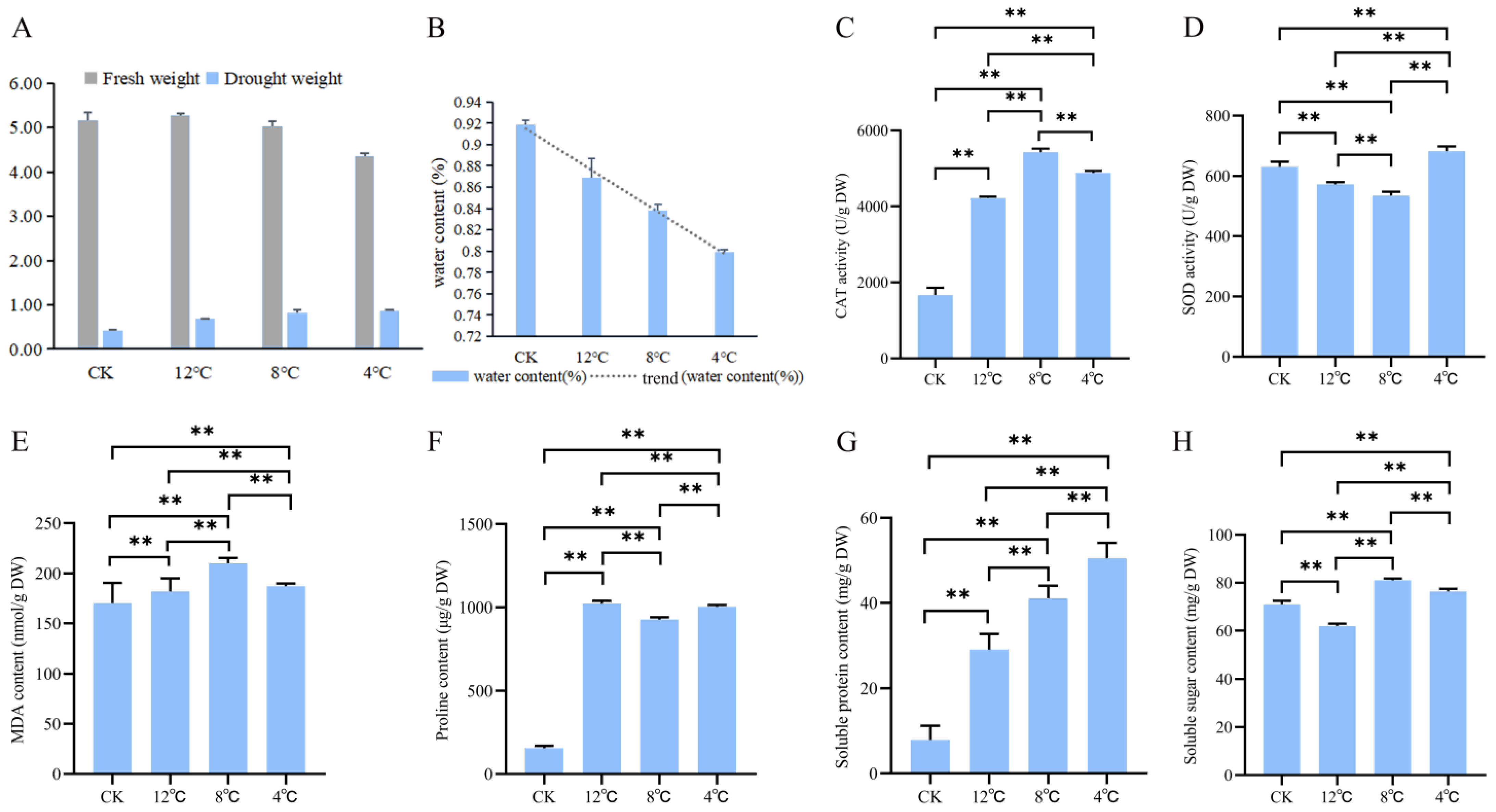
2.1. Biochemical Indicators Changes in Kohlrabi under Low-Temperature Stress
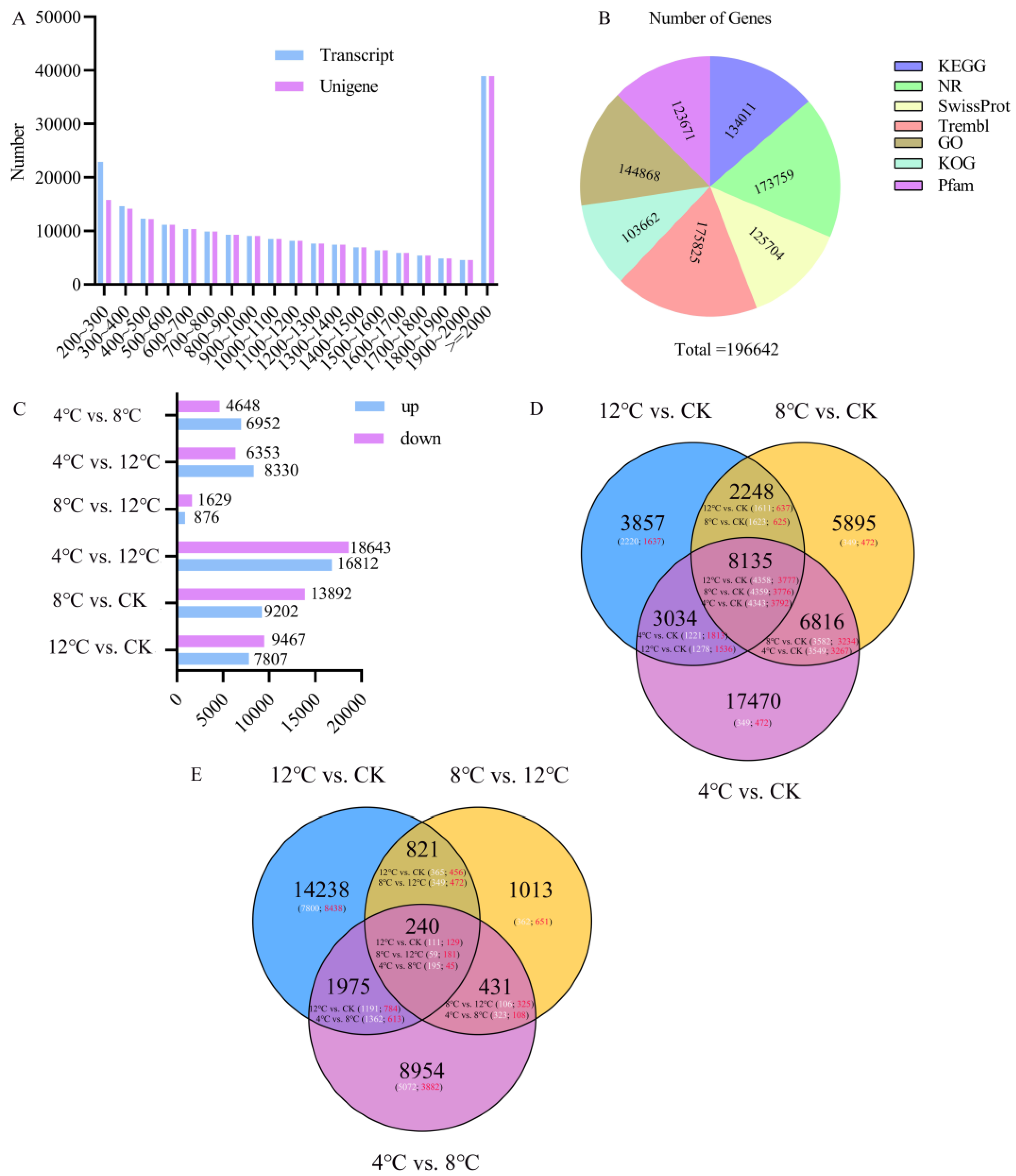
2.2. Transcriptome Data Summary and KEGG Enrichment Analysis
2.3. Availability Analysis of Transcriptome Data
2.4. Basic Genetic Analysis of Metabolite Networks under Low-Temperature Stress
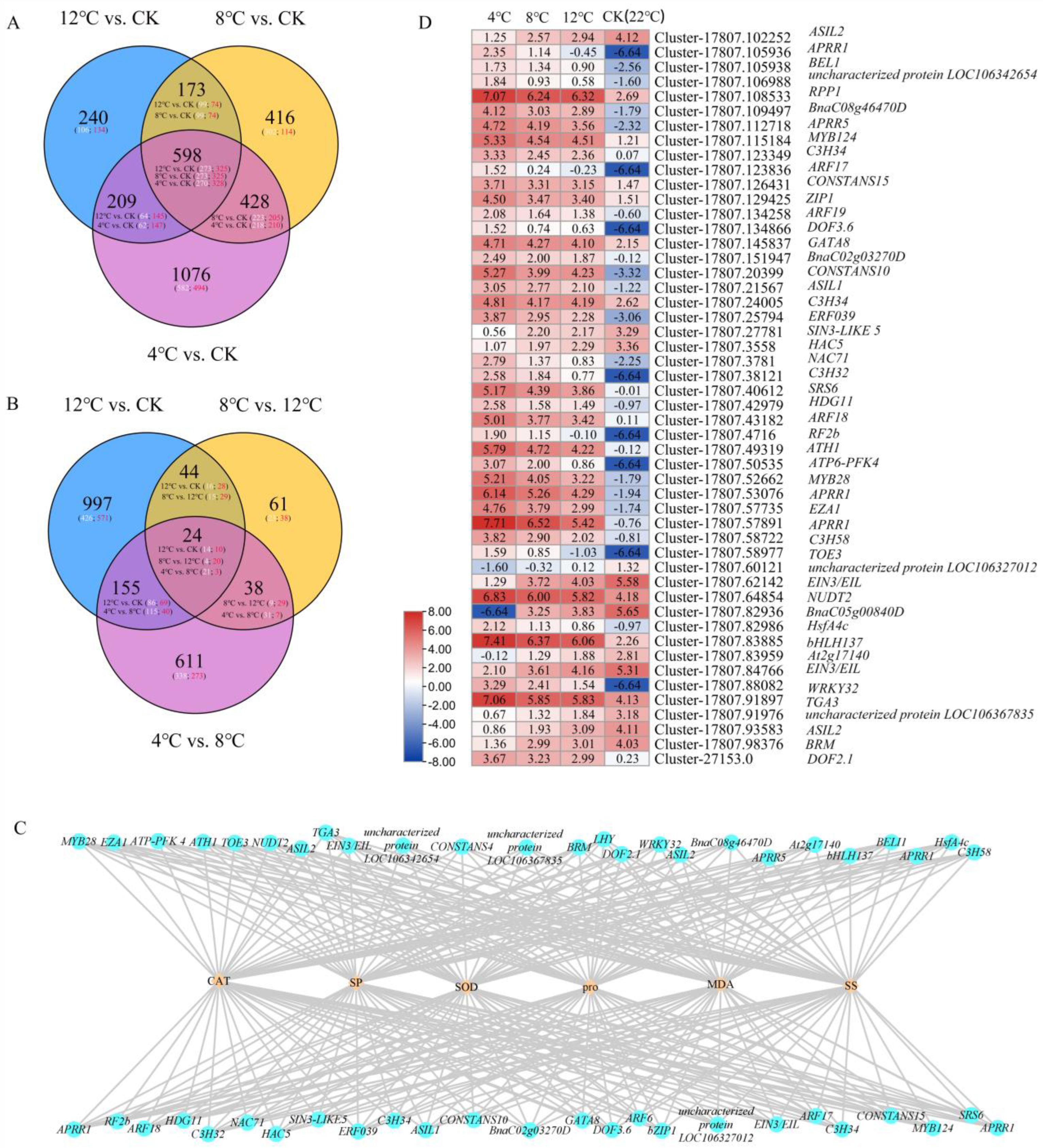
2.5. Analysis of Differentially Expressed Transcription Factor and Weighted Gene Co-Expression Network Analysis between Biochemical Indicators Related to Low Temperature and Differentially Expressed Transcription Factors
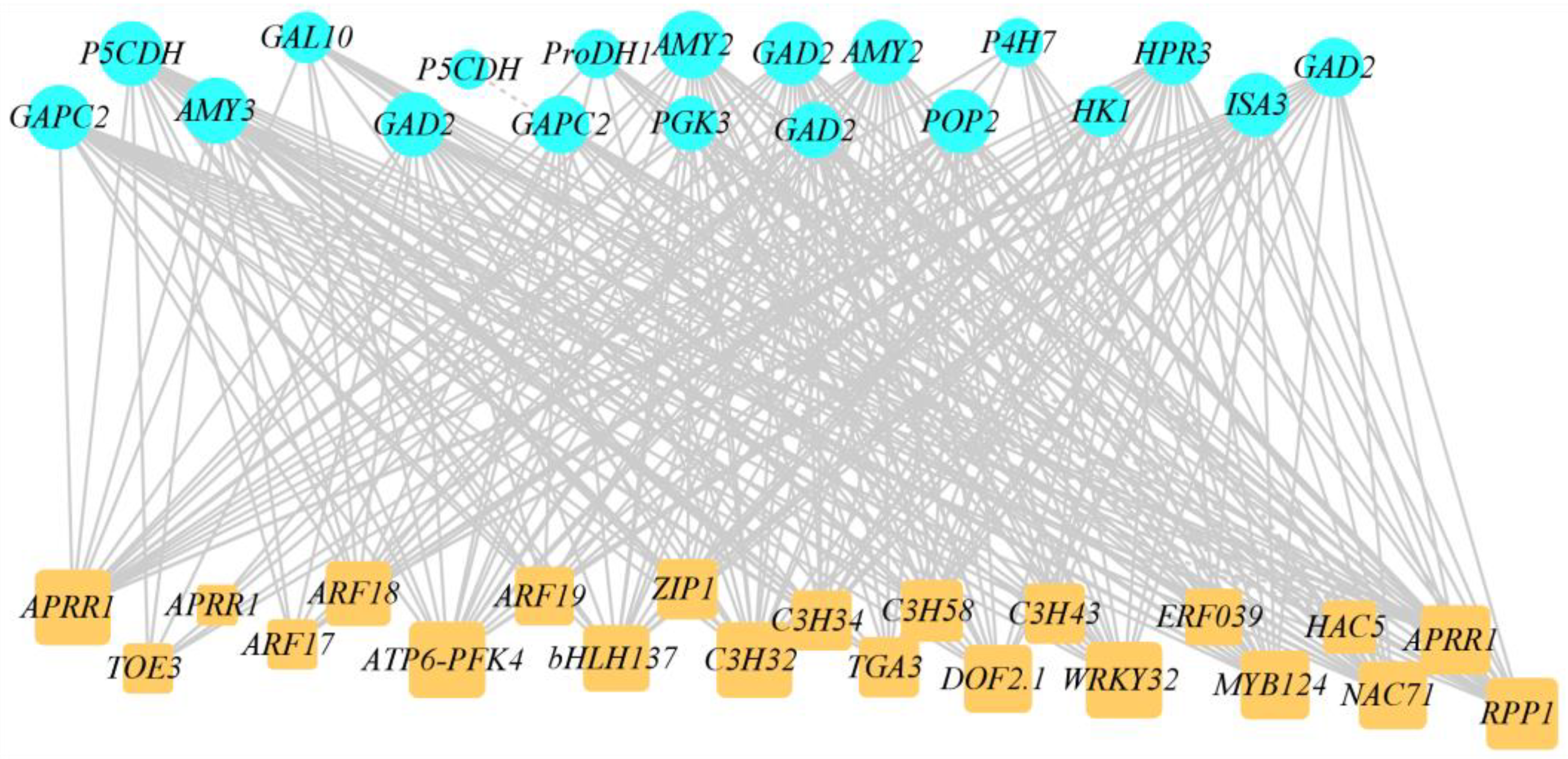
2.6. Joint Analysis of Low-Temperature-Related Differentially Expressed Transcription Factors and DEGs
2.7. BocARR-B Transcription Factor Gene Family Member Identification and Chromosomal Localization
2.8. Gene Structure and Conserved Motif Analysis of BocARR-B Transcription Factor Genes Family
2.9. Evolutionary Analysis of BocARR-B Transcription Factor Gene Family
2.10. Expression Analysis of BocARR-Bs via RT-qPCR and RNA Sequencing in Different Tissues and under Low-Temperature Stress
3. Discussion
4. Materials and Methods
4.1. Plant Material and Low-Temperature Stress Treatment
4.2. Measurement of Biochemical Indicators
4.3. Transcriptomic Analysis of Kohlrabi Leaves under Low-Temperature Stress Treatment
4.4. Availability Validation of RNA-Sequencing
4.5. Co-Expression Network Analysis of Discriminated Low-Temperature-Related DEGs
4.6. Identification and Chromosomal Distribution of BocARR-B Transcription Factor Family Members in Kohlrabi
4.7. Phylogenetic Analysis of BocARR-B Transcription Factor Family
4.8. Quantitative Real-Time PCR (RT-qPCR)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Soltan, H.A.H.; Osman, S.A.M.; Tantawy, I.A.A. The Role of New Transformants Phosphate Bio-Stimulates (PBS) Bacteria Inoculates on Growth. Yield and Quality of Kohlrabi Plants. J. Plant Sci. 2021, 12, 541–551. [Google Scholar] [CrossRef]
- Lim, T. Brassica oleracea (gongylodes group). In Edible Medicinal and Non Medicinal Plants; Springer: Dordrecht, The Netherlands, 2015; pp. 768–776. [Google Scholar] [CrossRef]
- Iradukunda, M.; Read, P.E. Influence of Fertilizer Rate on Swollen Stem Formation (“Bulbing”) and Vitamin C Content in Different Kohlrabi Cultivars. Ph.D. Thesis, University of Nebraska, Lincoln, NE, USA, 2022. [Google Scholar]
- Jahangir, M.; Kim, H.K.; Choi, Y.H.; Verpoorte, R. Health-affecting compounds in Brassicaceae. Compr. Rev. Food Sci. Food Saf. 2009, 8, 31–43. [Google Scholar] [CrossRef]
- Park, W.T.; Kim, J.K.; Park, S.; Lee, S.-W.; Li, X.; Kim, Y.B.; Uddin, M.R.; Park, N.I.; Kim, S.-J.; Park, S.U. Metabolic profiling of glucosinolates, anthocyanins, carotenoids, and other secondary metabolites in kohlrabi (Brassica oleracea var. gongylodes). J. Agric. Food Chem. 2012, 60, 8111–8116. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hu, Z.; Zhu, M.; Zhu, Z.; Wang, Z.; Tian, S.; Chen, G. Anthocyanin accumulation and molecular analysis of correlated genes in purple kohlrabi (Brassica oleracea var. gongylodes L.). J. Agric. Food Chem. 2015, 63, 4160–4169. [Google Scholar] [CrossRef]
- Prajapati, R.; Seong, S.H.; Kim, H.R.; Jung, H.A.; Choi, J.S. Isolation and Identification of Bioactive Compounds from the Tuber of Brassica oleracea var. gongylodes. Nat. Prod. Sci. 2020, 26, 214–220. [Google Scholar] [CrossRef]
- Golob, A.; Novak, T.; Marši’c, N.K.; Šircelj, H.; Stibilj, V.; Jerše, A.; Kroflič, A.; Germ, M. Biofortification with selenium and iodine changes morphological properties of Brassica oleracea L. var. gongylodes) and increases their contents in tubers. Plant Physiol. Biochem. 2020, 150, 234–243. [Google Scholar] [CrossRef]
- Sanghera, G.S.; Wani, S.H.; Hussain, W.; Singh, N.B. Engineering cold stress tolerance in crop plants. Curr. Genomics. 2011, 12, 30–43. [Google Scholar] [CrossRef]
- Jeon, J.; Kim, J. Cold stress signaling networks in Arabidopsis. J. Plant Biol. 2013, 56, 69–76. [Google Scholar] [CrossRef]
- Lyons, J.M. Chilling injury in plants. Ann. Rev. Plant Physiol. 1973, 24, 445–660. [Google Scholar] [CrossRef]
- Jouyban, Z.; Hasanzade, R.; Sharafi, S. Chilling stress in plants. Int. J. Agric. Crop Sci. 2013, 5, 2961–2968. [Google Scholar]
- Huang, B.-L.; Li, X.; Liu, P.; Ma, L.; Wu, W.; Zhang, X.; Li, Z.; Huang, B. Transcriptomic analysis of Eruca vesicaria subs. sativa lines with contrasting tolerance to polyethylene glycol-simulated drought stress. BMC Plant Biol. 2019, 19, 419. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Sun, F.; Zhang, N.; Rana, B.B.; Kharel, R.; Luo, P.; Si, H. RNA-seq provides insights into potato deubiquitinase responses to drought stress in seedling stage. Front. Plant Sci. 2023, 14, 1268448. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Wang, Y.; Dai, Y.; He, Y.; Li, C.X.; Mao, P.; Ma, X.R. The transcription factors of tall fescue in response to temperature stress. Plant Biol. J. 2021, 23, 89–99. [Google Scholar] [CrossRef]
- Dai, X.; Xu, Y.; Ma, Q.; Xu, W.; Wang, T.; Xue, Y.; Chong, K. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol. 2007, 143, 1739–1751. [Google Scholar] [CrossRef]
- Yang, X.; Zhao, T.; Rao, P.; Gao, K.; Yang, X.; Chen, Z.; An, X. Transcriptome profiling of Populus tomentosa under cold stress. Ind. Crops Prod. 2019, 135, 283–293. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef]
- Töpfer, N.; Kleessen, S.; Nikoloski, Z. Integration of metabolomics data into metabolic networks. Front. Plant Sci. 2015, 6, 49. [Google Scholar] [CrossRef]
- Thomashow, M.F. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef]
- Zhang, N.; Shi, J.; Zhao, H.; Jiang, J. Activation of small heat shock protein (SlHSP17.7) gene by cell wall invertase inhibitor (SlCIF1) gene involved in sugar metabolism in tomato. Gene 2018, 679, 90–99. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Kidokoro, S.; Yoneda, K.; Takasaki, H.; Takahashi, F.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Different cold-signaling pathways function in the responses to rapid and gradual decreases in temperature. Plant Cell 2017, 29, 760–774. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.-K. Abiotic stress responses in plants. Nat. Rev. Genet. 2021, 23, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Chen, R.; Chen, Z.; Lin, J.; Jin, X.; Ren, C.; Chen, Q.; Chen, F.; Yu, G.; Zhang, Y. The molecular characteristics of soybean ARR-B transcription factors. BIOCELL 2022, 46, 1575–1592. [Google Scholar] [CrossRef]
- Nguyen, K.H.; Ha, C.V.; Nishiyama, R.; Watanabe, Y.; Leyva-González, M.A. Arabidopsis type-B cytokinin response regulators ARR1, ARR10, and ARR12 negatively regulate plant responses to drought. Proc. Natl. Acad. Sci. USA 2016, 113, 3090–3095. [Google Scholar] [CrossRef]
- Jeon, J.; Kim, N.Y.; Kim, S.; Kang, N.Y.; Novák, O. A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. J. Biol. Chem. 2010, 285, 23371–23386. [Google Scholar] [CrossRef]
- Kang, N.Y.; Chuloh, C.; Jungmook, K. Inducible expression of Arabidopsis Response Regulator 22 (ARR22), a type-C ARR, in transgenic Arabidopsis enhances drought and freezing tolerance. PLoS ONE 2013, 8, e79248. [Google Scholar] [CrossRef]
- Zeng, J.J.; Zhu, X.D.; Haider, M.S.; Wang, X.C.; Zhang, C.; Wang, C. Genome-Wide Identification and Analysis of the Type-B Authentic Response Regulator Gene Family in Peach (Prunus persica). Cytogenet. Genome Res. 2017, 151, 41–49. [Google Scholar] [CrossRef]
- Wang, J.; Xia, J.; Song, Q. Genome-wide identification, genomic organization and expression profiles of SlARR-B gene family in tomato. J. Appl. Genet. 2020, 61, 391–404. [Google Scholar] [CrossRef]
- Yue, C.; Chen, Q.; Hu, J.; Li, C.; Luo, L.; Zeng, L. Genome-Wide Identification and Characterization of GARP Transcription Factor Gene Family Members Reveal Their Diverse Functions in Tea Plant (Camellia sinensis). Front. Plant Sci. 2022, 13, 947072. [Google Scholar] [CrossRef]
- Ahmad, A.; Selim, M.M.; Alderfasi, A.A.; Afzal, M. Effect of Drought Stress on Mungbean (Vigna radiata L.) under Arid Climatic Conditions of Saudi Arabia. In Ecosystems and Sustainable Development X; WIT Press: Southampton, UK, 2015; pp. 185–193. [Google Scholar] [CrossRef]
- Ball, L.; Accotto, G.P.; Bechtold, U.; Creissen, G.; Funck, D.; Jimenez, A.; Kular, B.; Leyland, N.; Mejia-Carranza, J.; Reynolds, H.; et al. Evidence for a direct link between glutathione biosynthesis and stress defense gene expression in Arabidopsis. Plant Cell 2004, 16, 2448–2462. [Google Scholar] [CrossRef]
- Vranova, E.; Inze, D.; Van Breusegem, F. Signal transduction during oxidative stress. J. Exp. Bot. 2002, 53, 1227–1236. [Google Scholar] [CrossRef] [PubMed]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Zalutskaya, Z.; Skryabina, U.S.; Ermilova, E.V. Generation of hydrogen peroxide and transcriptional regulation of antioxidant enzyme expression in Chlamydomonas reinhardtii under hypothermia. Russ. J. Plant Physiol. 2019, 66, 223–230. [Google Scholar] [CrossRef]
- Bychkov, I.A.; Kudryakova, N.V.; Kuznetsov, V.V.; Kusnetsov, V.V. Cold Stress Activates the Expression of Genes of the Chloroplast Transcription Apparatus in Arabidopsis thaliana Plants. Doklady. Biochem. Biophys. 2020, 494, 235–239. [Google Scholar] [CrossRef]
- Garratt, L.C.; Janagoudar, B.S.; Lowe, K.C.; Anthony, P.; Power, J.B.; Davey, M.R. Salinity tolerance and antioxidant status in cotton cultures. Free Radic. Biol. Med. 2002, 33, 502–511. [Google Scholar] [CrossRef]
- Samarina, L.S.; Malyukova, L.S.; Efremov, A.M.; Simonyan, T.A.; Matskiv, A.O.; Koninskaya, N.G.; Rakhmangulov, R.S.; Gvasaliya, M.V.; Malyarovskaya, V.I.; Ryndin, A.V.; et al. Physiological, biochemical and genetic responses of Caucasian tea (Camellia sinensis (L.) Kuntze) genotypes under cold and frost stress. PeerJ 2020, 8, e9787. [Google Scholar] [CrossRef]
- Rajendrakumar, C.S.V.; Reddy, B.V.B.; Reddy, A.R. Proline-protein interactions: Protection of structural and functional integrity of M4, lactate dehydrogenase. Biochem. Biophys. Res. Commun. 1994, 201, 957–963. [Google Scholar] [CrossRef]
- Anjum, F.; Rishi, V.; Ahmed, F. Compatibility of osmolytes with gibbs energy of stabilization of proteins. Biochim. Biophys. Acta 2000, 1476, 75–84. [Google Scholar] [CrossRef]
- Matysik, J.; Alia; Bhalu, B.; Mohanty, P. Molecular mechanisms of quenching of reaction oxygen species by proline under stress in plants. Curr. Sci. 2002, 82, 525–532. Available online: http://www.jstor.org/stable/24105959 (accessed on 13 July 2024).
- Jung, K.H.; An, G. Application of MapMan and RiceNet drives systematic analyses of the early heat stress transcriptome in rice seedlings. J. Plant Biol. 2012, 55, 436–449. [Google Scholar] [CrossRef]
- Vinocur, B.; Altman, A. Recent advances in engineering plant tolerance to abiotic stress: Achievements and limitations. Curr. Opin. Biotechnol. 2005, 16, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Cui, P.; Li, Y.; Cui, C.; Huo, Y.; Lu, G.; Yang, H. Proteomic and metabolic profile analysis of low-temperature storage responses in Ipomoea batata Lam. tuberous roots. BMC Plant Biol. 2020, 20, 435. [Google Scholar] [CrossRef] [PubMed]
- Van Den Ende, W.; Valluru, R. Sucrose, sucrosyl oligosaccharides, and oxidative stress: Scavenging and salvaging? J. Exp. Bot. 2009, 60, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Sheen, J. Dynamic and diverse sugar signaling. Curr. Opin. Plant Biol. 2016, 33, 116–125. [Google Scholar] [CrossRef]
- Rao, X.; Dixon, R.A. Co-expression networks for plant biology: Why and how. Acta Biochim. Biophys. Sin. 2019, 51, 981–988. [Google Scholar] [CrossRef]
- Ren, Y.; Han, R.; Ma, Y.; Li, X.; Deng, C.; Zhao, M.; Li, J.; Hou, Q.; Zhong, Q.; Shao, D. Transcriptomics Integrated with Metabolomics Unveil Carotenoids Accumulation and Correlated Gene Regulation in White and Yellow-Fleshed Turnip (Brassica rapa ssp. rapa). Genes 2022, 13, 953. [Google Scholar] [CrossRef]
- Hansen, B.O.; Vaid, N.; Musialak-Lange, M.; Janowski, M.; Mutwil, M. Elucidating gene function and function evolution through comparison of coexpression networks of plants. Front. Plant Sci. 2014, 5, 394. [Google Scholar] [CrossRef]
- Hirano, K.; Aya, K.; Morinaka, Y.; Nagamatsu, S.; Sato, Y.; Antonio, B.A.; Namiki, N. Survey of genes involved in rice secondary cell wall formation through a co-expression network. Plant Cell Physiol. 2013, 54, 1803–1821. [Google Scholar] [CrossRef]
- Chandran, A.K.N.; Jeong, H.Y.; Jung, K.-H.; Lee, C. Development of functional modules based on co-expression patterns for cell-wall biosynthesis related genes in rice. J. Plant Biol. 2016, 59, 1–15. [Google Scholar] [CrossRef]
- Hirano, K.; Kondo, M.; Aya, K.; Miyao, A.; Sato, Y.; Antonio, B.A.; Namiki, N. Identification of transcription factors involved in rice secondary cell wall formation. Plant Cell Physiol. 2013, 54, 1791–1802. [Google Scholar] [CrossRef]
- Rao, X.; Chen, X.; Shen, H.; Ma, Q.; Li, G.; Tang, Y.; Pena, M. Gene regulatory networks for lignin biosynthesis in switchgrass (Panicum virgatum). Plant Biotechnol. J. 2019, 17, 580–593. [Google Scholar] [CrossRef] [PubMed]
- Rao, X.; Shen, H.; Pattathil, S.; Hahn, M.G.; Gelineo-Albersheim, I.; Mohnen, D.; Pu, Y. Dynamic changes in transcriptome and cell wall composition underlying brassinosteroid-mediated lignification of switchgrass suspension cells. Biotechnol. Biofuels 2017, 10, 266. [Google Scholar] [CrossRef]
- Chen, X.; Ma, Q.; Rao, X.; Tang, Y.; Wang, Y.; Li, G.; Zhang, C. Genomescale identification of cell-wall-related genes in switchgrass through comparative genomics and computational analyses of transcriptomic data. Bioenergy Res. 2015, 9, 172–180. [Google Scholar] [CrossRef]
- Shi, R.; Wang, J.P.; Lin, Y.C.; Li, Q.; Sun, Y.H.; Chen, H.; Sederoff, R.R. Tissue and cell-type co-expression networks of transcription factors and wood component genes in Populus trichocarpa. Planta 2017, 245, 927–938. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Li, C.-H.; Huang, J. Systematic identification of cell-wall related genes in Populus based on analysis of functional modules in co-expression network. PLoS ONE 2014, 9, e95176. [Google Scholar] [CrossRef] [PubMed]
- D’Agostino, I.B. Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 2000, 124, 1706–1717. [Google Scholar] [CrossRef]
- Gong, X.; Yan, B.; Hu, J.; Yang, C.; Li, Y.; Liu, J.; Liao, W. Transcriptome profiling of rubber tree (Hevea brasiliensis) discovers candidate regulators of the cold stress response. Genes. Genom. 2018, 40, 1181–1197. [Google Scholar] [CrossRef]
- Peng, X.; Wu, Q.; Teng, L.; Tang, F.; Pi, Z.; Shen, S. Transcriptional regulation of the paper mulberry under cold stress as revealed by a comprehensive analysis of transcription factors. BMC Plant Biol. 2015, 15, 108. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full length transcriptome assembly from RNA Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Lucas, A. Amap: Another Multidimensional Analysis Package. 2022. Available online: https://cran.r-project.org/web/packages/amap/index.html (accessed on 23 December 2023).
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bian, S.; Zhao, M.; Zhang, H.; Ren, Y. Differentially Expressed Genes Identification of Kohlrabi Seedlings (Brassica oleracea var. caulorapa L.) under Polyethylene Glycol Osmotic Stress and AP2/ERF Transcription Factor Family Analysis. Plants 2024, 13, 1167. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


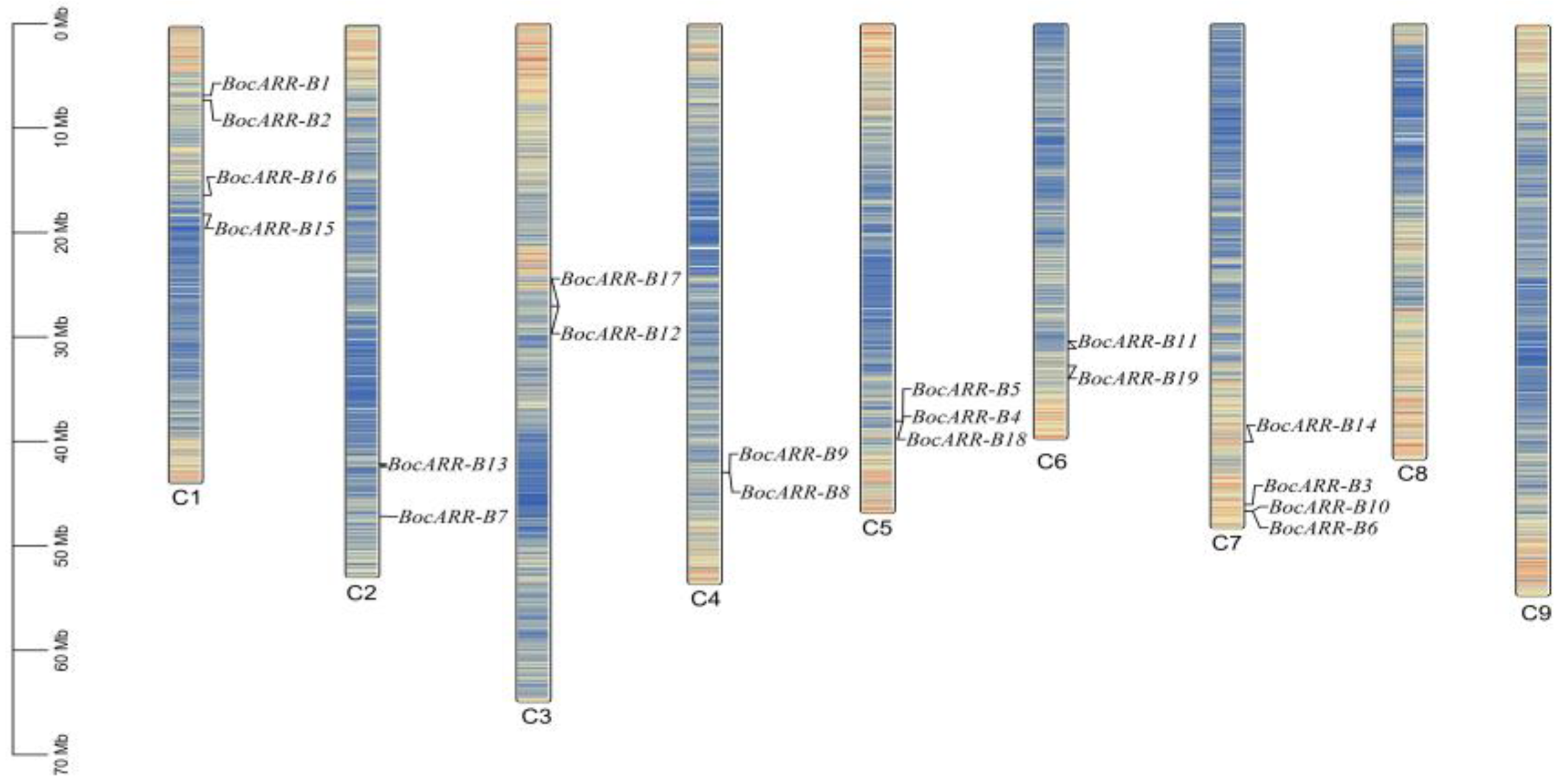


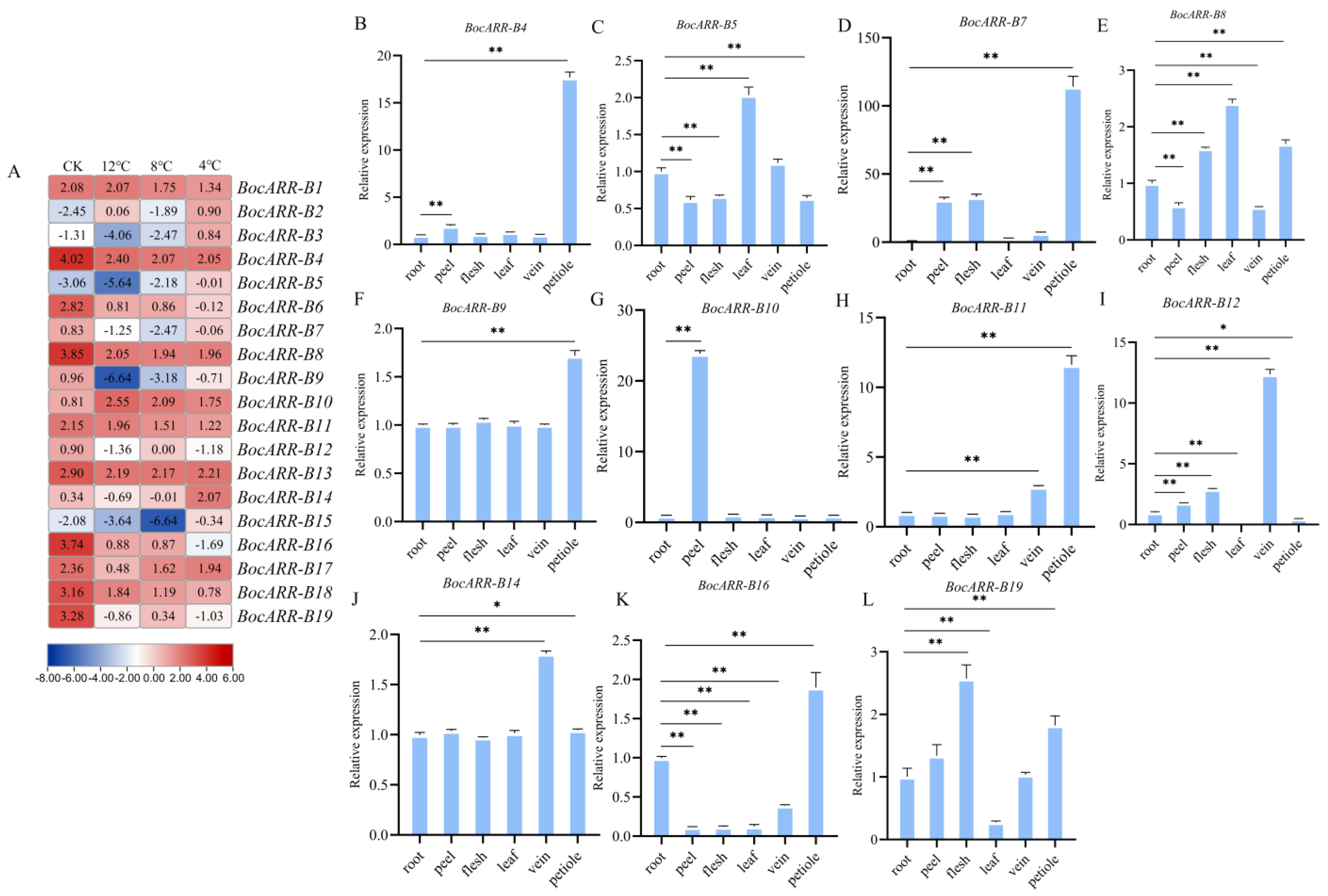
| Gene ID | Sequence ID | Accession Number of NCBI Reference Sequence | Length of cDNA | Number of Amino Acids | Molecular Weight | Theoretical pI | Aliphatic Index |
|---|---|---|---|---|---|---|---|
| BocARR-B1 | Cluster-17807.114153 | XM_013763325.1 | 1518 | 505 | 56,405.73 | 6.33 | 75.03 |
| BocARR-B2 | Cluster-17807.114154 | XM_022718524.2 | 1491 | 496 | 55,418.6 | 6.24 | 75.6 |
| BocARR-B3 | Cluster-17807.84395 | XM_013746126.1 | 1896 | 631 | 68,971.17 | 6.08 | 70.25 |
| BocARR-B4 | Cluster-17807.71963 | XM_013816288.3 | 1899 | 632 | 69,032.27 | 6.17 | 69.68 |
| BocARR-B5 | Cluster-17807.7188 | XM_013760760.1 | 1131 | 376 | 41,499.48 | 8.74 | 89.15 |
| BocARR-B6 | Cluster-17807.138306 | XM_048748308.1 | 1314 | 437 | 48,873.92 | 8.87 | 88.1 |
| BocARR-B7 | Cluster-17807.90061 | XM_013774622.1 | 1941 | 646 | 70,765.03 | 6.38 | 69.69 |
| BocARR-B8 | Cluster-17807.24114 | XM_013774622.1 | 1941 | 646 | 70,941.29 | 6.48 | 69.4 |
| BocARR-B9 | Cluster-17807.138684 | XM_013775692.1 | 1746 | 581 | 63,895.5 | 6.06 | 74.17 |
| BocARR-B10 | Cluster-17807.138685 | XM_013775692.1 | 1734 | 577 | 63,465.09 | 6.06 | 74.68 |
| BocARR-B11 | Cluster-17807.122837 | XM_013731190.1 | 2040 | 679 | 74,037.8 | 5.94 | 72.77 |
| BocARR-B12 | Cluster-17807.96507 | XM_013731190.1 | 2043 | 680 | 73,913.57 | 5.87 | 73.51 |
| BocARR-B13 | Cluster-17807.119682 | XM_013731190.1 | 2040 | 679 | 74,002.75 | 5.94 | 73.34 |
| BocARR-B14 | Cluster-17807.22246 | XM_013736811.1 | 1608 | 535 | 60,256.55 | 5.28 | 80.5 |
| BocARR-B15 | Cluster-18118.0 | XM_013736309.1 | 1506 | 501 | 56,851.62 | 5.08 | 81.08 |
| BocARR-B16 | Cluster-17807.71962 | XM_013743001.1 | 1803 | 600 | 65,834.76 | 6.21 | 70.63 |
| BocARR-B17 | Cluster-17807.116306 | XM_013743960.1 | 1767 | 588 | 65,877.4 | 5.78 | 77.6 |
| BocARR-B18 | Cluster-17807.137797 | XM_013852208.3 | 1809 | 602 | 66,358.36 | 6.47 | 71.51 |
| BocARR-B19 | Cluster-17807.18348 | XM_013852209.3 | 1806 | 601 | 66,152.09 | 6.38 | 70.67 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bian, S.; Shao, D.; Zhao, Q.; Li, Q.; Ren, Y. Transcriptome-Based Screening of Candidate Low-Temperature-Associated Genes and Analysis of the BocARR-B Transcription Factor Gene Family in Kohlrabi (Brassica oleracea L. var. caulorapa L.). Int. J. Mol. Sci. 2024, 25, 9261. https://doi.org/10.3390/ijms25179261
Bian S, Shao D, Zhao Q, Li Q, Ren Y. Transcriptome-Based Screening of Candidate Low-Temperature-Associated Genes and Analysis of the BocARR-B Transcription Factor Gene Family in Kohlrabi (Brassica oleracea L. var. caulorapa L.). International Journal of Molecular Sciences. 2024; 25(17):9261. https://doi.org/10.3390/ijms25179261
Chicago/Turabian StyleBian, Shuanling, Dengkui Shao, Qingsheng Zhao, Quanhui Li, and Yanjing Ren. 2024. "Transcriptome-Based Screening of Candidate Low-Temperature-Associated Genes and Analysis of the BocARR-B Transcription Factor Gene Family in Kohlrabi (Brassica oleracea L. var. caulorapa L.)" International Journal of Molecular Sciences 25, no. 17: 9261. https://doi.org/10.3390/ijms25179261
APA StyleBian, S., Shao, D., Zhao, Q., Li, Q., & Ren, Y. (2024). Transcriptome-Based Screening of Candidate Low-Temperature-Associated Genes and Analysis of the BocARR-B Transcription Factor Gene Family in Kohlrabi (Brassica oleracea L. var. caulorapa L.). International Journal of Molecular Sciences, 25(17), 9261. https://doi.org/10.3390/ijms25179261





