Envelope Protein-Targeting Zika Virus Entry Inhibitors
Abstract
1. Introduction
2. ZIKV Replication and Life Cycle
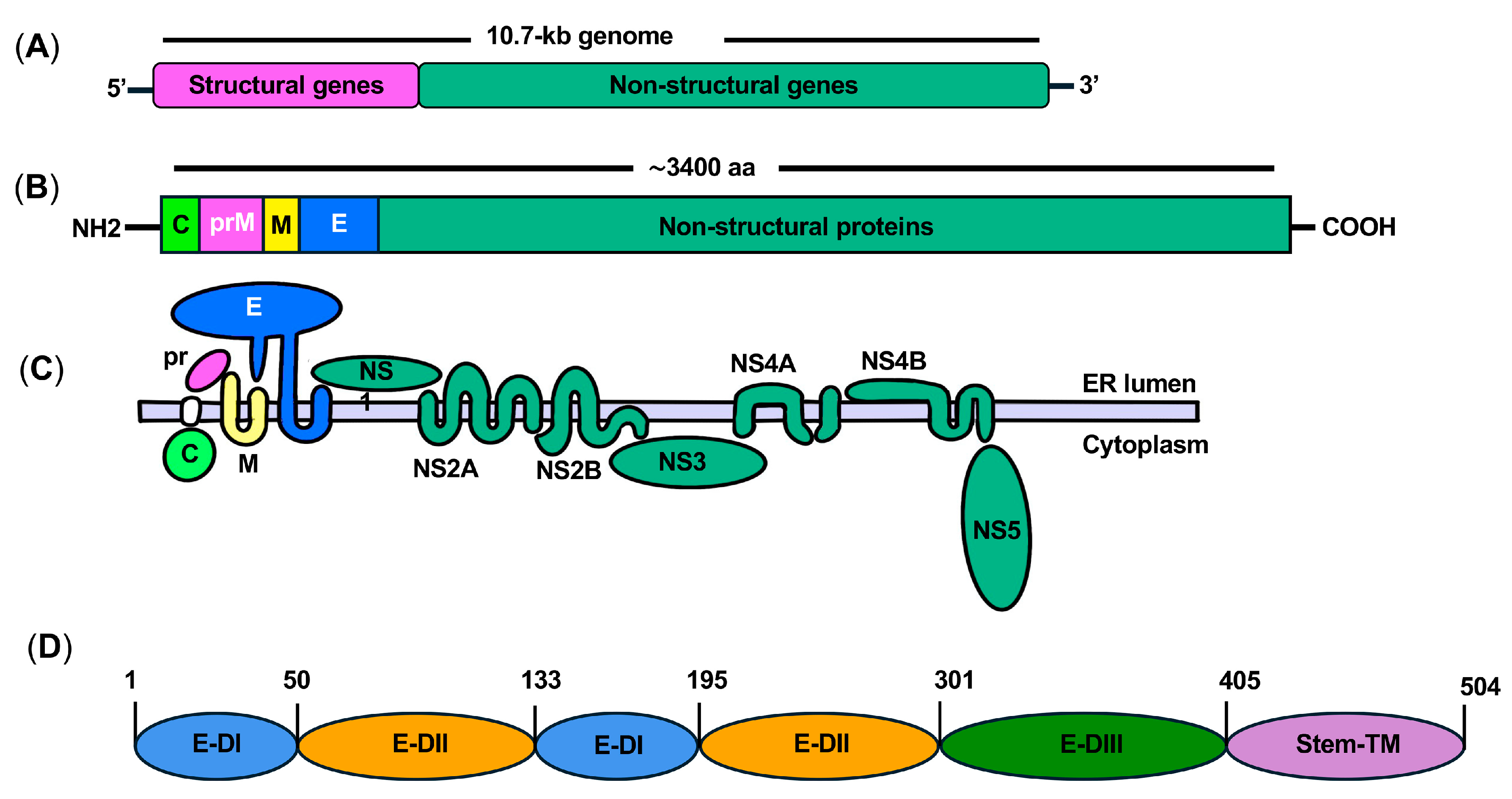
3. ZIKV Genome and Proteins
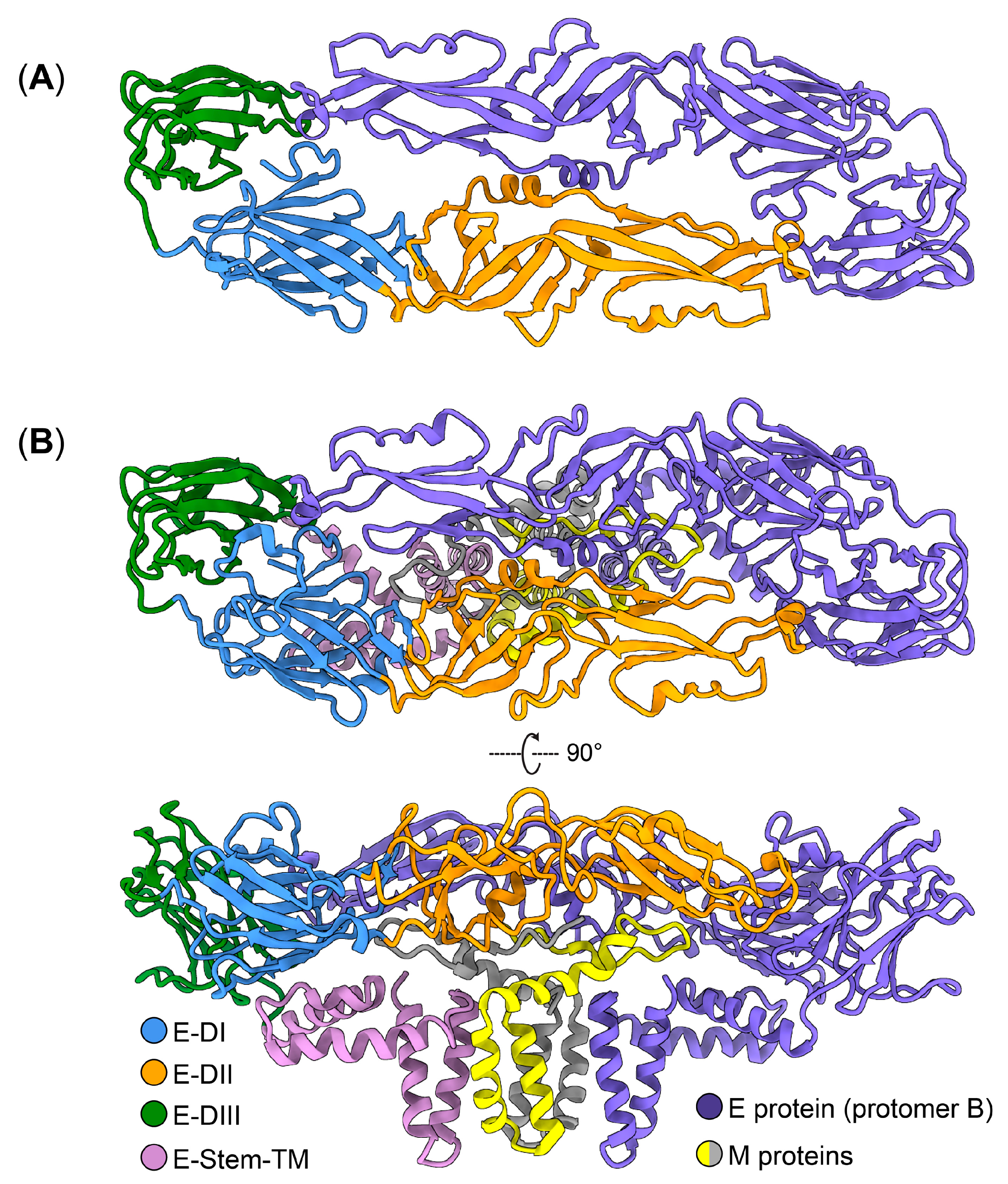
4. Structure and Function of ZIKV E Protein
5. Inhibitors of ZIKV E Protein-Mediated Entry
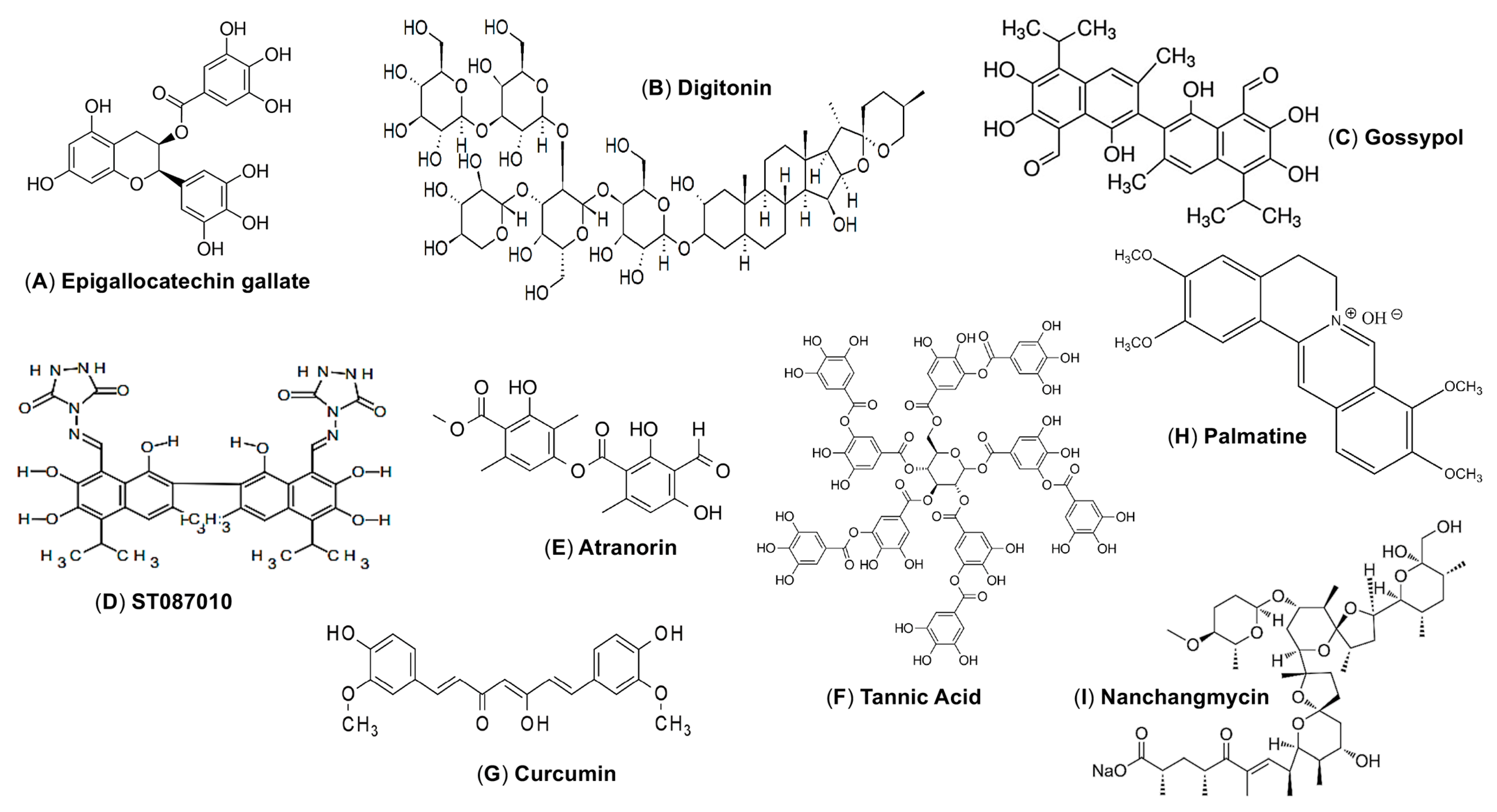
5.1. E Protein-Targeting Natural Products as ZIKV Entry Inhibitors
| E-binding Affinity (EC50) | Antiviral Activity (IC50) (ZIKV Strain) | Cytotoxicity (CC50) | Selectivity Index (SI) | Protection in Animals | Ref. | |
|---|---|---|---|---|---|---|
| Epigallocatechin gallate (EGCG) | ~119.94 kcal/mol | 21.4 µM (ZIKVBR) | >200 µM | 9 | ND | [60] |
| Curcumin | ND | 1.9 µM (HD78788) | 11.6 µM | 6 | ND | [67] |
| Nanchangmycin | ND | 0.1–0.4 µM (MR766 and Mex2-81) | ~10 µM | ND | ND | [68] |
| Gossypol | 7.12 µM | 3.8 µM (PAN2016) 4.4 µM (R116265) 4.2 µM (PAN2015) 0.7 µM (FLR) 3.1 µM (R103451) 4.6 µM (PRVABC59) | 14.5 μM | 4.3 | There is no protective efficacy in mice | [62,63] |
| ST087010 | 4.95 µM | 3.2 µM (PAN2016) 2.9 µM (R116265) 3.5 µM (PAN2015) 2.8 µM (FLR) 3.6 µM (R103451) 2.4 µM (PRVABC59) | 49.6 μM | 17.1 | Protects Ifnar1−/− mice from infection of ZIKV (R103451 and PAN2026 strains) | [63] |
| Palmatine | ND | 80 μM (IC90) (PRVABC59) | >100 µM | ND | ND | [69] |
| Digitonin | ND | 4.31 µM (PAN2016) 6.52 µM (R116265) 5.0 µM (PAN2015) 3.34 µM (FLR) 4.3 µM (R103451) | 56.29 µM | ND | ND | [62] |
| Atranorin | ND | 10.39 µM (SZ01/2016) | >50 µM | >4 | ND | [65] |
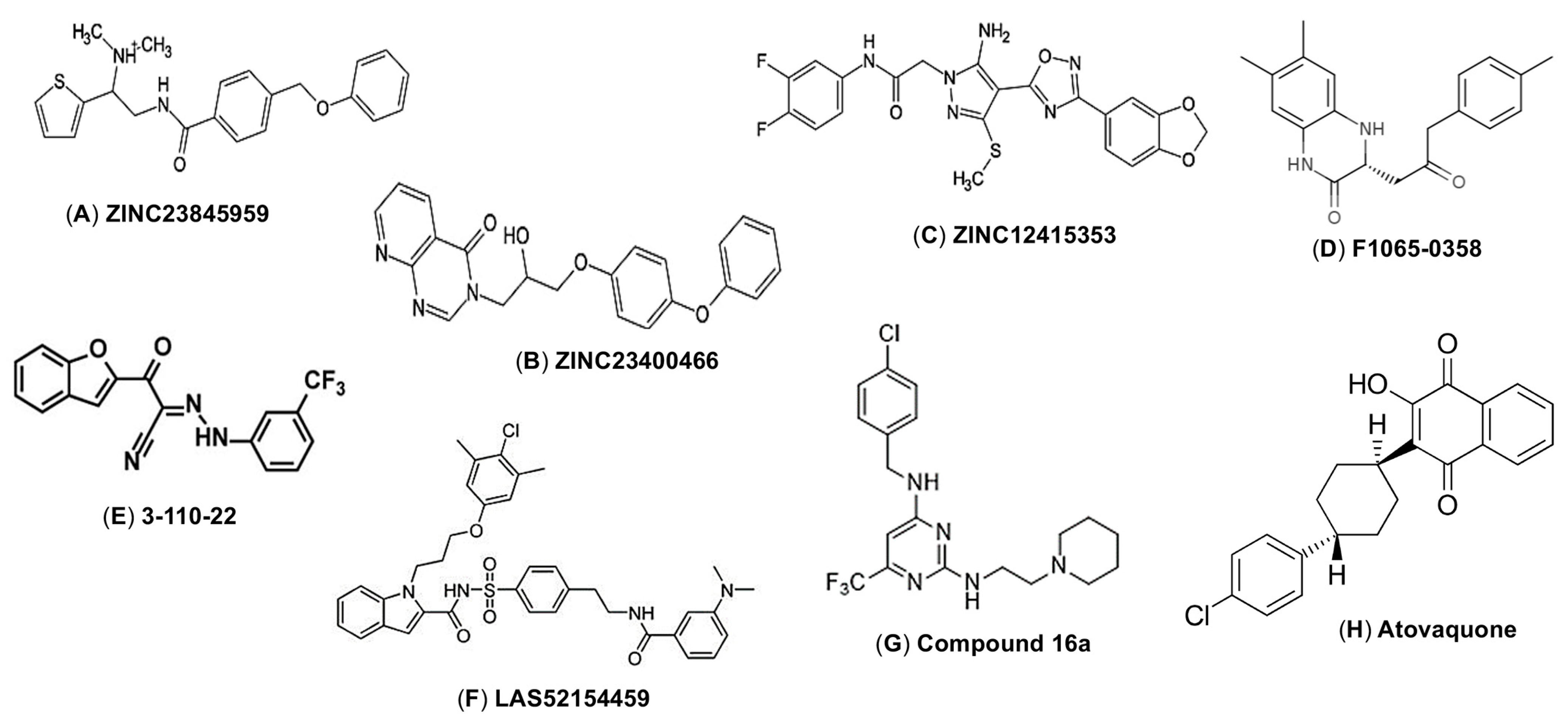
| F1065-0358 | ~8.314 kcal/mol (Docking score) | 14 µM | >200 µM | >14 | ND | [70] |
| ZINC23845959 | 9.35 Kcal/mol (Docking score) | 3–5 µM (MR766) | >25 µM | ND | ND | [71] |
| ZINC23400466 | 4.77 Kcal/mol (Docking score) | 3–5 µM (MR766) | >25 µM | ND | ND | [71] |
| ZINC12415353 | 6.15 Kcal/mol (Docking score) | 3–5 µM (MR766) | >25 µM | ND | ND | [71] |
| 3-110-22 | 2.1 μM | 4.2 µM (IC90) (PF13/251013-18) | 37.5 µM | ~9 | ND | [72,73] |
| 3-110-2 | ND | 4.8 µM (IC90) (PF13/251013-18) | ND | ND | ND | |
| 3-149-3 | ND | 14.9 µM (IC90) (PF13/251013-18) | ND | ND | ND | |
| LAS52154459 | ND | 5.13 µM (IC90) (PF13/251013-18) | 98.11 µM | 19 | ND | [74] |
| Compound 16a | ND | 2.61 µM (IC90) (INEVH116141) | 23.83 µM | 9.13 | ND | [75] |
| Atovaquone | ND | 2.1 μM IC90 (MR766) | ND | ND | ND | [76] |
5.2. E Protein-Targeting Small Molecules as ZIKV Entry Inhibitors
6. Host Factors That Modify the ZIKV E Protein
7. Potential Challenges of ZIKV E-Targeting Entry Inhibitors
8. An Overview of ZIKV Vaccines Based on the E Protein
9. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dick, G.W.A.; Kitchen, S.F.; Haddow, A.J. Zika Virus (I). Isolations and Serological Specificity. Trans. R. Soc. Trop. Med. Hyg. 1952, 46, 509–520. [Google Scholar] [CrossRef]
- Duffy, M.R.; Chen, T.H.; Hancock, W.T.; Powers, A.M.; Kool, J.L.; Lanciotti, R.S.; Pretrick, M.; Marfel, M.; Holzbauer, S.; Dubray, C.; et al. Zika Virus Outbreak on Yap Island, Federated States of Micronesia. N. Engl. J. Med. 2009, 360, 2536–2543. [Google Scholar] [CrossRef]
- Musso, D.; Nilles, E.J.; Cao-Lormeau, V.-M. Rapid Spread of Emerging Zika Virus in the Pacific Area. Clin. Microbiol. Infect. 2014, 20, O595–O596. [Google Scholar] [CrossRef] [PubMed]
- Lowe, R.; Barcellos, C.; Brasil, P.; Cruz, O.G.; Honório, N.A.; Kuper, H.; Carvalho, M.S. The Zika Virus Epidemic in Brazil: From Discovery to Future Implications. Int. J. Environ. Res. Public Health 2018, 15, 96. [Google Scholar] [CrossRef] [PubMed]
- Gulland, A. Zika Virus Is a Global Public Health Emergency, Declares WHO. BMJ 2016, 352, i657. [Google Scholar] [CrossRef]
- Boyer, S.; Calvez, E.; Chouin-Carneiro, T.; Diallo, D.; Failloux, A.-B. An Overview of Mosquito Vectors of Zika Virus. Microbes Infect. 2018, 20, 646–660. [Google Scholar] [CrossRef] [PubMed]
- Gregory, C.J.; Oduyebo, T.; Brault, A.C.; Brooks, J.T.; Chung, K.W.; Hills, S.; Kuehnert, M.J.; Mead, P.; Meaney-Delman, D.; Rabe, I.; et al. Modes of Transmission of Zika Virus. J. Infect. Dis. 2017, 216, S875–S883. [Google Scholar] [CrossRef]
- Masmejan, S.; Musso, D.; Vouga, M.; Pomar, L.; Dashraath, P.; Stojanov, M.; Panchaud, A.; Baud, D. Zika Virus. Pathogens 2020, 9, 898. [Google Scholar] [CrossRef]
- Li, C.; Deng, Y.Q.; Zu, S.; Quanquin, N.; Shang, J.; Tian, M.; Ji, X.; Zhang, N.N.; Dong, H.L.; Xu, Y.P.; et al. Zika Virus Shedding in the Stool and Infection through the Anorectal Mucosa in Mice. Emerg. Microbes Infect. 2018, 7, 169. [Google Scholar] [CrossRef]
- Atkinson, B.; Hearn, P.; Afrough, B.; Lumley, S.; Carter, D.; Aarons, E.J.; Simpson, A.J.; Brooks, T.J.; Hewson, R. Detection of Zika Virus in Semen. Emerg. Infect. Dis. 2016, 22, 940. [Google Scholar] [CrossRef]
- Gourinat, A.C.; O’Connor, O.; Calvez, E.; Goarant, C.; Dupont-Rouzeyrol, M. Detection of Zika Virus in Urine. Emerg. Infect. Dis. 2015, 21, 84–86. [Google Scholar] [CrossRef] [PubMed]
- Regla-Nava, J.A.; Viramontes, K.M.; Vozdolska, T.; Huynh, A.T.; Villani, T.; Gardner, G.; Johnson, M.; Ferro, P.J.; Shresta, S.; Kim, K. Detection of Zika Virus in Mouse Mammary Gland and Breast Milk. PLoS Negl. Trop. Dis. 2019, 13, e0007080. [Google Scholar] [CrossRef]
- Besnard, M.; Lastère, S.; Teissier, A.; Cao-Lormeau, V.M.; Musso, D. Evidence of Perinatal Transmission of Zika Virus, French Polynesia, December 2013 and February 2014. Eurosurveillance 2014, 19, 20751. [Google Scholar] [CrossRef]
- Carod-Artal, F.J. Neurological complications of Zika virus infection. Expert Rev. Anti Infect. Ther. 2018, 16, 399–410. [Google Scholar] [CrossRef]
- Reynolds, M.R.; Jones, A.M.; Petersen, E.E.; Lee, E.H.; Rice, M.E.; Bingham, A.; Ellington, S.R.; Evert, N.; Reagan-Steiner, S.; Oduyebo, T.; et al. Vital Signs: Update on Zika Virus–Associated Birth Defects and Evaluation of All U.S. Infants with Congenital Zika Virus Exposure—U.S. Zika Pregnancy Registry, 2016. Morb. Mortal. Wkly. Rep. 2017, 66, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Smoots, A.N.; Olson, S.M.; Cragan, J.; Delaney, A.; Roth, N.M.; Godfred-Cato, S.; Jones, A.M.; Nahabedian, J.F.; Fornoff, J.; Sandidge, T.; et al. Population-Based Surveillance for Birth Defects Potentially Related to Zika Virus Infection—22 States and Territories, January 2016-June 2017. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Paixao Enny, S.; Cardim Luciana, L.; Costa Maria, C.N.; Brickley Elizabeth, B.; de Carvalho-Sauer Rita, C.O.; Carmo Eduardo, H.; Andrade Roberto, F.S.; Rodrigues Moreno, S.; Veiga Rafael, V.; Costa Larissa, C.; et al. Mortality from Congenital Zika Syndrome—Nationwide Cohort Study in Brazil. N. Engl. J. Med. 2022, 386, 757–767. [Google Scholar] [CrossRef]
- Cao, B.; Diamond, M.S.; Mysorekar, I.U. Maternal-Fetal Transmission of Zika Virus: Routes and Signals for Infection. J. Interferon Cytokine Res. 2017, 37, 287–294. [Google Scholar] [CrossRef]
- Troumani, Y.; Touhami, S.; Jackson, T.L.; Ventura, C.V.; Stanescu-Segall, D.M.; Errera, M.-H.; Rousset, D.; Bodaghi, B.; Cartry, G.; David, T.; et al. Association of Anterior Uveitis with Acute Zika Virus Infection in Adults. JAMA Ophthalmol. 2021, 139, 95–102. [Google Scholar] [CrossRef]
- Weaver, S.C.; Costa, F.; Garcia-Blanco, M.A.; Ko, A.I.; Ribeiro, G.S.; Saade, G.; Shi, P.-Y.; Vasilakis, N. Zika Virus: History, Emergence, Biology, and Prospects for Control. Antivir. Res. 2016, 130, 69–80. [Google Scholar] [CrossRef]
- Siu, R.; Bukhari, W.; Todd, A.; Gunn, W.; Huang, Q.S.; Timmings, P. Acute Zika Infection with Concurrent Onset of Guillain-Barré Syndrome. Neurology 2016, 87, 1623–1624. [Google Scholar] [CrossRef]
- Gyawali, N.; Bradbury, R.S.; Taylor-Robinson, A.W. The Global Spread of Zika Virus: Is Public and Media Concern Justified in Regions Currently Unaffected? Infect. Dis. Poverty 2016, 5, 37. [Google Scholar] [CrossRef] [PubMed]
- Basu, R.; Tumban, E. Zika Virus on a Spreading Spree: What We Now Know That Was Unknown in the 1950’s. Virol. J. 2016, 13, 165. [Google Scholar] [CrossRef] [PubMed]
- Pan American Health Organization. Zika: A Silent Virus Requiring Enhanced Surveillance and Control. Available online: https://www.paho.org/en/news/1-9-2023-zika-silent-virus-requiring-enhanced-surveillance-and-control (accessed on 23 August 2024).
- World Health Organization. WHO to Identify Pathogens That Could Cause Future Outbreaks and Pandemics. Available online: https://www.who.int/news/item/21-11-2022-who-to-identify-pathogens-that-could-cause-future-outbreaks-and-pandemics (accessed on 23 August 2024).
- Jernej, M.; Misa, K.; Nataša, T.; Mara, P.; Mateja, P.-P.; Jerica, M.; Marko, K.; Katarina, R.R.; Tina, V.V.; Vesna, F.V.; et al. Zika Virus Associated with Microcephaly. N. Engl. J. Med. 2016, 374, 951–958. [Google Scholar] [CrossRef]
- Kuno, G.; Chang, G.J.; Tsuchiya, K.R.; Karabatsos, N.; Cropp, C.B. Phylogeny of the Genus Flavivirus. J. Virol. 1998, 72, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Agumadu, V.C.; Ramphul, K. Zika Virus: A Review of Literature. Cureus 2018, 10, e3025. [Google Scholar] [CrossRef]
- Fourié, T.; Grard, G.; Leparc-Goffart, I.; Briolant, S.; Fontaine, A. Variability of Zika Virus Incubation Period in Humans. Open Forum Infect. Dis. 2018, 5, ofy261. [Google Scholar] [CrossRef]
- Lessler, J.; Ott, C.T.; Carcelen, A.C.; Konikoff, J.M.; Williamson, J.; Bi, Q.; Kucirka, L.M.; Cummings, D.A.; Reich, N.G.; Chaisson, L.H. Times to Key Events in Zika Virus Infection and Implications for Blood Donation: A Systematic Review. Bull. World Health Organ. 2016, 94, 841–849. [Google Scholar] [CrossRef]
- Bhatnagar, J.; Rabeneck, D.B.; Martines, R.B.; Reagan-Steiner, S.; Ermias, Y.; Estetter, L.B.C.; Suzuki, T.; Ritter, J.; Keating, M.K.; Hale, G.; et al. Zika Virus RNA Replication and Persistence in Brain and Placental Tissue. Emerg. Infect. Dis. 2017, 23, 405–414. [Google Scholar] [CrossRef]
- Baz, M.; Boivin, G. Antiviral Agents in Development for Zika Virus Infections. Pharmaceuticals 2019, 12, 101. [Google Scholar] [CrossRef]
- Agrelli, A.; de Moura, R.R.; Crovella, S.; Brandão, L.A.C. ZIKA Virus Entry Mechanisms in Human Cells. Infect. Genet. Evol. 2019, 69, 22–29. [Google Scholar] [CrossRef]
- van Leur, S.W.; Heunis, T.; Munnur, D.; Sanyal, S. Pathogenesis and Virulence of Flavivirus Infections. Virulence 2021, 12, 2814–2838. [Google Scholar] [CrossRef]
- Pielnaa, P.; Al-Saadawe, M.; Saro, A.; Dama, M.F.; Zhou, M.; Huang, Y.; Huang, J.; Xia, Z. Zika Virus-Spread, Epidemiology, Genome, Transmission Cycle, Clinical Manifestation, Associated Challenges, Vaccine and Antiviral Drug Development. Virology 2020, 543, 34–42. [Google Scholar] [CrossRef]
- Sirohi, D.; Kuhn, R.J. Zika Virus Structure, Maturation, and Receptors. J. Infect. Dis. 2017, 216, S935–S944. [Google Scholar] [CrossRef] [PubMed]
- Cox, B.D.; Stanton, R.A.; Schinazi, R.F. Predicting Zika Virus Structural Biology: Challenges and Opportunities for Intervention. Antivir. Chem. Chemother. 2015, 24, 118–126. [Google Scholar] [CrossRef]
- Sotcheff, S.; Routh, A. Understanding Flavivirus Capsid Protein Functions: The Tip of the Iceberg. Pathogens 2020, 9, 42. [Google Scholar] [CrossRef] [PubMed]
- Valente, A.P.; Moraes, A.H. Zika Virus Proteins at an Atomic Scale: How Does Structural Biology Help Us to Understand and Develop Vaccines and Drugs against Zika Virus Infection? J. Venom. Anim. Toxins Trop. Dis. 2019, 25, e20190013. [Google Scholar] [CrossRef]
- Bernatchez, J.A.; Tran, L.T.; Li, J.; Luan, Y.; Siqueira-Neto, J.L.; Li, R. Drugs for the Treatment of Zika Virus Infection. J. Med. Chem. 2020, 63, 470–489. [Google Scholar] [CrossRef] [PubMed]
- Golubeva, V.A.; Nepomuceno, T.C.; de Gregoriis, G.; Mesquita, R.D.; Li, X.; Dash, S.; Garcez, P.P.; Suarez-Kurtz, G.; Izumi, V.; Koomen, J.; et al. Network of Interactions between ZIKA Virus Non-Structural Proteins and Human Host Proteins. Cells 2020, 9, 153. [Google Scholar] [CrossRef]
- Brown, W.C.; Akey, D.L.; Konwerski, J.R.; Tarrasch, J.T.; Skiniotis, G.; Kuhn, R.J.; Smith, J.L. Extended Surface for Membrane Association in Zika Virus NS1 Structure. Nat. Struct. Mol. Biol. 2016, 23, 865–867. [Google Scholar] [CrossRef]
- Zhang, X.; Xie, X.; Zou, J.; Xia, H.; Shan, C.; Chen, X.; Shi, P.Y. Genetic and Biochemical Characterizations of Zika Virus NS2A Protein. Emerg. Microbes Infect. 2019, 8, 585–602. [Google Scholar] [CrossRef]
- Xia, H.; Luo, H.; Shan, C.; Muruato, A.E.; Nunes, B.T.D.; Medeiros, D.B.A.; Zou, J.; Xie, X.; Giraldo, M.I.; Vasconcelos, P.F.C.; et al. An Evolutionary NS1 Mutation Enhances Zika Virus Evasion of Host Interferon Induction. Nat. Commun. 2018, 9, 414. [Google Scholar] [CrossRef]
- Best, S.M. The Many Faces of the Flavivirus NS5 Protein in Antagonism of Type I Interferon Signaling. J. Virol. 2017, 91, e01970-16. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Yi, G.; Du, F.; Chuang, Y.C.; Vaughan, R.C.; Sankaran, B.; Kao, C.C.; Li, P. Structure and Function of the Zika Virus Full-Length NS5 Protein. Nat. Commun. 2017, 8, 14762. [Google Scholar] [CrossRef]
- Dai, L.; Song, J.; Lu, X.; Deng, Y.-Q.; Musyoki, A.M.; Cheng, H.; Zhang, Y.; Yuan, Y.; Song, H.; Haywood, J.; et al. Structures of the Zika Virus Envelope Protein and Its Complex with a Flavivirus Broadly Protective Antibody. Cell Host Microbe 2016, 19, 696–704. [Google Scholar] [CrossRef]
- Sirohi, D.; Chen, Z.; Sun, L.; Klose, T.; Pierson, T.C.; Rossmann, M.G.; Kuhn, R.J. The 3.8 Å Resolution Cryo-EM Structure of Zika Virus. Science 2016, 352, 467–470. [Google Scholar] [CrossRef]
- Yang, C.; Gong, R.; de Val, N. Development of Neutralizing Antibodies against Zika Virus Based on Its Envelope Protein Structure. Virol. Sin. 2019, 34, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Rinkenberger, N.; Schoggins, J.W. Comparative Analysis of Viral Entry for Asian and African Lineages of Zika Virus. Virology 2019, 533, 59–67. [Google Scholar] [CrossRef]
- Bhardwaj, U.; Pandey, N.; Rastogi, M.; Singh, S.K. Gist of Zika Virus Pathogenesis. Virology 2021, 560, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Mercer, J.; Schelhaas, M.; Helenius, A. Virus Entry by Endocytosis. Annu. Rev. Biochem. 2010, 79, 803–833. [Google Scholar] [CrossRef] [PubMed]
- Barba-Spaeth, G.; Dejnirattisai, W.; Rouvinski, A.; Vaney, M.C.; Medits, I.; Sharma, A.; Simon-Lorière, E.; Sakuntabhai, A.; Cao-Lormeau, V.-M.; Haouz, A.; et al. Structural Basis of Potent Zika-Dengue Virus Antibody Cross-Neutralization. Nature 2016, 536, 48–53. [Google Scholar] [CrossRef]
- Saiz, J.C.; Martín-Acebes, M.A. The Race to Find Antivirals for Zika Virus. Antimicrob. Agents Chemother. 2017, 61, e00411-17. [Google Scholar] [CrossRef]
- Li, K.; Ji, Q.; Jiang, S.; Zhang, N. Advancement in the Development of Therapeutics Against Zika Virus Infection. Front. Cell. Infect. Microbiol. 2022, 12, 946957. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Murali, A.; Singh, S.K.; Giri, R. Epigallocatechin Gallate, an Active Green Tea Compound Inhibits the Zika Virus Entry into Host Cells via Binding the Envelope Protein. Int. J. Biol. Macromol. 2017, 104, 1046–1054. [Google Scholar] [CrossRef] [PubMed]
- Isaacs, C.E.; Wen, G.Y.; Xu, W.; Jia, J.H.; Rohan, L.; Corbo, C.; Di Maggio, V.; Jenkins, E.C.; Hillier, S. Epigallocatechin Gallate Inactivates Clinical Isolates of Herpes Simplex Virus. Antimicrob. Agents Chemother. 2008, 52, 962–970. [Google Scholar] [CrossRef]
- Nance, C.L.; Siwak, E.B.; Shearer, W.T. Preclinical Development of the Green Tea Catechin, Epigallocatechin Gallate, as an HIV-1 Therapy. J. Allergy Clin. Immunol. 2009, 123, 459–465. [Google Scholar] [CrossRef]
- Calland, N.; Albecka, A.; Belouzard, S.; Wychowski, C.; Duverlie, G.; Descamps, V.; Hober, D.; Dubuisson, J.; Rouillé, Y.; Séron, K. (−)-Epigallocatechin-3-Gallate Is a New Inhibitor of Hepatitis C Virus Entry. Hepatology 2012, 55, 720. [Google Scholar] [CrossRef]
- Carneiro, B.M.; Batista, M.N.; Braga, A.C.S.; Nogueira, M.L.; Rahal, P. The Green Tea Molecule EGCG Inhibits Zika Virus Entry. Virology 2016, 496, 215–218. [Google Scholar] [CrossRef] [PubMed]
- Chu, K.O.; Wang, C.C.; Chu, C.Y.; Choy, K.W.; Pang, C.P.; Rogers, M.S. Uptake and Distribution of Catechins in Fetal Organs Following in Utero Exposure in Rats. Hum. Reprod. 2007, 22, 280–287. [Google Scholar] [CrossRef]
- Gao, Y.; Tai, W.; Wang, N.; Li, X.; Jiang, S.; Debnath, A.K.; Du, L.; Chen, S. Identification of Novel Natural Products as Effective and Broad-Spectrum Anti-Zika Virus Inhibitors. Viruses 2019, 11, 1019. [Google Scholar] [CrossRef]
- Gao, Y.; Tai, W.; Wang, X.; Jiang, S.; Debnath, A.K.; Du, L.; Chen, S. A Gossypol Derivative Effectively Protects against Zika and Dengue Virus Infection without Toxicity. BMC Biol. 2022, 20, 143. [Google Scholar] [CrossRef] [PubMed]
- Vu, T.H.; Lamer, A.C.L.; Lalli, C.; Boustie, J.; Samson, M.; Lohézic-Le Dévéhat, F.; Le Seyec, J. Depsides: Lichen Metabolites Active against Hepatitis C Virus. PLoS ONE 2015, 10, e0120405. [Google Scholar] [CrossRef]
- Huang, G.; Wang, H.; Wang, X.; Yang, T.; Zhang, X.; Feng, C.; Zhao, W.; Tang, W. Atranorin Inhibits Zika Virus Infection in Human Glioblastoma Cell Line SNB-19 via Targeting Zika Virus Envelope Protein. Phytomedicine 2024, 125, 155343. [Google Scholar] [CrossRef]
- Priya, S.; Kumar, N.S.; Hemalatha, S. Antiviral Phytocompounds Target Envelop Protein to Control Zika Virus. Comput. Biol. Chem. 2018, 77, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Mounce, B.C.; Cesaro, T.; Carrau, L.; Vallet, T.; Vignuzzi, M. Curcumin Inhibits Zika and Chikungunya Virus Infection by Inhibiting Cell Binding. Antivir. Res. 2017, 142, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Rausch, K.; Hackett, B.A.; Weinbren, N.L.; Reeder, S.M.; Sadovsky, Y.; Hunter, C.A.; Schultz, D.C.; Coyne, C.B.; Cherry, S. Screening Bioactives Reveals Nanchangmycin as a Broad Spectrum Antiviral Active against Zika Virus. Cell Rep. 2017, 18, 804–815. [Google Scholar] [CrossRef]
- Ho, Y.J.; Lu, J.W.; Huang, Y.L.; Lai, Z.Z. Palmatine Inhibits Zika Virus Infection by Disrupting Virus Binding, Entry, and Stability. Biochem. Biophys. Res. Commun. 2019, 518, 732–738. [Google Scholar] [CrossRef]
- Sharma, N.; Prosser, O.; Kumar, P.; Tuplin, A.; Giri, R. Small Molecule Inhibitors Possibly Targeting the Rearrangement of Zika Virus Envelope Protein. Antivir. Res. 2020, 182, 104876. [Google Scholar] [CrossRef]
- Telehany, S.M.; Humby, M.S.; McGee, T.D., Jr.; Riley, S.P.; Jacobs, A.; Rizzo, R.C. Identification of Zika Virus Inhibitors Using Homology Modeling and Similarity-Based Screening to Target Glycoprotein E. Biochemistry 2020, 59, 3709–3724. [Google Scholar] [CrossRef]
- Li, P.-C.; Jang, J.; Hsia, C.-Y.; Groomes, P.V.; Lian, W.; de Wispelaere, M.; Pitts, J.D.; Wang, J.; Kwiatkowski, N.; Gray, N.S.; et al. Small Molecules Targeting the Flavivirus E Protein with Broad-Spectrum Activity and Antiviral Efficacy in Vivo. ACS Infect. Dis. 2019, 5, 460–472. [Google Scholar] [CrossRef] [PubMed]
- de Wispelaere, M.; Lian, W.; Potisopon, S.; Li, P.C.; Jang, J.; Ficarro, S.B.; Clark, M.J.; Zhu, X.; Kaplan, J.B.; Pitts, J.D.; et al. Inhibition of Flaviviruses by Targeting a Conserved Pocket on the Viral Envelope Protein. Cell Chem. Biol. 2018, 25, 1006–1016.e8. [Google Scholar] [CrossRef] [PubMed]
- Pitts, J.; Hsia, C.Y.; Lian, W.; Wang, J.; Pfeil, M.-P.; Kwiatkowski, N.; Li, Z.; Jang, J.; Gray, N.S.; Yang, P.L. Identification of Small Molecule Inhibitors Targeting the Zika Virus Envelope Protein. Antivir. Res. 2019, 164, 147–153. [Google Scholar] [CrossRef]
- Gallo, F.N.; Marquez, A.B.; Fidalgo, D.M.; Dana, A.; Dellarole, M.; García, C.C.; Bollini, M. Antiviral Drug Discovery: Pyrimidine Entry Inhibitors for Zika and Dengue Viruses. Eur. J. Med. Chem. 2024, 272, 116465. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Ichinohe, T.; Watanabe, A.; Kobayashi, A.; Zhang, R.; Song, J.; Kawaguchi, Y.; Matsuda, Z.; Inoue, J.-I. The Antimalarial Compound Atovaquone Inhibits Zika and Dengue Virus Infection by Blocking E Protein-Mediated Membrane Fusion. Viruses 2020, 12, 1475. [Google Scholar] [CrossRef] [PubMed]
- Kumari, N.; Kulkarni, A.A.; Lin, X.; McLean, C.; Ammosova, T.; Ivanov, A.; Hipolito, M.; Nekhai, S.; Nwulia, E. Inhibition of HIV-1 by Curcumin A, a Novel Curcumin Analog. Drug Des. Devel. Ther. 2015, 9, 5051–5060. [Google Scholar] [CrossRef] [PubMed]
- Palanichamy, K.; Joshi, A.; Mehmetoglu-Gurbuz, T.; Bravo, M.F.; Shlain, M.A.; Schiro, F.; Naeem, Y.; Garg, H.; Braunschweig, A.B. Anti-Zika Activity of a Library of Synthetic Carbohydrate Receptors. J. Med. Chem. 2019, 62, 4110–4119. [Google Scholar] [CrossRef]
- Lin, M.; Zhao, Z.; Yang, Z.; Meng, Q.; Tan, P.; Xie, W.; Qin, Y.; Wang, R.F.; Cui, J. USP38 Inhibits Type I Interferon Signaling by Editing TBK1 Ubiquitination through NLRP4 Signalosome. Mol. Cell 2016, 64, 267–281. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Q.; Hu, D.; Gao, D.; Wang, W.; Wu, K.; Wu, J. USP38 Inhibits Zika Virus Infection by Removing Envelope Protein Ubiquitination. Viruses 2021, 13, 2029. [Google Scholar] [CrossRef]
- Linder, M.E.; Deschenes, R.J. Palmitoylation: Policing Protein Stability and Traffic. Nat. Rev. Mol. Cell Biol. 2007, 8, 74–84. [Google Scholar] [CrossRef]
- Hu, D.; Zou, H.; Chen, W.; Li, Y.; Luo, Z.; Wang, X.; Guo, D.; Meng, Y.; Liao, F.; Wang, W.; et al. ZDHHC11 Suppresses Zika Virus Infections by Palmitoylating the Envelope Protein. Viruses 2023, 15, 144. [Google Scholar] [CrossRef]
- Prasad, V.M.; Miller, A.S.; Klose, T.; Sirohi, D.; Buda, G.; Jiang, W.; Kuhn, R.J.; Rossmann, M.G. Structure of the Immature Zika Virus at 9 Å Resolution. Nat. Struct. Mol. Biol. 2017, 24, 184–186. [Google Scholar] [CrossRef] [PubMed]
- Rey, F.A.; Stiasny, K.; Vaney, M.C.; Dellarole, M.; Heinz, F.X. The Bright and the Dark Side of Human Antibody Responses to Flaviviruses: Lessons for Vaccine Design. EMBO Rep. 2018, 19, 206–224. [Google Scholar] [CrossRef]
- Contreras, M.; Stuart, J.B.; Levoir, L.M.; Belmont, L.; Goo, L. Defining the Impact of Flavivirus Envelope Protein Glycosylation Site Mutations on Sensitivity to Broadly Neutralizing Antibodies. mBio 2024, 15, e0304823. [Google Scholar] [CrossRef]
- Carbaugh, D.L.; Baric, R.S.; Lazear, H.M. Envelope Protein Glycosylation Mediates Zika Virus Pathogenesis. J. Virol. 2019, 93, e00113-19. [Google Scholar] [CrossRef] [PubMed]
- Ishida, K.; Yagi, H.; Kato, Y.; Morita, E. N-Linked Glycosylation of Flavivirus E Protein Contributes to Viral Particle Formation. PLoS Pathog. 2023, 19, e1011681. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Jiménez, F.; Pérez-Olais, J.H.; Raymond, C.; King, B.J.; McClure, C.P.; Urbanowicz, R.A.; Ball, J.K. Challenges on the Development of a Pseudotyping Assay for Zika Glycoproteins. J. Med. Microbiol. 2021, 70, 001413. [Google Scholar] [CrossRef] [PubMed]
- Majowicz, S.A.; Narayanan, A.; Moustafa, I.M.; Bator, C.M.; Hafenstein, S.L.; Jose, J. Zika Virus M Protein Latches and Locks the E Protein from Transitioning to an Immature State after prM Cleavage. NPJ Viruses 2023, 1, 4. [Google Scholar] [CrossRef]
- da Queiroz, E.R.S.; de Medronho, R.A. Overlap between Dengue, Zika and Chikungunya Hotspots in the City of Rio de Janeiro. PLoS ONE 2022, 17, e0273980. [Google Scholar] [CrossRef]
- Brito, A.F.; Machado, L.C.; Oidtman, R.J.; Siconelli, M.J.L.; Tran, Q.M.; Fauver, J.R.; de Carvalho, R.D.O.; Dezordi, F.Z.; Pereira, M.R.; de Castro-Jorge, L.A.; et al. Lying in Wait: The Resurgence of Dengue Virus after the Zika Epidemic in Brazil. Nat. Commun. 2021, 12, 2619. [Google Scholar] [CrossRef]
- Cooper, C.D.; Addison-Smith, I.; Guzman, H.V. Quantitative Electrostatic Force Tomography for Virus Capsids in Interaction with an Approaching Nanoscale Probe. Nanoscale 2022, 14, 12232–12237. [Google Scholar] [CrossRef]
- Zandi, R.; Dragnea, B.; Travesset, A.; Podgornik, R. On Virus Growth and Form. Phys. Rep. 2020, 847, 1–102. [Google Scholar] [CrossRef]
- Park, H.; Tajkhorshid, E. Probing Druggability of the Zika Virus Envelope Proteins Using a Novel Computational Approach. Biophys. J. 2021, 120, 176a. [Google Scholar] [CrossRef]
- Alaofi, A.L. Probing the Flexibility of Zika Virus Envelope Protein DIII Epitopes Using Molecular Dynamics Simulations. Mol. Simul. 2020, 46, 541–547. [Google Scholar] [CrossRef]
- Peng, Z.Y.; Yang, S.; Lu, H.Z.; Wang, L.M.; Li, N.; Zhang, H.T.; Xing, S.Y.; Du, Y.N.; Deng, S.-Q. A Review on Zika Vaccine Development. Pathog. Dis. 2024, 82, ftad036. [Google Scholar] [CrossRef] [PubMed]
- Buitrago-Pabón, A.L.; Ruiz-Sáenz, S.; Jiménez-Alberto, A.; Aparicio-Ozores, G.; Castelán-Vega, J.A.; Ribas-Aparicio, R.M. An Update on Zika Virus Vaccine Development and New Research Approaches. Microbiol. Res. 2024, 15, 667–692. [Google Scholar] [CrossRef]
- Richner, J.M.; Himansu, S.; Dowd, K.A.; Butler, S.L.; Salazar, V.; Fox, J.M.; Julander, J.G.; Tang, W.W.; Shresta, S.; Pierson, T.C.; et al. Modified mRNA Vaccines Protect against Zika Virus Infection. Cell 2017, 168, 1114–1125.e10. [Google Scholar] [CrossRef]
- Schrauf, S.; Tschismarov, R.; Tauber, E.; Ramsauer, K. Current Efforts in the Development of Vaccines for the Prevention of Zika and Chikungunya Virus Infections. Front. Immunol. 2020, 11, 592. [Google Scholar] [CrossRef]
- López-Camacho, C.; Abbink, P.; Larocca, R.A.; Dejnirattisai, W.; Boyd, M.; Badamchi-Zadeh, A.; Wallace, Z.R.; Doig, J.; Velazquez, R.S.; Neto, R.D.L.; et al. Rational Zika Vaccine Design via the Modulation of Antigen Membrane Anchors in Chimpanzee Adenoviral Vectors. Nat. Commun. 2018, 9, 2441. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roy, A.; Liu, Q.; Yang, Y.; Debnath, A.K.; Du, L. Envelope Protein-Targeting Zika Virus Entry Inhibitors. Int. J. Mol. Sci. 2024, 25, 9424. https://doi.org/10.3390/ijms25179424
Roy A, Liu Q, Yang Y, Debnath AK, Du L. Envelope Protein-Targeting Zika Virus Entry Inhibitors. International Journal of Molecular Sciences. 2024; 25(17):9424. https://doi.org/10.3390/ijms25179424
Chicago/Turabian StyleRoy, Abhijeet, Qian Liu, Yang Yang, Asim K. Debnath, and Lanying Du. 2024. "Envelope Protein-Targeting Zika Virus Entry Inhibitors" International Journal of Molecular Sciences 25, no. 17: 9424. https://doi.org/10.3390/ijms25179424
APA StyleRoy, A., Liu, Q., Yang, Y., Debnath, A. K., & Du, L. (2024). Envelope Protein-Targeting Zika Virus Entry Inhibitors. International Journal of Molecular Sciences, 25(17), 9424. https://doi.org/10.3390/ijms25179424







