Molecular Characterization of Odorant-Binding Protein Genes Associated with Host-Seeking Behavior in Oides leucomelaena
Abstract
1. Introduction
2. Results
2.1. Library Assembly
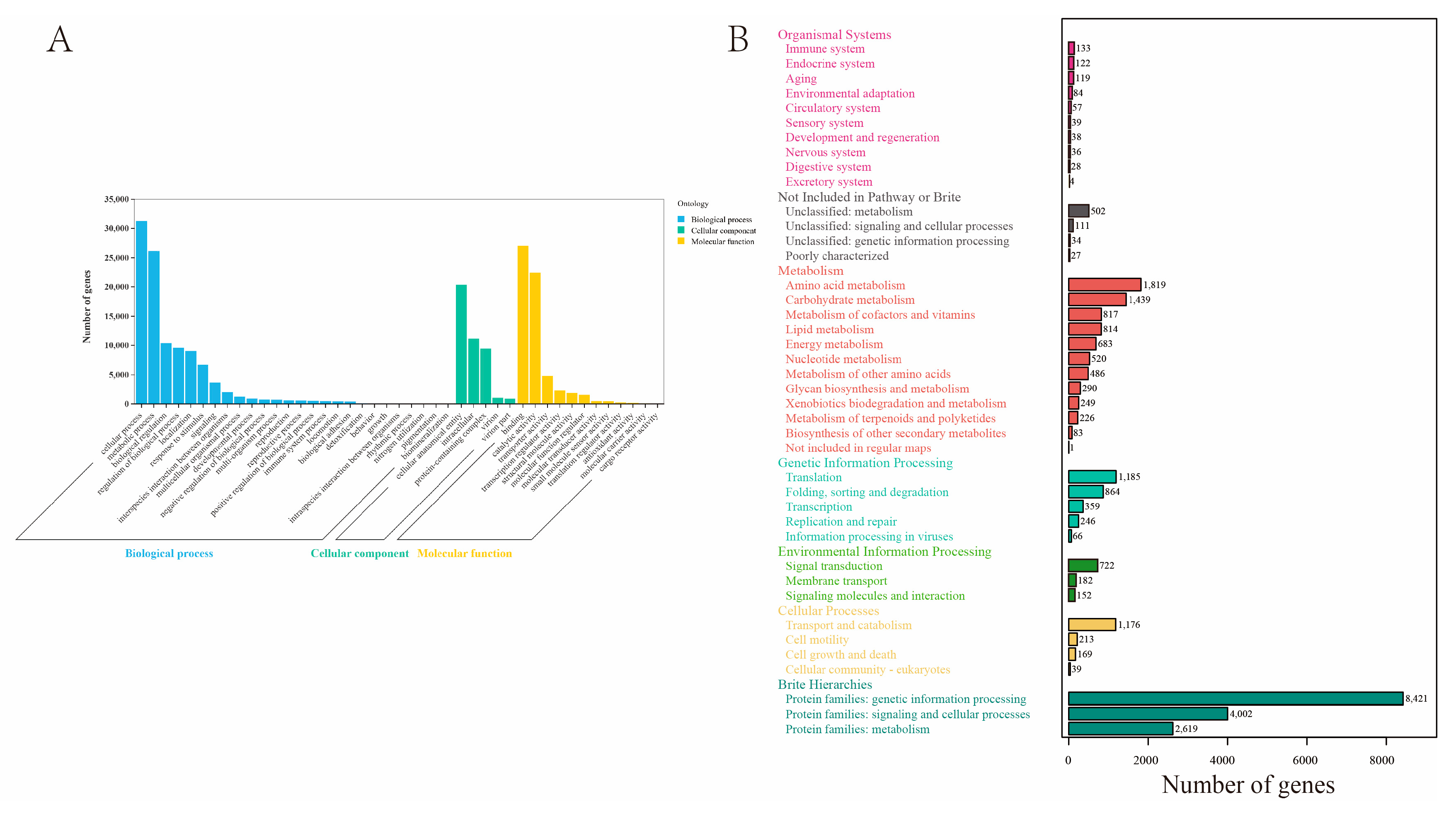
2.2. Transcript Annotation
2.3. Chemosensory-Related Proteins
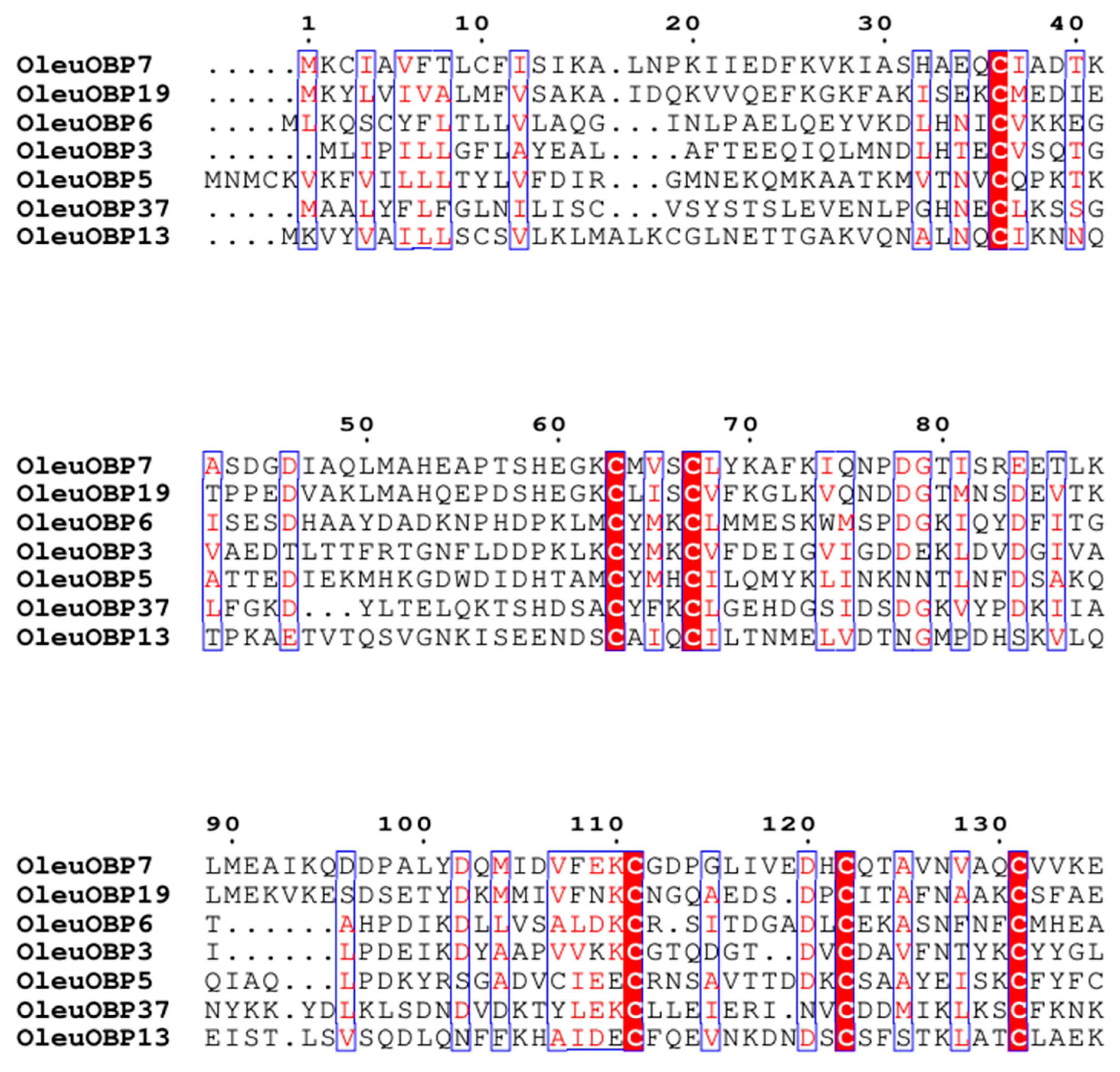
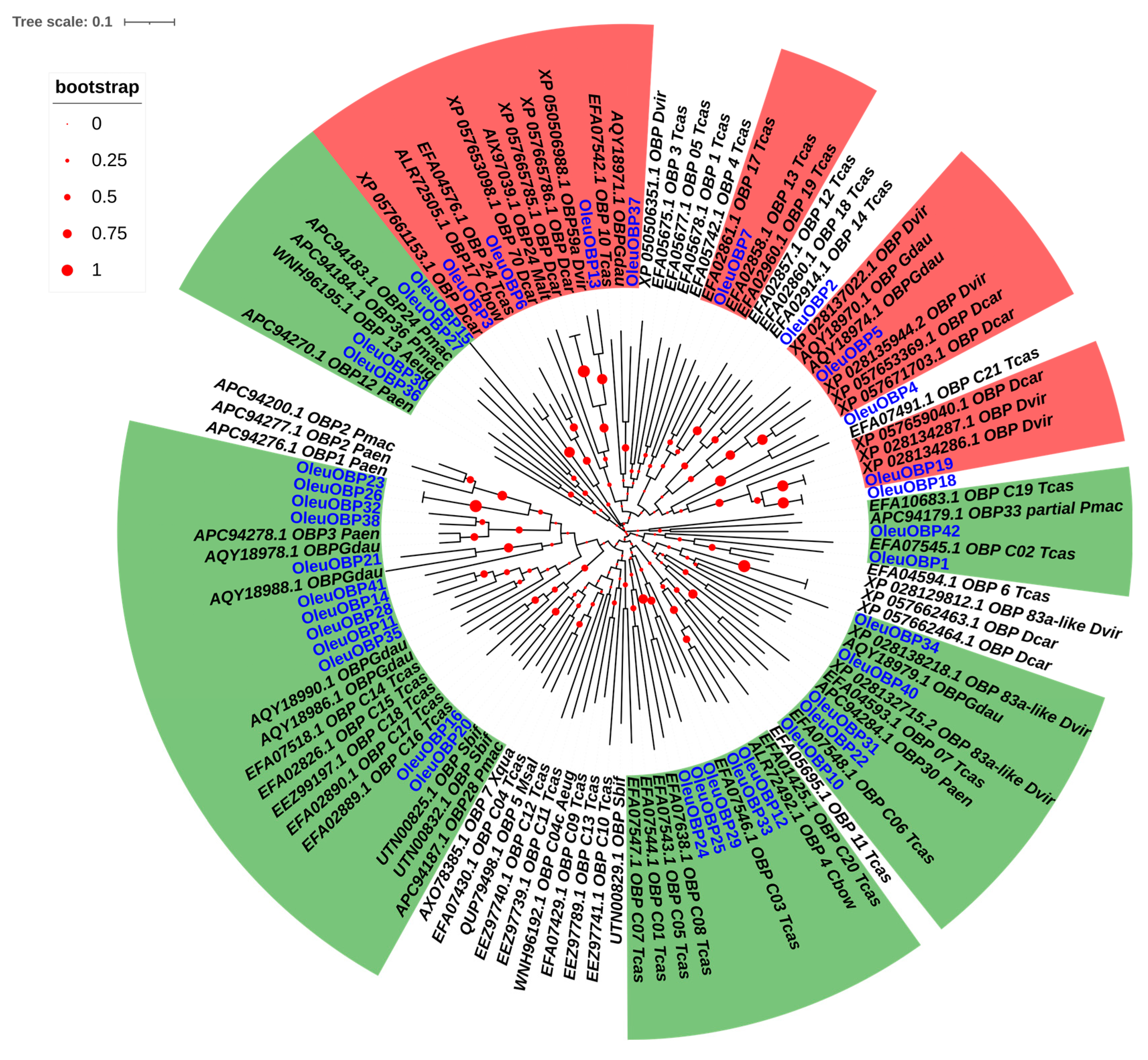
2.4. Sequence Comparison and Phylogenetic Tree Analysis of OBPs
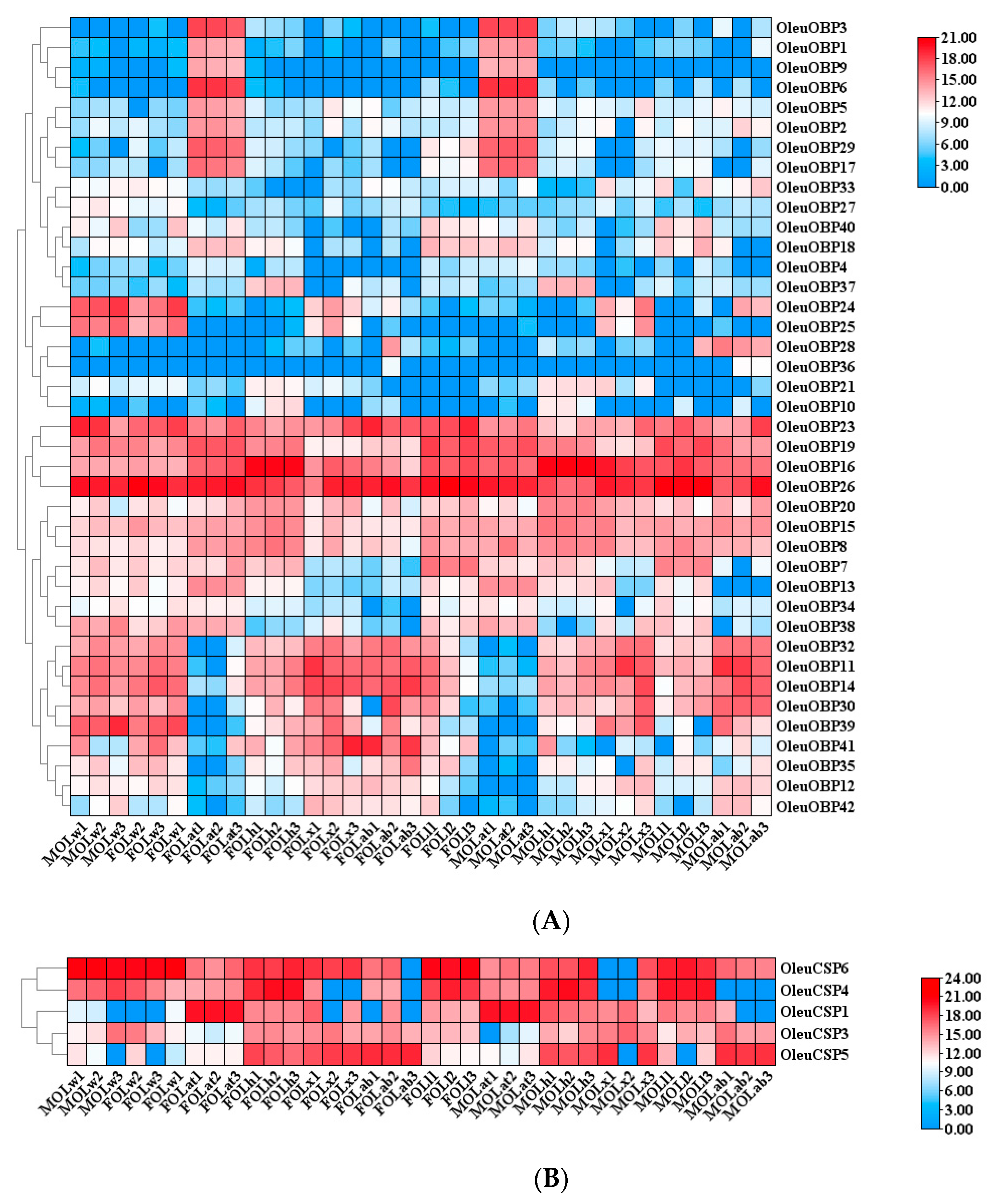
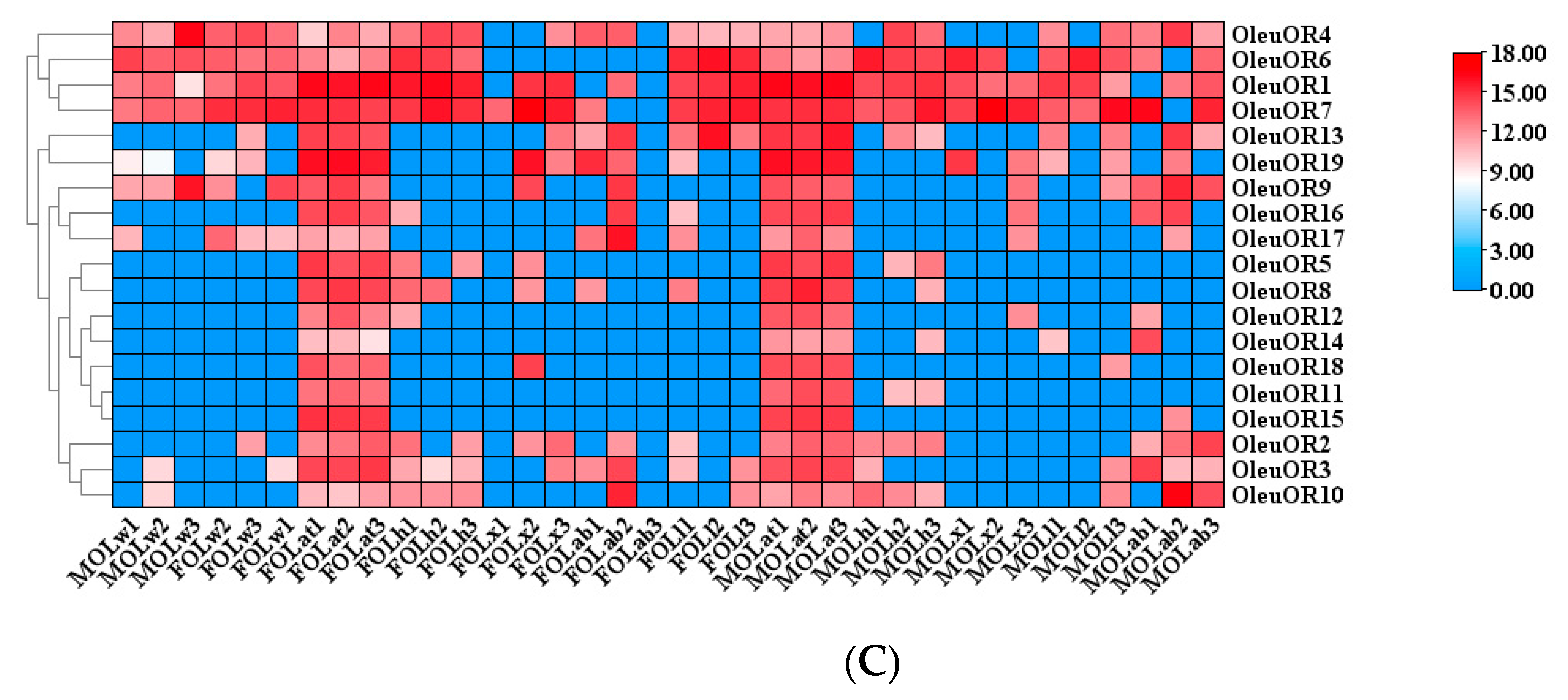
2.5. Antennal-Specific OBP
2.6. β-Caryophyllene Is a Molecule with High Affinity for Antennae-Specific Proteins
3. Discussion
4. Materials and Methods
4.1. Insect Collection
4.2. RNA Sequencing and Annotation
4.3. Gene Identification and Sequence Analysis
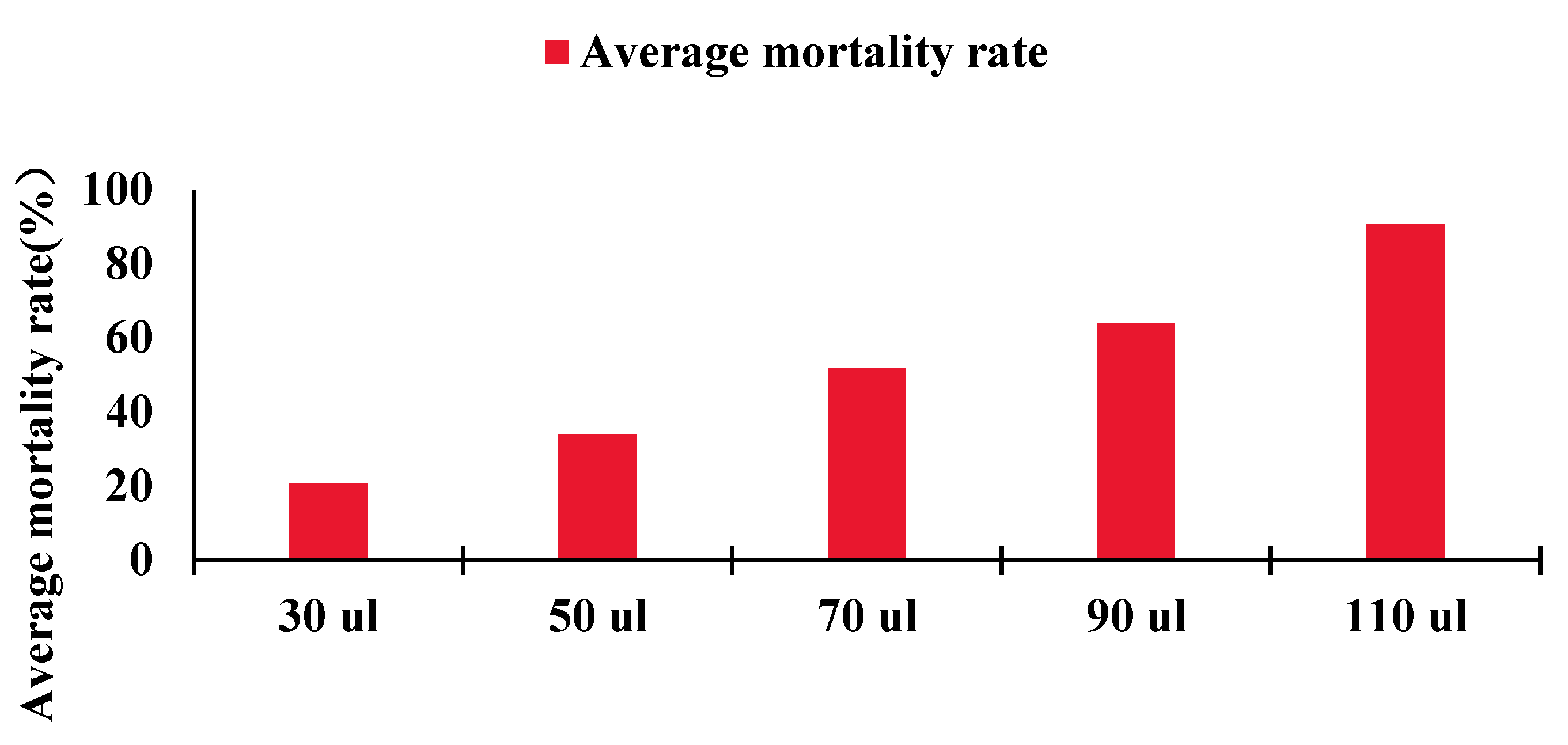
4.4. Compound Fumigation Experiment
4.5. Specific Expression of OBP Genes
4.6. Molecular Docking
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zou, Q.; Huang, Y.; Zhang, W.; Lu, C.; Yuan, J. A Comprehensive Review of the Pharmacology, Chemistry, Traditional Uses and Quality Control of Star Anise (Illicium verum Hook. F.): An Aromatic Medicinal Plant. Molecules 2023, 28, 7378. [Google Scholar] [CrossRef]
- Sharafan, M.; Jafernik, K.; Ekiert, H.; Kubica, P.; Kocjan, R.; Blicharska, E.; Szopa, A. Illicium verum (Star Anise) and Trans -Anethole as Valuable Raw Materials for Medicinal and Cosmetic Applications. Molecules 2022, 27, 650. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Shen, J.; Ding, C.; Yang, X. A Review of Chinese Species of the Genus Oides Weber, 1801 (Coleoptera: Chrysomelidae: Galerucinae). Insects 2024, 15, 114. [Google Scholar] [CrossRef] [PubMed]
- Ji, M.; Ning, D.; Chen, H.; Han, M.; Feng, Z. The Occurrence and Control of Oides leucomelaena Weise. Anhui Agric. Sci. Bull. 2007, 13, 142. [Google Scholar]
- Huang, N.; Chang, M.; Peng, S.; Wu, Y. Life History and Spatial Distribution of Oides duporti Laboisserie in Guangxi Province. J. Anhui Agric. Sci. 2022, 50, 2. [Google Scholar]
- Wu, X. Integrated Pest Management Methods for Oides leucomelaena Weise. Guangxi For. Sci. 2001, 1, 34. [Google Scholar]
- Benton, R. Evolution, Multigene Family Evolution: Perspectives from Insect Chemoreceptors. Trends Ecol. Evol. 2015, 30, 590–600. [Google Scholar] [CrossRef]
- Abendroth, J.A.; Moural, T.W.; Wei, H.; Zhu, F. Roles of Iinsect Odorant Binding Proteins in Communication and Xenobiotic Adaptation. Front. Insect. Sci. 2023, 3, 1274197. [Google Scholar] [CrossRef]
- Wang, X.; Verschut, T.A.; Billeter, J.C.; Maan, M.E. Evolution, Seven Questions on the Chemical Ecology and Neurogenetics of Resource-Mediated Speciation. Front. Ecol. Evol. 2021, 9, 640486. [Google Scholar]
- Li, J.; Wang, X.; Zhang, L. Sex Pheromones and Olfactory Proteins in Antheraea Moths: A. pernyi and A. polyphemus (Lepidoptera: Saturniidae). Arch. Insect. Biochem. Physiol. 2020, 105, e21729. [Google Scholar] [CrossRef]
- Kaleem Ullah, R.M.; Jia, B.; Liang, S.; Sikandar, A.; Gao, F.; Wu, H. Uncovering the Chemosensory System of a Subterranean Termite, Odontotermes formosanus (Shiraki) (Isoptera: Termitidae): Revealing the Chemosensory Genes and Gene Expression Patterns. Insects 2023, 14, 883. [Google Scholar] [CrossRef] [PubMed]
- Lizana, P.; Machuca, J.; Larama, G.; Quiroz, A.; Mutis, A.; Venthur, H. Mating-based Regulation and Ligand Binding of an Odorant-Binding Protein Support the Inverse Sexual Communication of the Greater Wax Moth, Galleria mellonella (Lepidoptera: Pyralidae). Insect. Mol. Biol. 2020, 29, 337–351. [Google Scholar] [CrossRef]
- Wang, J.; Gao, P.; Luo, Y.; Tao, J. Characterization and Expression Profiling of Odorant-Binding Proteins in Anoplophora glabripennis Motsch. Gene 2019, 693, 25–36. [Google Scholar] [CrossRef]
- Lechuga-Paredes, P.; Segura-León, O.L.; Cibrián-Tovar, J.; Torres-Huerta, B.; Velázquez-González, J.C.; Cruz-Jaramillo, J.L. Odorant-Binding and Chemosensory Proteins in Anthonomus eugenii (Coleoptera: Curculionidae) and Their Tissue Expression. Int. J. Mol. Sci. 2023, 24, 3406. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.N.; Grosse-Wilde, E.; Keeling, C.I.; Bengtsson, J.M.; Yuen, M.M.S.; Li, M.; Hillbur, Y.; Bohlmann, J.; Hansson, B.S.; Schlyter, F. Antennal Transcriptome Analysis of the Chemosensory Gene Families in the Tree Killing Bark Beetles, Ips typographus and Dendroctonus ponderosae (Coleoptera: Curculionidae: Scolytinae). BMC Genom. 2013, 14, 198. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Wang, H.; Shi, J.; Wang, X.; Zheng, H.; Wong, G.K.-S.; Clark, T.; Wang, W.; Wang, J.; Kang, L. Identification and Characterization of Insect-Specific Proteins by Genome Data Analysis. BMC Genom. 2007, 8, 93. [Google Scholar] [CrossRef]
- Webster, M.T.; Beaurepaire, A.; Neumann, P.; Stolle, E. Population Genomics for Insect Conservation. Annu. Rev. Anim. Biosci. 2023, 11, 115–140. [Google Scholar] [CrossRef]
- Denell, R.; Shippy, T. Comparative Insect Developmental Genetics: Phenotypes without Mutants. Bioessays 2001, 23, 379–382. [Google Scholar] [CrossRef]
- Behura, S.K. Insect Phylogenomics. Insect. Mol. Biol. 2015, 24, 403–411. [Google Scholar] [CrossRef]
- Brito, N.F.; Moreira, M.F.; Melo, A.C.A. A Look Inside Odorant-Binding Proteins in Insect Chemoreception. J. Insect. Physiol. 2016, 95, 51–65. [Google Scholar] [CrossRef]
- Venthur, H.; Zhou, J.-J. Odorant Receptors and Odorant-Binding Proteins as Insect Pest Control Targets: A Comparative Analysis. Front. Physiol. 2018, 9, 1163. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jin, Y.; Chen, Q.; Wen, M.; Zhao, H.; Duan, H.; Ren, B. Selectivity and Ligand-Based Molecular Modeling of an Odorant-Binding Protein from the Leaf Beetle Ambrostoma quadriimpressum (Coleoptera: Chrysomelidae) in Relation to Habitat-Related Volatiles. Sci. Rep. 2017, 7, 15374. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Niimura, Y.; Nozawa, M. The Evolution of Animal Chemosensory Receptor Gene Repertoires: Roles of Chance and Necessity. Nat. Rev. Genet. 2008, 9, 951–963. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.N.; Christer, L.F.; Newcomb, R.D. Evolution, Insect Olfaction and the Evolution of Receptor Tuning. Front. Ecol. Evol. 2015, 3, 53. [Google Scholar] [CrossRef]
- Li, L.; Zhou, Y.T.; Tan, Y.; Zhou, X.R.; Pang, B.P. Identification of Odorant-Binding Protein Genes in Galeruca daurica (Coleoptera: Chrysomelidae) and Analysis of Their Expression Profiles. Bull. Entomol. Res. 2017, 107, 550–561. [Google Scholar] [CrossRef]
- Tang, Q.-F.; Shen, C.; Zhang, Y.; Yang, Z.-P.; Han, R.-R.; Wang, J. Antennal Transcriptome Analysis of the Maize Weevil Sitophilus zeamais: Identification and Tissue Expression Profiling of Candidate Odorant- Binding Protein Genes. Arch. Insect. Biochem. Physiol 2019, 101, e21542. [Google Scholar] [CrossRef]
- Andersson, M.N.; Keeling, C.I.; Mitchell, R.F. Genomic Content of Chemosensory Genes Correlates with Host Range in Wood-Boring Beetles (Dendroctonus ponderosae, Agrilus planipennis, and Anoplophora glabripennis). BMC Genom. 2019, 20, 690. [Google Scholar] [CrossRef]
- Han, X.; Weng, M.; Shi, W.; Wen, Y.; Long, Y.; Hu, X.; Ji, G.; Zhu, Y.; Wen, X.; Zhang, F.; et al. The Neurotranscriptome of Monochamus alternatus. Int. J. Mol. Sci. 2024, 25, 4553. [Google Scholar] [CrossRef]
- Han, W.-K.; Tang, F.-X.; Yan, Y.-Y.; Wang, Y.; Zhang, Y.-X.; Yu, N.; Wang, K.; Liu, Z.-W. An OBP Gene Highly Expressed in Non-Chemosensory Tissues Affects the Phototaxis and Reproduction of Spodoptera frugiperda. Insect. Mol. Biol. 2024, 33, 81–90. [Google Scholar] [CrossRef]
- Pei, H.; Xie, G.; Yao, X.; Wang, S.; Yan, J.; Dai, L.; Wang, Y. Exploring the Binding Affinity and Characteristics of DcitOBP9 in citrus psyllids. Gene 2024, 923, 148551. [Google Scholar] [CrossRef]
- Kuang, Y.; Shangguan, C.; Yuan, S.; Zhang, Q.; Qiu, Z.; Gao, L.; Yu, X. Candidate Odorant-Binding Protein and Chemosensory Protein Genes in the Turnip Aphid Lipaphis erysimi. Arch. Insect. Biochem. Physiol. 2023, 113, e22022. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Merchant, A.; Zhao, Z.; Zhang, Y.; Zhang, J.; Zhang, Q.; Wang, Q.; Zhou, X.; Li, X. Characterization of MaltOBP1, a Minus-C Odorant-Binding Protein, From the Japanese Pine Sawyer Beetle, Monochamus alternatus Hope (Coleoptera: Cerambycidae). Front. Physiol. 2020, 11, 212. [Google Scholar] [CrossRef]
- Mitchell, R.F.; Schneider, T.M.; Schwartz, A.M.; Andersson, M.N.; Mckenna, D.D. The Diversity and Evolution of Odorant Receptors in Beetles (Coleoptera). Insect. Mol. Biol. 2020, 29, 77–91. [Google Scholar] [CrossRef]
- Andersson, M.N.; Larsson, M.C.; Schlyter, F. Specificity and Redundancy in the Olfactory System of the Bark Beetle Ips typographus: Single-Cell Responses to Ecologically Relevant Odors. J. Insect. Physiol. 2009, 55, 556–567. [Google Scholar] [CrossRef] [PubMed]
- De Groot, A.C.; Schmidt, E. Essential Oils, part III: Chemical Composition. Dermatitis 2016, 27, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Lyu, F.; Hai, X.; Wang, Z. A Review of the Host Plant Location and Recognition Mechanisms of Asia n Longhorn Beetle. Insects 2023, 14, 292. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.; Guo, Y.; Xie, Y.; Ren, Y.; Bi, Y.; Wang, F.; Fang, Q.; Ye, G. Rice Volatile Compound (E)-β-caryophyllene Induced by Rice Dwarf Virus (RDV) Attracts the Natural Enemy Cyrtorhinus lividipennis to Prey on RDV Insect Vectors. Pest. Manag. Sci. 2024, 80, 874–884. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a Full-Length Transcriptome without a Genome from RNA-Seq Data. Nat. Biotechnol. 2013, 29, 644. [Google Scholar]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-Throughput Functional Annotation and Data Mining with the Blast2GO Suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Larralde, M.; Zeller, G. PyHMMER: A Python Library Binding to HMMER for Efficient Sequence Analysis. Bioinformatics 2023, 39, btad214. [Google Scholar] [CrossRef]
- Gertz, E.M.; Yu, Y.-K.; Agarwala, R.; Schäffer, A.A.; Altschul, S.F. Composition-Based Statistics and Translated Nucleotide Searches: Improving the TBLASTN Module of BLAST. BMC Biol. 2006, 4, 41. [Google Scholar] [CrossRef] [PubMed]
- Sudhir, K.; Glen, S.; Tamura, K. Evolution, MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870. [Google Scholar]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Norgan, A.P.; Coffman, P.K.; Kocher, J.-P.A.; Katzmann, D.J.; Sosa, C.P. Multilevel Parallelization of AutoDock 4.2. J. Cheminform. 2011, 3, 12. [Google Scholar] [CrossRef]

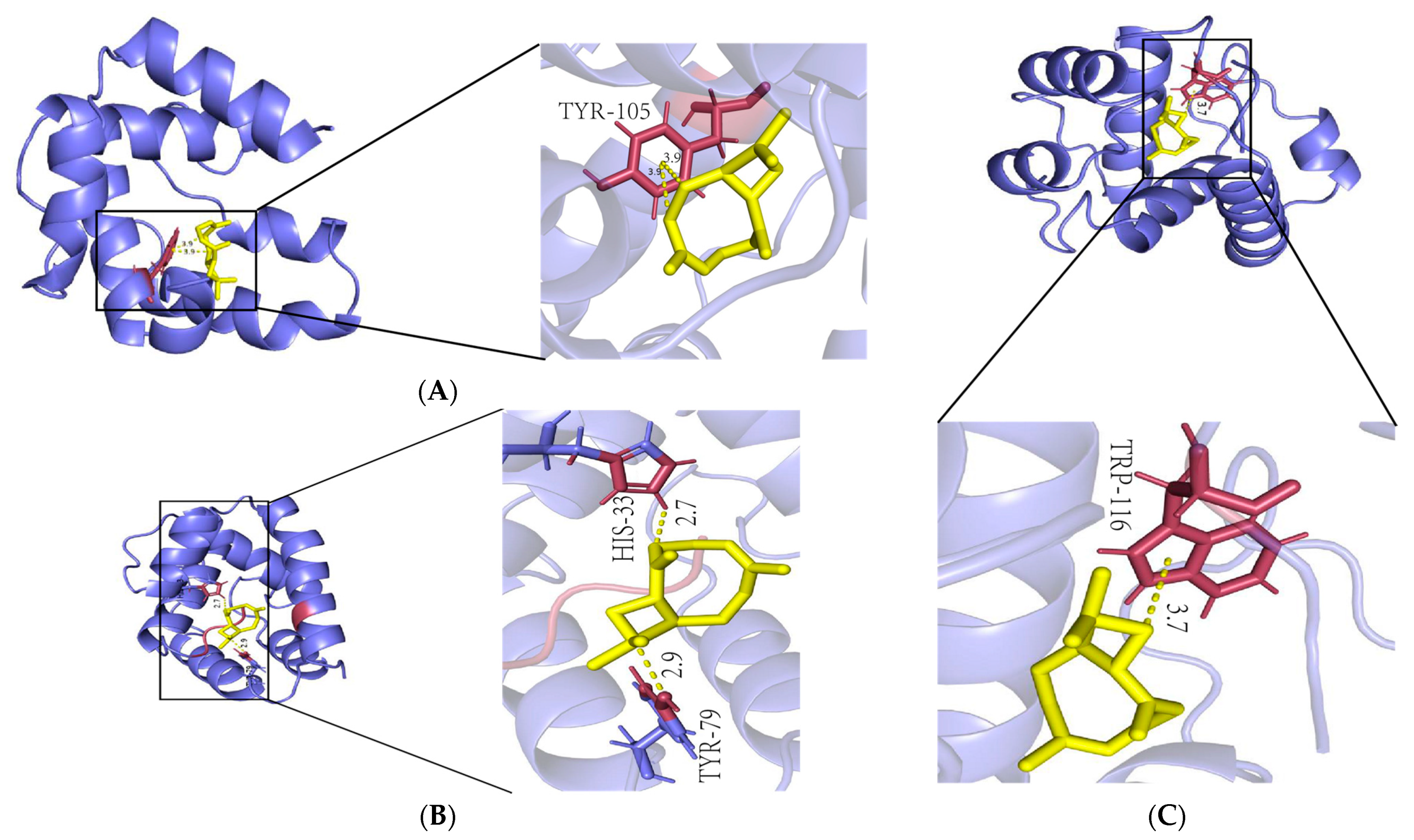
| Protein | Ligand | Binding Energy (kJ/mol) |
|---|---|---|
| OleuOBP3 | (Z)-anethole | −17.40544 |
| OleuOBP5 | (Z)-anethole | −20.69296 |
| OleuOBP6 | (Z)-anethole | −21.10336 |
| OleuOBP3 | β-caryophyllene | −24.12168 |
| OleuOBP5 | β-caryophyllene | −21.03352 |
| OleuOBP6 | β-caryophyllene | −31.7984 |
| OleuOBP3 | 3-carene | −22.5936 |
| OleuOBP5 | 3-carene | −17.9912 |
| OleuOBP6 | 3-carene | −25.5224 |
| OleuOBP3 | 4-allylanisole | −23.012 |
| OleuOBP5 | 4-allylanisole | −16.736 |
| OleuOBP6 | 4-allylanisole | −16.736 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, N.; Li, K.; Ma, H.; Hu, L.; Yang, Y.; Liu, L. Molecular Characterization of Odorant-Binding Protein Genes Associated with Host-Seeking Behavior in Oides leucomelaena. Int. J. Mol. Sci. 2024, 25, 9436. https://doi.org/10.3390/ijms25179436
Zhao N, Li K, Ma H, Hu L, Yang Y, Liu L. Molecular Characterization of Odorant-Binding Protein Genes Associated with Host-Seeking Behavior in Oides leucomelaena. International Journal of Molecular Sciences. 2024; 25(17):9436. https://doi.org/10.3390/ijms25179436
Chicago/Turabian StyleZhao, Ning, Kai Li, Huifen Ma, Lianrong Hu, Yingxue Yang, and Ling Liu. 2024. "Molecular Characterization of Odorant-Binding Protein Genes Associated with Host-Seeking Behavior in Oides leucomelaena" International Journal of Molecular Sciences 25, no. 17: 9436. https://doi.org/10.3390/ijms25179436
APA StyleZhao, N., Li, K., Ma, H., Hu, L., Yang, Y., & Liu, L. (2024). Molecular Characterization of Odorant-Binding Protein Genes Associated with Host-Seeking Behavior in Oides leucomelaena. International Journal of Molecular Sciences, 25(17), 9436. https://doi.org/10.3390/ijms25179436






