Antennal Transcriptome Screening and Identification of Chemosensory Proteins in the Double-Spine European Spruce Bark Beetle, Ips duplicatus (Coleoptera: Scolytinae)
Abstract
1. Introduction
2. Results
2.1. De Novo Antennal Transcriptome Sequencing and Assembly
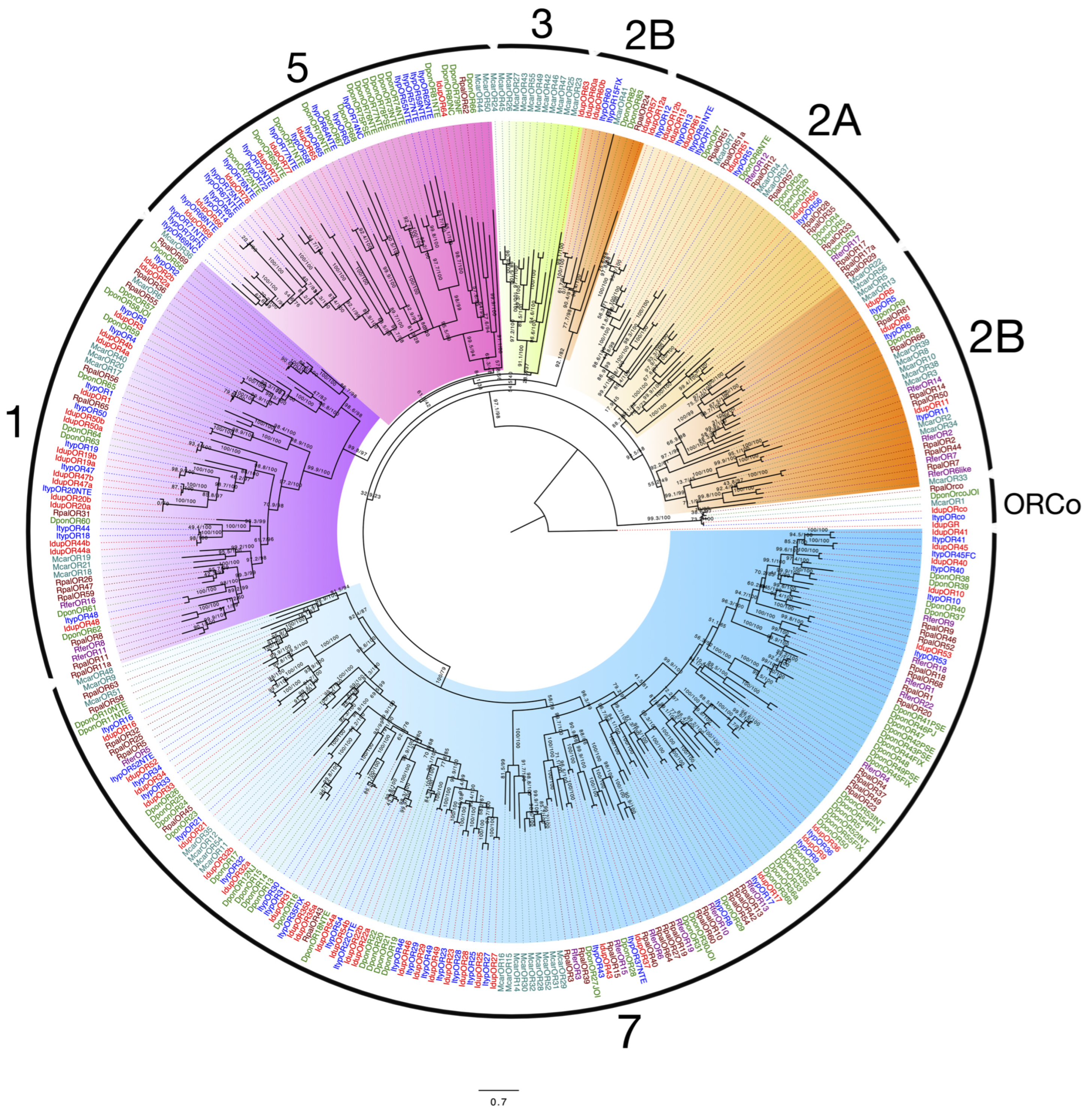
2.2. Odorant Receptors in I. duplicatus
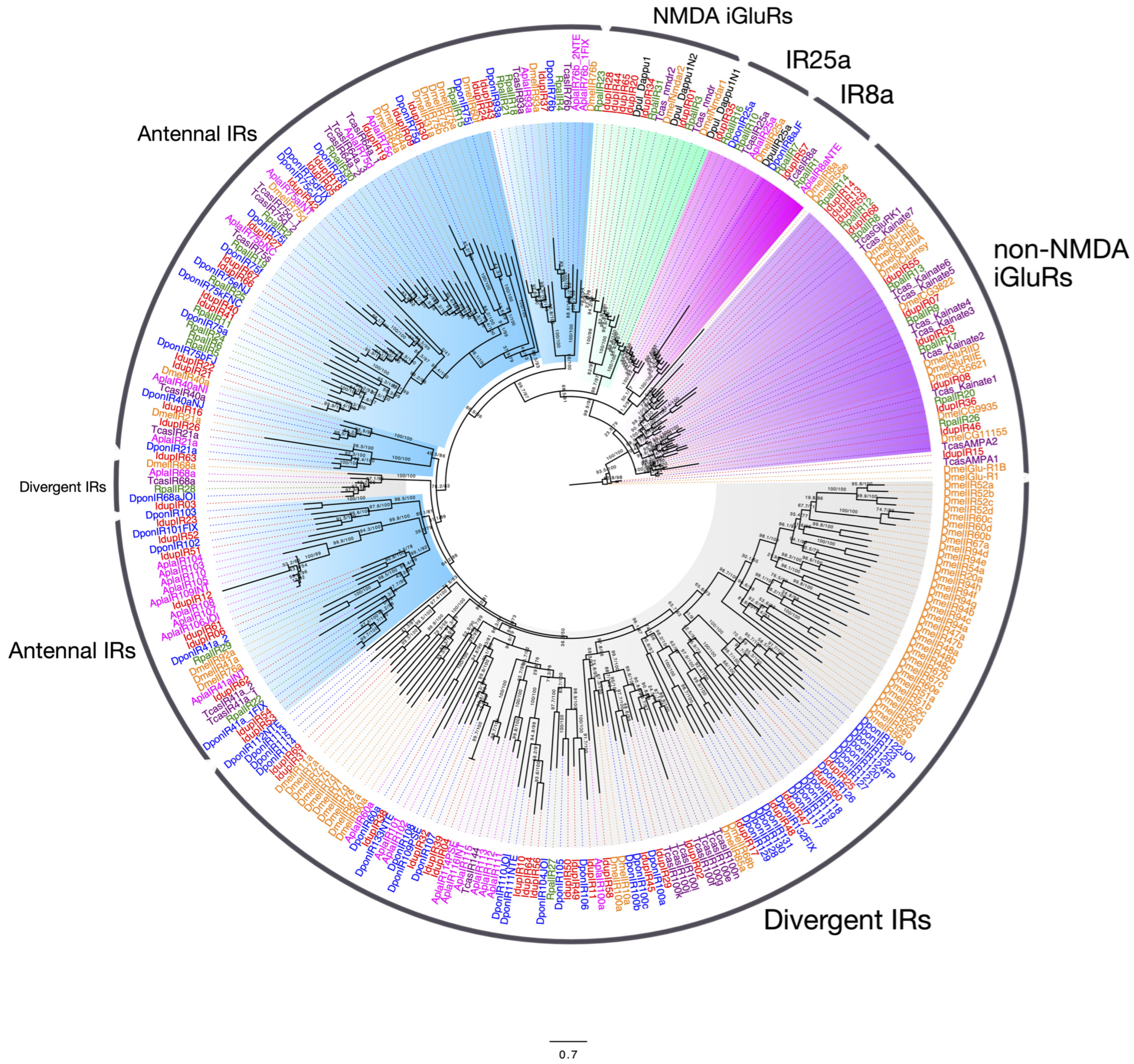
2.3. Ionotropic Receptors and iGulR Family Receptors in I. duplicatus
2.4. Gustatory Receptors in I. duplicatus
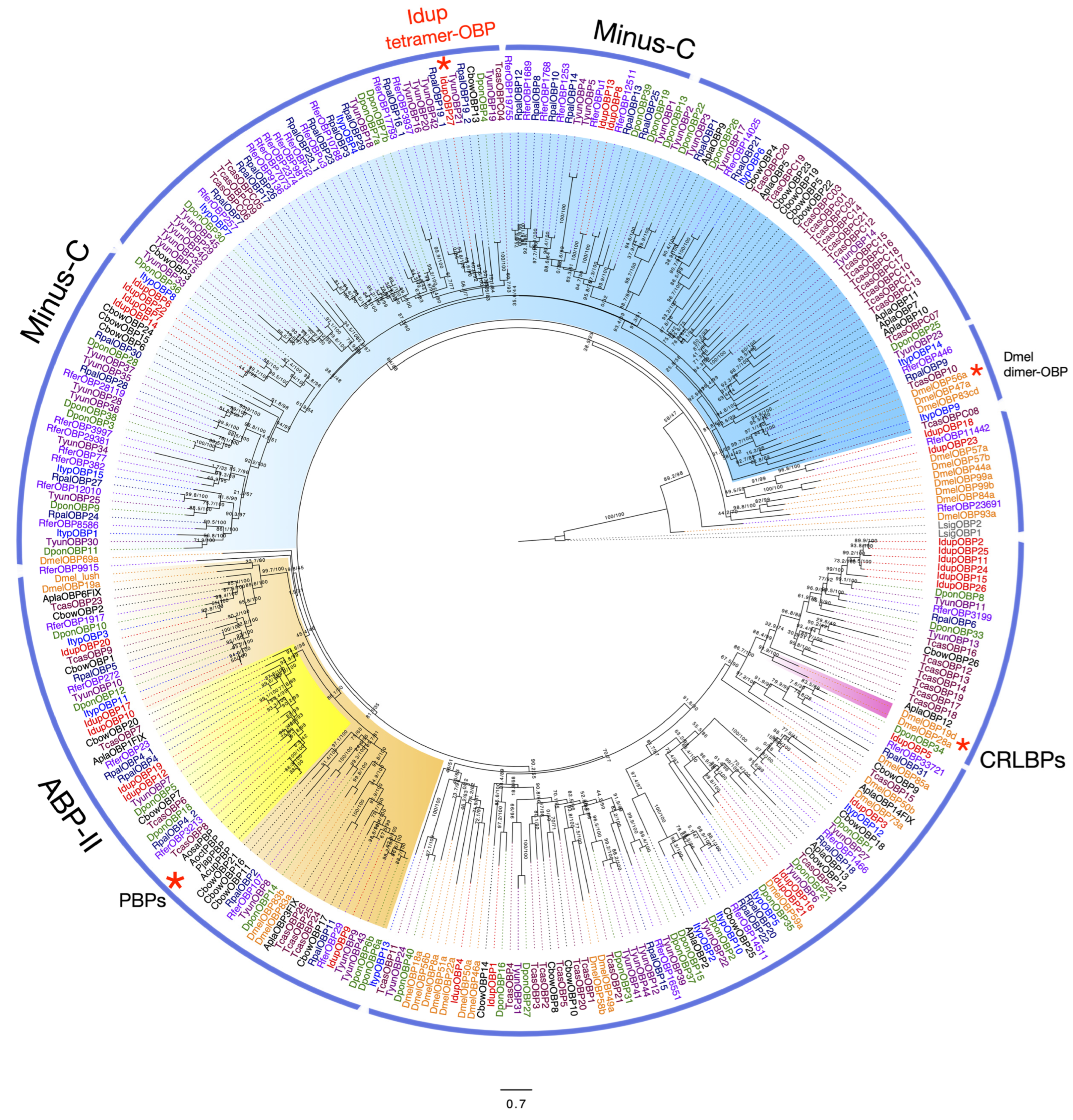
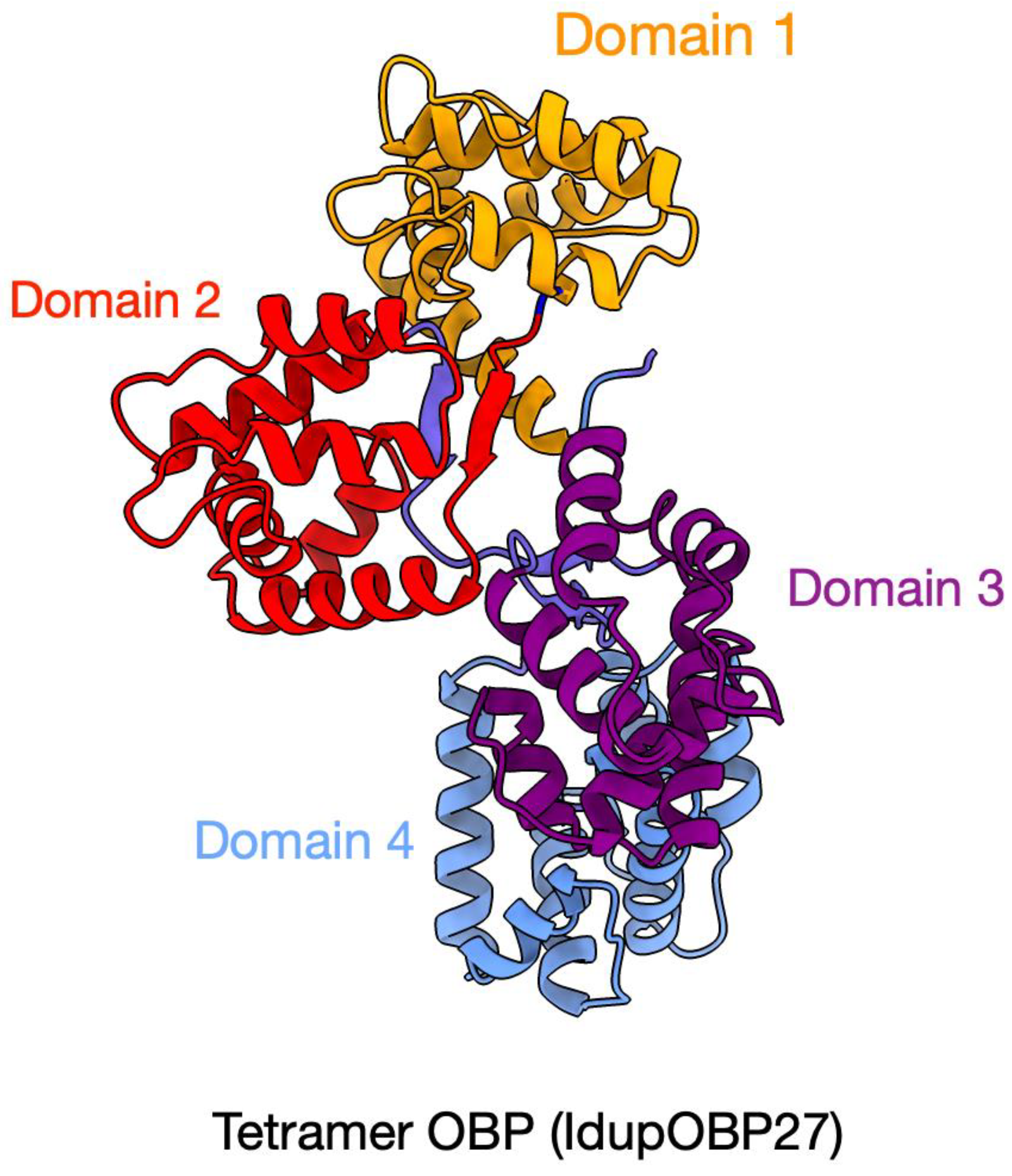
2.5. Odorant-Binding Proteins in I. duplicatus
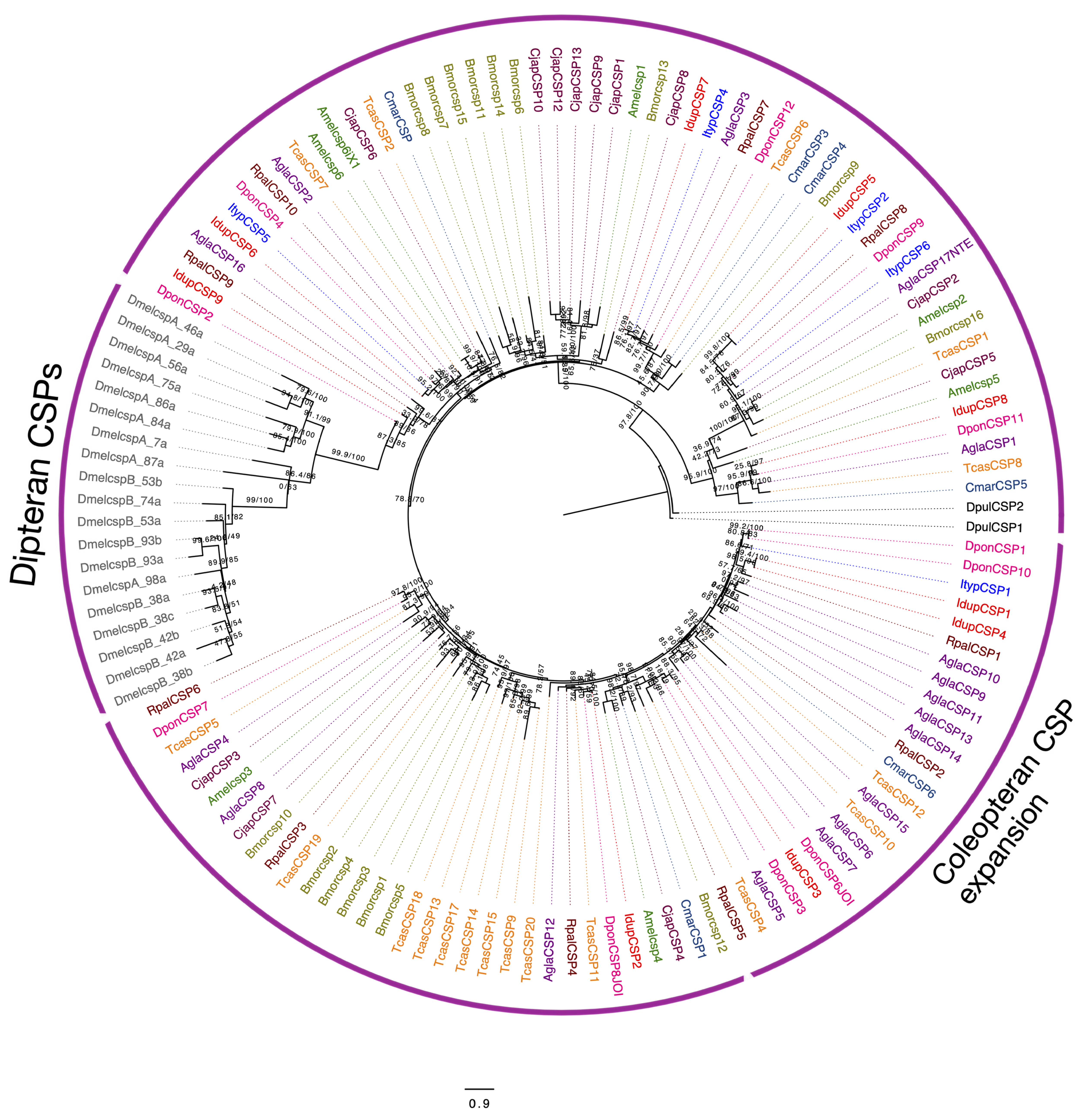
2.6. Chemosensory Proteins (CSPs) in I. duplicatus
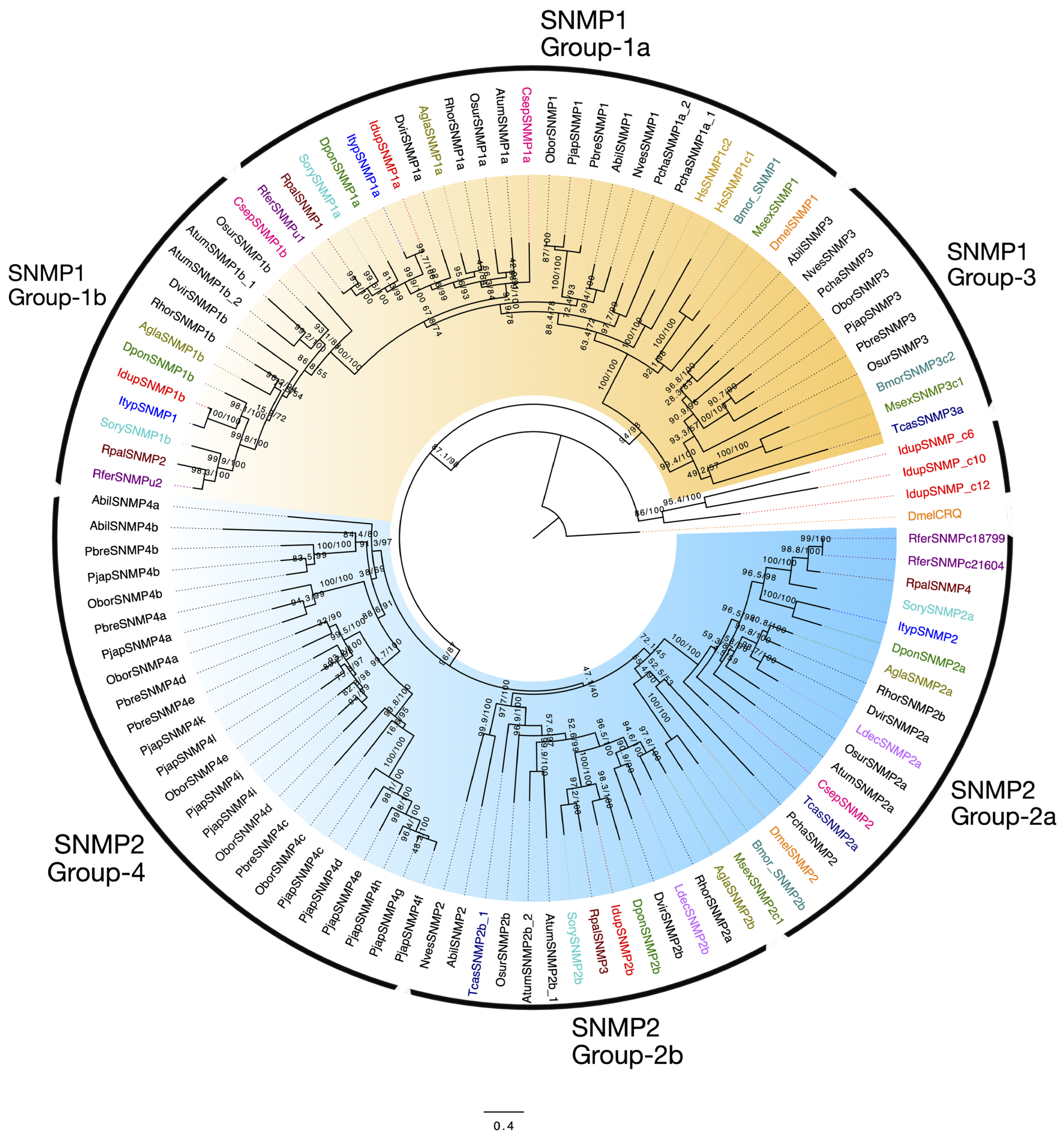
2.7. Sensory Neuron Membrane Proteins (SNMPs) in I. duplicatus
3. Discussion
4. Materials and Methods
4.1. Insect Collection and Antennal Tissue Dissection
4.2. RNA Extraction and Sequencing
4.3. Transcriptome Assembly and Gene Annotation
4.4. Phylogenetic Analysis of Candidate Chemosensory Proteins
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wermelinger, B.; Mathis, D.S.; Knížek, M.; Forster, B. Tracking the Spread of the Northern Bark Beetle (Ips duplicatus [Sahlb.]) in Europe and First Records from Switzerland and Liechtenstein. Alp. Entomol. 2020, 4, 179–184. [Google Scholar] [CrossRef]
- Jeger, M.; Bragard, C.; Caffier, D.; Candresse, T.; Chatzivassiliou, E.; Dehnen-Schmutz, K.; Gilioli, G.; Jaques Miret, J.A.; MacLeod, A.; Navajas Navarro, M.; et al. Pest Categorisation of Ips duplicatus. EFSA J. 2017, 15, e05040. [Google Scholar] [CrossRef] [PubMed]
- Davídková, M.; Kleinová, L.; Doležal, P. Overwintering Migration of the Double-Spined Spruce Bark Beetle Ips duplicatus (Sahlberg, 1836) (Coleoptera; Curculionidae). Forests 2023, 14, 131. [Google Scholar] [CrossRef]
- Holuša, J.; Lukášová, K.; Lubojacký, J. Comparison of Seasonal Flight Activity of Ips typographus and Ips duplicatus. Sci. Agric. Bohem. 2012, 2012, 109–115. [Google Scholar]
- Zimová, S.; Resnerová, K.; Vanická, H.; Horák, J.; Trombik, J.; Kacprzyk, M.; Lindelöw, Å.; Duduman, M.L.; Holuša, J. Infection Levels of the Microsporidium Larssoniella duplicati in Populations of the Invasive Bark Beetle Ips duplicatus: From Native to New Outbreak Areas. Forests 2019, 10, 131. [Google Scholar] [CrossRef]
- Holusa, J.; Lubojacky, J.; Knizek, M. Distribution of the Double-Spined Spruce Bark Beetle Ips duplicatus in the Czech Republic: Spreading in 1997–2009. Phytoparasitica 2010, 38, 435–443. [Google Scholar] [CrossRef]
- Kavčič, A.; Devetak, Z.; Piškur, B.; Groznik, E.; De Groot, M. First Record of the Northern Spruce Bark Beetle, Ips duplicatus (Sahlberg, 1836), in Slovenia. BioInvasions Rec. 2023, 12, 699–710. [Google Scholar] [CrossRef]
- Toth, D.; Maitah, M.; Maitah, K.; Jarolínová, V. The Impacts of Calamity Logging on the Development of Spruce Wood Prices in Czech Forestry. Forests 2020, 11, 283. [Google Scholar] [CrossRef]
- Duduman, M.L.; Beránková, K.; Jakuš, R.; Hradecký, J.; Jirošová, A. Efficiency and Sustainability of Ips duplicatus (Coleoptera: Curculionidae) Pheromone Dispensers with Different Designs. Forests 2022, 13, 511. [Google Scholar] [CrossRef]
- Holuša, J.; Grodzki, W.; Lukašová, K.; Lubojacký, J. Pheromone Trapping of the Double-Spined Bark Beetle Ips duplicatus (Coleoptera: Curculionidae, Scolytinae): Seasonal Variation in Abundance. Folia For. Pol. Ser. A 2013, 55, 3–9. [Google Scholar] [CrossRef]
- Byers, J.A.; Schlyter, F.; Birgersson, G.; Francke, W. E-Myrcenol In Ips duplicatus: An Aggregation Pheromone Component New for Bark Beetles. Experientia 1990, 46, 1209–1211. [Google Scholar] [CrossRef]
- Schlyter, F.; Birgersson, G.; Byers, J.A.; Bakke, A. The Aggregation Pheromone of Ips duplicatus and Its Role in Competitive Interactions with I. typographus (Coleoptera: Scolytidae). Chemoecology 1992, 3, 103–112. [Google Scholar] [CrossRef]
- Schlyter, F.; Zhang, Q.H.; Liu, G.T.; Ji, L.Z. A Successful Case of Pheromone Mass Trapping of the Bark Beetle Ips duplicatus in a Forest Island, Analysed by 20-Year Time-Series Data. Integr. Pest. Manag. Rev. 2001, 6, 185–196. [Google Scholar] [CrossRef]
- Zhang, Q.-H.; Schlyter, F.; Liu, G.-T.; Sheng, M.-L.; Birgersson, G. Electrophysiological and Behavioral Responses of Ips duplicatus to Aggregation Pheromone in Inner Mongolia, China: Amitinol as a Potential Pheromone Component. J. Chem. Ecol. 2007, 33, 1303–1315. [Google Scholar] [CrossRef] [PubMed]
- Duduman, M.L. Field Response of the Northern Spruce Bark Beetle Ips duplicatus (Sahlberg) (Coleoptera: Curculionidae, Scolytinae) to Different Combinations of Synthetic Pheromone with (−)-α-Pinene and (+)-Limonene. Agric. For. Entomol. 2014, 16, 102–109. [Google Scholar] [CrossRef]
- Šotola, V.; Holuša, J.; Kuželka, K.; Kula, E. Felled and Lure Trap Trees with Uncut Branches Are Only Weakly Attractive to the Double-Spined Bark Beetle, Ips duplicatus. Forests 2021, 12, 941. [Google Scholar] [CrossRef]
- Hansson, B.S.; Stensmyr, M.C. Evolution of Insect Olfaction. Neuron 2011, 72, 698–711. [Google Scholar] [CrossRef] [PubMed]
- de Bruyne, M.; Baker, T.C. Odor Detection in Insects: Volatile Codes. J. Chem. Ecol. 2008, 34, 882–897. [Google Scholar] [CrossRef] [PubMed]
- Clyne, P.J.; Warr, C.G.; Freeman, M.R.; Lessing, D.; Kim, J.; Carlson, J.R. A Novel Family of Divergent Seven-Transmembrane Proteins: Candidate Odorant Receptors in Drosophila. Neuron 1999, 22, 327–338. [Google Scholar] [CrossRef]
- Gao, Q.; Chess, A. Identification of Candidate Drosophila Olfactory Receptors from Genomic DNA Sequence. Genomics 1999, 60, 31–39. [Google Scholar] [CrossRef]
- Vosshall, L.B.; Stocker, R.F. Molecular Architecture of Smell and Taste in Drosophila. Annu. Rev. Neurosci. 2007, 30, 505–533. [Google Scholar] [CrossRef] [PubMed]
- Benton, R.; Vannice, K.S.; Vosshall, L.B. An Essential Role for a CD36-Related Receptor in Pheromone Detection in Drosophila. Nature 2007, 450, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Benton, R.; Vannice, K.S.; Gomez-Diaz, C.; Vosshall, L.B. Variant Ionotropic Glutamate Receptors as Chemosensory Receptors in Drosophila. Cell 2009, 136, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Scalzotto, M.; Ng, R.; Cruchet, S.; Saina, M.; Armida, J.; Su, C.Y.; Benton, R. Pheromone Sensing in Drosophila Requires Support Cell-Expressed Osiris 8. BMC Biol. 2022, 20, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Robertson, H.M. The Insect Chemoreceptor Superfamily Is Ancient in Animals. Chem. Senses 2015, 40, 609–614. [Google Scholar] [CrossRef]
- Thoma, M.; Missbach, C.; Jordan, M.D.; Grosse-Wilde, E.; Newcomb, R.D.; Hansson, B.S. Transcriptome Surveys in Silverfish Suggest a Multistep Origin of the Insect Odorant Receptor Gene Family. Front. Ecol. Evol. 2019, 7, 281. [Google Scholar] [CrossRef]
- del Mármol, J.; Yedlin, M.A.; Ruta, V. The Structural Basis of Odorant Recognition in Insect Olfactory Receptors. Nature 2021, 597, 126–131. [Google Scholar] [CrossRef]
- Zhao, J.; Chen, A.Q.; Ryu, J.; del Mármol, J. Structural Basis of Odor Sensing by Insect Heteromeric Odorant Receptors. Science 2024, 384, 1460–1467. [Google Scholar] [CrossRef]
- Wang, Y.; Qiu, L.; Wang, B.; Guan, Z.; Dong, Z.; Zhang, J.; Cao, S.; Yang, L.; Wang, B.; Gong, Z.; et al. Structural Basis for Odorant Recognition of the Insect Odorant Receptor OR-Orco Heterocomplex. Science 2024, 384, 1453–1460. [Google Scholar] [CrossRef]
- Zhou, X.; Rokas, A.; Berger, S.L.; Liebig, J.; Ray, A.; Zwiebel, L.J. Chemoreceptor Evolution in Hymenoptera and Its Implications for the Evolution of Eusociality. Genome Biol. Evol. 2015, 7, 2407–2416. [Google Scholar] [CrossRef]
- Yuvaraj, J.K.; Roberts, R.E.; Sonntag, Y.; Hou, X.Q.; Grosse-Wilde, E.; Machara, A.; Zhang, D.D.; Hansson, B.S.; Johanson, U.; Löfstedt, C.; et al. Putative Ligand Binding Sites of Two Functionally Characterized Bark Beetle Odorant Receptors. BMC Biol. 2021, 19, 16. [Google Scholar] [CrossRef]
- Croset, V.; Rytz, R.; Cummins, S.F.; Budd, A.; Brawand, D.; Kaessmann, H.; Gibson, T.J.; Benton, R. Ancient Protostome Origin of Chemosensory Ionotropic Glutamate Receptors and the Evolution of Insect Taste and Olfaction. PLoS Genet. 2010, 6, e1001064. [Google Scholar] [CrossRef] [PubMed]
- Clyne, P.J.; Warr, C.G.; Carlson, J.R. Candidate Taste Receptors in Drosophila. Science 2000, 287, 1830–1834. [Google Scholar] [CrossRef]
- Scott, K.; Brady, R.; Cravchik, A.; Morozov, P.; Rzhetsky, A.; Zuker, C.; Axel, R.; Brady, R., Jr.; Cravchik, A.; Morozov, P.; et al. A Chemosensory Gene Family Encoding Candidate Gustatory and Olfactory Receptors in Drosophila. Cell 2001, 104, 661–673. [Google Scholar] [CrossRef]
- Weiss, L.A.; Dahanukar, A.; Kwon, J.Y.; Banerjee, D.; Carlson, J.R. The Molecular and Cellular Basis of Bitter Taste in Drosophila. Neuron 2011, 69, 258–272. [Google Scholar] [CrossRef] [PubMed]
- Montell, C. A Taste of the Drosophila Gustatory Receptors. Curr. Opin. Neurobiol. 2009, 19, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Leal, W.S. Odorant Reception in Insects: Roles of Receptors, Binding Proteins, and Degrading Enzymes. Annu. Rev. Entomol. 2013, 58, 373–391. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Diaz, C.; Bargeton, B.; Abuin, L.; Bukar, N.; Reina, J.H.; Bartoi, T.; Graf, M.; Ong, H.; Ulbrich, M.H.; Masson, J.F.; et al. A CD36 Ectodomain Mediates Insect Pheromone Detection via a Putative Tunnelling Mechanism. Nat. Commun. 2016, 7, 11866. [Google Scholar] [CrossRef]
- Lautenschlager, C.; Leal, W.S.; Clardy, J. Bombyx mori Pheromone-Binding Protein Binding Non-Pheromone Ligands: Implications for Pheromone Recognition. Structure 2007, 15, 1148–1154. [Google Scholar] [CrossRef]
- Pelosi, P.; Iovinella, I.; Felicioli, A.; Dani, F.R.; Kadarkarai, M.E. Soluble Proteins of Chemical Communication: An Overview across Arthropods. Front. Physiol. 2014, 5, 320. [Google Scholar] [CrossRef]
- Venthur, H.; Mutis, A.; Zhou, J.J.; Quiroz, A. Ligand Binding and Homology Modelling of Insect Odorant-Binding Proteins. Physiol. Entomol. 2014, 39, 183–198. [Google Scholar] [CrossRef]
- Pelosi, P.; Iovinella, I.; Zhu, J.; Wang, G.; Dani, F.R. Beyond Chemoreception: Diverse Tasks of Soluble Olfactory Proteins in Insects. Biol. Rev. 2018, 93, 184–200. [Google Scholar] [CrossRef] [PubMed]
- Vogt, R.G.; Miller, N.E.; Litvack, R.; Fandino, R.A.; Sparks, J.; Staples, J.; Friedman, R.; Dickens, J.C. The Insect SNMP Gene Family. Insect Biochem. Mol. Biol. 2009, 39, 448–456. [Google Scholar] [CrossRef] [PubMed]
- Rogers, M.E.; Sun, M.; Lerner, M.R.; Vogt, R.G. Snmp-1, a Novel Membrane Protein of Olfactory Neurons of the Silk Moth Antheraea polyphemus with Homology to the CD36 Family of Membrane Proteins. J. Biol. Chem. 1997, 272, 14792–14799. [Google Scholar] [CrossRef] [PubMed]
- Cassau, S.; Krieger, J. Evidence for a Role of SNMP2 and Antennal Support Cells in Sensillum Lymph Clearance Processes of Moth Pheromone-Responsive Sensilla. Insect Biochem. Mol. Biol. 2024, 164, 104046. [Google Scholar] [CrossRef]
- Cassau, S.; Sander, D.; Karcher, T.; Laue, M.; Hause, G.; Breer, H.; Krieger, J. The Sensilla-Specific Expression and Subcellular Localization of SNMP1 and SNMP2 Reveal Novel Insights into Their Roles in the Antenna of the Desert Locust Schistocerca gregaria. Insects 2022, 13, 579. [Google Scholar] [CrossRef]
- Johny, J.; Nihad, M.; Alharbi, H.A.; AlSaleh, M.A.; Antony, B. Silencing Sensory Neuron Membrane Protein RferSNMPu1 Impairs Pheromone Detection in the Invasive Asian Palm Weevil. Sci. Rep. 2024, 14, 16541. [Google Scholar] [CrossRef]
- Gonzalez, F.; Johny, J.; Walker, W.B.; Guan, Q.; Mfarrej, S.; Jakše, J.; Montagné, N.; Jacquin-Joly, E.; Alqarni, A.A.; Al-Saleh, M.A.; et al. Antennal Transcriptome Sequencing and Identification of Candidate Chemoreceptor Proteins from an Invasive Pest, the American Palm Weevil, Rhynchophorus palmarum. Sci. Rep. 2021, 11, 8334. [Google Scholar] [CrossRef]
- Antony, B.; Johny, J.; Montagné, N.; Jacquin-Joly, E.; Capoduro, R.; Cali, K.; Persaud, K.; Al-Saleh, M.A.; Pain, A. Pheromone Receptor of the Globally Invasive Quarantine Pest of the Palm Tree, the Red Palm Weevil (Rhynchophorus ferrugineus). Mol. Ecol. 2021, 30, 2025–2039. [Google Scholar] [CrossRef]
- Powell, D.; Groβe-Wilde, E.; Krokene, P.; Roy, A.; Chakraborty, A.; Löfstedt, C.; Vogel, H.; Andersson, M.N.; Schlyter, F. A Highly-Contiguous Genome Assembly of the Eurasian Spruce Bark Beetle, Ips typographus, Provides Insight into a Major Forest Pest. Commun. Biol. 2021, 4, 1059. [Google Scholar] [CrossRef]
- Roberts, R.E.; Biswas, T.; Yuvaraj, J.K.; Grosse-Wilde, E.; Powell, D.; Hansson, B.S.; Löfstedt, C.; Andersson, M.N. Odorant Receptor Orthologues in Conifer-Feeding Beetles Display Conserved Responses to Ecologically Relevant Odours. Mol. Ecol. 2022, 31, 3693–3707. [Google Scholar] [CrossRef]
- Hou, X.-Q.; Yuvaraj, J.K.; Roberts, R.E.; Zhang, D.-D.; Unelius, C.R.; Löfstedt, C.; Andersson, M.N. Functional Evolution of a Bark Beetle Odorant Receptor Clade Detecting Monoterpenoids of Different Ecological Origins. Mol. Biol. Evol. 2021, 38, 4934–4947. [Google Scholar] [CrossRef]
- Andersson, M.N.; Grosse-Wilde, E.; Keeling, C.I.; Bengtsson, J.M.; Yuen, M.M.S.S.; Li, M.; Hillbur, Y.; Bohlmann, J.; Hansson, B.S.; Schlyter, F. Antennal Transcriptome Analysis of the Chemosensory Gene Families in the Tree Killing Bark Beetles, Ips typographus and Dendroctonus ponderosae (Coleoptera: Curculionidae: Scolytinae). BMC Genom. 2013, 14, 198. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, R.F.; Schneider, T.M.; Schwartz, A.M.; Andersson, M.N.; McKenna, D.D. The Diversity and Evolution of Odorant Receptors in Beetles (Coleoptera). Insect Mol. Biol. 2020, 29, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, R.F.; Hughes, D.T.; Luetje, C.W.; Millar, J.G.; Soriano-Agaton, F.; Hanks, L.M.; Robertson, H.M. Sequencing and Characterizing Odorant Receptors of the Cerambycid Beetle Megacyllene caryae. Insect Biochem. Mol. Biol. 2012, 42, 499–505. [Google Scholar] [CrossRef]
- Andersson, M.N.; Keeling, C.I.; Mitchell, R.F. Genomic Content of Chemosensory Genes Correlates with Host Range in Wood-Boring Beetles (Dendroctonus ponderosae, Agrilus planipennis, and Anoplophora glabripennis). BMC Genom. 2019, 20, 690. [Google Scholar] [CrossRef] [PubMed]
- Joseph, R.M.; Carlson, J.R. Drosophila Chemoreceptors: A Molecular Interface Between the Chemical World and the Brain. Trends Genet. 2015, 31, 683–695. [Google Scholar] [CrossRef]
- Delventhal, R.; Carlson, J.R. Bitter Taste Receptors Confer Diverse Functions to Neurons. Elife 2016, 5, e11181. [Google Scholar] [CrossRef]
- Chahda, J.S.; Soni, N.; Sun, J.S.; Ebrahim, S.A.M.; Weiss, B.L.; Carlson, J.R. The Molecular and Cellular Basis of Olfactory Response to Tsetse Fly Attractants. PLoS Genet. 2019, 15, e1008005. [Google Scholar] [CrossRef]
- Dahanukar, A.; Foster, K.; van der Goes van Naters, W.M.; Carlson, J.R. A Gr Receptor Is Required for Response to the Sugar Trehalose in Taste Neurons of Drosophila. Nat. Neurosci. 2001, 4, 1182–1186. [Google Scholar] [CrossRef] [PubMed]
- Bray, S.; Amrein, H. A Putative Drosophila Pheromone Receptor Expressed in Male-Specific Taste Neurons Is Required for Efficient Courtship. Neuron 2003, 39, 1019–1029. [Google Scholar] [CrossRef]
- Andrews, J.C.; Fernández, M.P.; Yu, Q.; Leary, G.P.; Leung, A.K.W.; Kavanaugh, M.P.; Kravitz, E.A.; Certel, S.J. Octopamine Neuromodulation Regulates Gr32a-Linked Aggression and Courtship Pathways in Drosophila Males. PLoS Genet. 2014, 10, e1004356. [Google Scholar] [CrossRef] [PubMed]
- Hekmat-Scafe, D.S.; Scafe, C.R.; McKinney, A.J.; Tanouye, M.A. Genome-Wide Analysis of the Odorant-Binding Protein Gene Family in Drosophila melanogaster. Genome Res. 2002, 12, 1357–1369. [Google Scholar] [CrossRef]
- Sánchez-Gracia, A.; Rozas, J. Divergent Evolution and Molecular Adaptation in the Drosophila Odorant-Binding Protein Family: Inferences from Sequence Variation at the OS-E and OS-F Genes. BMC Evol. Biol. 2008, 8, 323. [Google Scholar] [CrossRef] [PubMed]
- Venthur, H.; Zhou, J.-J. Odorant Receptors and Odorant-Binding Proteins as Insect Pest Control Targets: A Comparative Analysis. Front. Physiol. 2018, 9, 1163. [Google Scholar] [CrossRef]
- Große-Wilde, E.; Svatoš, A.; Krieger, J. A Pheromone-Binding Protein Mediates the Bombykol-Induced Activation of a Pheromone Receptor in vitro. Chem. Senses 2006, 31, 547–555. [Google Scholar] [CrossRef]
- Antony, B.; Johny, J.; Aldosari, S.A. Silencing the Odorant Binding Protein RferOBP1768 Reduces the Strong Preference of Palm Weevil for the Major Aggregation Pheromone Compound Ferrugineol. Front. Physiol. 2018, 9, 252. [Google Scholar] [CrossRef]
- Wojtasek, H.; Hansson, B.S.; Leal, W.S. Attracted or Repelled?—A Matter of Two Neurons, One Pheromone Binding Protein, and a Chiral Center. Biochem. Biophys. Res. Commun. 1998, 250, 217–222. [Google Scholar] [CrossRef]
- Pelosi, P.; Zhou, J.; Ban, L.P.; Calvello, M. Soluble Proteins in Insect Chemical Communication. Cell. Mol. Life Sci. 2006, 63, 1658–1676. [Google Scholar] [CrossRef] [PubMed]
- Schiebe, C.; Blaženec, M.; Jakuš, R.; Unelius, C.R.; Schlyter, F. Semiochemical Diversity Diverts Bark Beetle Attacks from Norway Spruce Edges. J. Appl. Entomol. 2011, 135, 726–737. [Google Scholar] [CrossRef]
- Antony, B.; Soffan, A.; Jakše, J.; Abdelazim, M.M.; Aldosari, S.A.; Aldawood, A.S.; Pain, A. Identification of the Genes Involved in Odorant Reception and Detection in the Palm Weevil Rhynchophorus ferrugineus, an Important Quarantine Pest, by Antennal Transcriptome Analysis. BMC Genom. 2016, 17, 69. [Google Scholar] [CrossRef] [PubMed]
- Johny, J.; Diallo, S.; Lukšan, O.; Shewale, M.; Kalinová, B.; Hanus, R.; Große-Wilde, E. Conserved Orthology in Termite Chemosensory Gene Families. Front. Ecol. Evol. 2023, 10, 1065947. [Google Scholar] [CrossRef]
- Yuvaraj, J.K.; Roberts, R.E.; Hansson, B.S.; Andersson, M.N. Eurasian Spruce Bark Beetle Detects Anti-Attractant Lanierone Using a Highly Expressed Specialist Odorant Receptor, Present in Several Functional Sensillum Types. Res. Sq. 2024, in press. [Google Scholar] [CrossRef]
- Wang, X.; Wang, S.; Yi, J.; Li, Y.; Liu, J.; Wang, J.; Xi, J. Three Host Plant Volatiles, Hexanal, Lauric Acid, and Tetradecane, Are Detected by an Antenna-Biased Expressed Odorant Receptor 27 in the Dark Black Chafer Holotrichia parallela. J. Agric. Food Chem. 2020, 68, 7316–7323. [Google Scholar] [CrossRef]
- Antony, B.; Montagné, N.; Comte, A.; Mfarrej, S.; Jakše, J.; Capoduro, R.; Shelke, R.; Cali, K.; AlSaleh, M.A.; Persaud, K.; et al. Deorphanizing an Odorant Receptor Tuned to Palm Tree Volatile Esters in the Asian Palm Weevil Sheds Light on the Mechanisms of Palm Tree Selection. Insect Biochem. Mol. Biol. 2024, 169, 104129. [Google Scholar] [CrossRef]
- Ji, T.; Xu, Z.; Jia, Q.; Wang, G.; Hou, Y. Non-Palm Plant Volatile α-Pinene Is Detected by Antenna-Biased Expressed Odorant Receptor 6 in the Rhynchophorus ferrugineus (Olivier) (Coleoptera: Curculionidae). Front. Physiol. 2021, 12, 701545. [Google Scholar] [CrossRef]
- Brajon, L.; Comte, A.; Capoduro, R.; Meslin, C.; Antony, B.; Al-Saleh, M.A.; Pain, A.; Jacquin-Joly, E.; Montagné, N. A Conserved Pheromone Receptor in the American and the Asian Palm Weevils Is Also Activated by Host Plant Volatiles. Curr. Res. Insect Sci. 2024, 6, 100090. [Google Scholar] [CrossRef]
- Anderson, A.R.; Newcomb, R.D. Olfactory Genomics and Biotechnology in Insect Control. In Insect Pheromone Biochemistry and Molecular Biology, 2nd ed.; Blomquist, G.J., Vogt, R.G., Eds.; Elsevier: London, UK, 2021; pp. 645–674. ISBN 978-0-12-819628-1. [Google Scholar]
- Bohbot, J.D.; Vernick, S. The Emergence of Insect Odorant Receptor-Based Biosensors. Biosensors 2020, 10, 26. [Google Scholar] [CrossRef]
- Hoddle, M.; Antony, B.; El-Shafie, H.; Chamorro, L.; Milosavljević, I.; Bernhard, L.; Faleiro, R. Taxonomy, Biology, Symbionts, Omics, and Management of Rhynchophorus Palm Weevils (Coleoptera: Curculionidae: Dryophthorinae). Annu. Rev. Entomol. 2024, 69, 449–479. [Google Scholar] [CrossRef] [PubMed]
- Arntsen, C.; Guillemin, J.; Audette, K.; Stanley, M. Tastant-Receptor Interactions: Insights from the Fruit Fly. Front. Nutr. 2024, 11, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.-Y.; Li, Z.-B.; Zhao, N.; Song, Q.-S.; Zhu, J.-Y.; Yang, B. Identification and Characterization of Chemosensory Gene Families in the Bark Beetle, Tomicus yunnanensis. Comp. Biochem. Physiol. Part D Genom. Proteom. 2018, 25, 73–85. [Google Scholar] [CrossRef]
- Zhou, J.J.; Huang, W.; Zhang, G.A.; Pickett, J.A.; Field, L.M. “Plus-C” Odorant-Binding Protein Genes in Two Drosophila Species and the Malaria Mosquito Anopheles gambiae. Gene 2004, 327, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Rihani, K.; Ferveur, J.F.; Briand, L. The 40-Year Mystery of Insect Odorant-Binding Proteins. Biomolecules 2021, 11, 509. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, P.; Zhu, J.; Knoll, W. Odorant-Binding Proteins as Sensing Elements for Odour Monitoring. Sensors 2018, 18, 3248. [Google Scholar] [CrossRef]
- Tegoni, M.; Campanacci, V.; Cambillau, C. Structural Aspects of Sexual Attraction and Chemical Communication in Insects. Trends Biochem. Sci. 2004, 29, 257–264. [Google Scholar] [CrossRef]
- Vogt, R.G. Biochemical Diversity of Odor Detection. In Insect Pheromone Biochemistry and Molecular Biology; Blomquist, G., Vogt, R., Eds.; Elsevier: San Diego, CA, USA, 2003; pp. 391–445. ISBN 978-0-12-107151-6. [Google Scholar]
- German, P.F.; van der Poel, S.; Carraher, C.; Kralicek, A.V.; Newcomb, R.D. Insights into Subunit Interactions within the Insect Olfactory Receptor Complex Using FRET. Insect Biochem. Mol. Biol. 2013, 43, 138–145. [Google Scholar] [CrossRef]
- Nichols, Z.; Vogt, R.G. The SNMP/CD36 Gene Family in Diptera, Hymenoptera and Coleoptera: Drosophila melanogaster, D. pseudoobscura, Anopheles gambiae, Aedes aegypti, Apis mellifera, and Tribolium castaneum. Insect Biochem. Mol. Biol. 2008, 38, 398–415. [Google Scholar] [CrossRef]
- Rogers, M.E.; Steinbrecht, R.A.; Vogt, R.G. Expression of SNMP-1 in Olfactory Neurons and Sensilla of Male and Female Antennae of the Silkmoth Antheraea polyphemus. Cell Tissue Res. 2001, 303, 433–446. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarise Analysis Results for Multiple Tools and Samples in a Single Report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for Clustering the next-Generation Sequencing Data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Dippel, S.; Kollmann, M.; Oberhofer, G.; Montino, A.; Knoll, C.; Krala, M.; Rexer, K.; Frank, S.; Kumpf, R.; Schachtner, J.; et al. Morphological and Transcriptomic Analysis of a Beetle Chemosensory System Reveals a Gnathal Olfactory Center. BMC Biol. 2016, 14, 90. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sun, L.; Cao, D.; Walker, W.B.; Zhang, Y.; Wang, G. Identification of Candidate Olfactory Genes in Leptinotarsa decemlineata by Antennal Transcriptome Analysis. Front. Ecol. Evol. 2015, 3, 60. [Google Scholar] [CrossRef]
- Frickey, T.; Lupas, A. CLANS: A Java Application for Visualizing Protein Families Based on Pairwise Similarity. Bioinformatics 2004, 20, 3702–3704. [Google Scholar] [CrossRef] [PubMed]
- Hallgren, J.; Tsirigos, K.D.; Damgaard Pedersen, M.; Juan, J.; Armenteros, A.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM Predicts Alpha and Beta Transmembrane Proteins Using Deep Neural Networks. bioRxiv 2022. bioRxiv:2022.04.08.487609. [Google Scholar]
- Tsirigos, K.D.; Peters, C.; Shu, N.; Käll, L.; Elofsson, A. The TOPCONS Web Server for Consensus Prediction of Membrane Protein Topology and Signal Peptides. Nucleic Acids Res. 2015, 43, W401–W407. [Google Scholar] [CrossRef]
- Vogt, R.G.; Große-Wilde, E.; Zhou, J.J. The Lepidoptera Odorant Binding Protein Gene Family: Gene Gain and Loss within the GOBP/PBP Complex of Moths and Butterflies. Insect Biochem. Mol. Biol. 2015, 62, 142–153. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Ren, D.; Zhao, L.; Jiang, F.; Song, J.; Wang, X.; Kang, L. Identification of Odorant-Binding Proteins (OBPs) and Functional Analysis of Phase-Related OBPs in the Migratory Locust. Front. Physiol. 2018, 9, 984. [Google Scholar] [CrossRef]
- Felsenstein, J. Evolutionary Trees from DNA Sequences: A Maximum Likelihood Approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Saina, M.; Busengdal, H.; Sinigaglia, C.; Petrone, L.; Oliveri, P.; Rentzsch, F.; Benton, R. A Cnidarian Homologue of an Insect Gustatory Receptor Functions in Developmental Body Patterning. Nat. Commun. 2015, 6, 6243. [Google Scholar] [CrossRef] [PubMed]
- Maïbèche-Coisne, M.; Nikonov, A.A.; Ishida, Y.; Jacquin-Joly, E.; Leal, W.S. Pheromone Anosmia in a Scarab Beetle Induced by in vivo Inhibition of a Pheromone-Degrading Enzyme. Proc. Natl. Acad. Sci. USA 2004, 101, 11459–11464. [Google Scholar] [CrossRef]
- Missbach, C.; Vogel, H.; Hansson, B.S.; Große-Wilde, E. Identification of Odorant Binding Proteins and Chemosensory Proteins in Antennal Transcriptomes of the Jumping Bristletail Lepismachilis y-signata and the Firebrat Thermobia domestica: Evidence for an Independent OBP-OR Origin. Chem. Senses 2015, 40, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.-J.; Robertson, G.; He, X.; Dufour, S.; Hooper, A.M.; Pickett, J.A.; Keep, N.H.; Field, L.M. Characterisation of Bombyx mori Odorant-Binding Proteins Reveals That a General Odorant-Binding Protein Discriminates Between Sex Pheromone Components. J. Mol. Biol. 2009, 389, 529–545. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.J.; Li, G.C.; Zhu, J.Y.; Liu, N.Y. Genome-Based Analysis Reveals a Novel SNMP Group of the Coleoptera and Chemosensory Receptors in Rhaphuma horsfieldi. Genomics 2020, 112, 2713–2728. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2017, 20, 1160–1166. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. TrimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast Selection of Best-Fit Models of Protein Evolution. Bioinformatics 2017, 27, 1164–1165. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R.; Teeling, E. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Shimodaira, H.; Hasegawa, M. Multiple Comparisons of Log-Likelihoods with Applications to Phylogenetic Inference. Mol. Biol. Evol. 1999, 16, 1114–1116. [Google Scholar] [CrossRef]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 Predicts All Five Types of Signal Peptides Using Protein Language Models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The Conserved Domain Database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for Structure Building and Analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef]

| IDUP_AM1 | IDUP_AM2 | IDUP_AF1 | IDUP_AF2 | All Combined | |
|---|---|---|---|---|---|
| Total raw reads | 40,749,080 | 47,677,670 | 39,480,112 | 40,689,528 | |
| Total transcripts | 105,416 | 121,285 | 91,822 | 98,264 | 204,588 |
| Total genes | 61,659 | 70,337 | 50,264 | 54,391 | 125,878 |
| GC content | 39.89 | 39.82 | 38.96 | 39.26 | 39.21 |
| N50 length | 2101 | 2027 | 2455 | 2036 | 2317 |
| Average length | 1049.33 | 1011.11 | 1244.16 | 1048.26 | 1027.03 |
| Complete BUSCOs (insecta_odb10) | 93.71% | 94.37% | 95.17% | 94% | 99.71% |
| BUSCOs fragmented % | 4.24 | 3.58 | 3.22 | 2.37 | 0.15 |
| % mapped to genome * | 88.56% | 88.30% | 87.97% | 88.63% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Johny, J.; Große-Wilde, E.; Kalinová, B.; Roy, A. Antennal Transcriptome Screening and Identification of Chemosensory Proteins in the Double-Spine European Spruce Bark Beetle, Ips duplicatus (Coleoptera: Scolytinae). Int. J. Mol. Sci. 2024, 25, 9513. https://doi.org/10.3390/ijms25179513
Johny J, Große-Wilde E, Kalinová B, Roy A. Antennal Transcriptome Screening and Identification of Chemosensory Proteins in the Double-Spine European Spruce Bark Beetle, Ips duplicatus (Coleoptera: Scolytinae). International Journal of Molecular Sciences. 2024; 25(17):9513. https://doi.org/10.3390/ijms25179513
Chicago/Turabian StyleJohny, Jibin, Ewald Große-Wilde, Blanka Kalinová, and Amit Roy. 2024. "Antennal Transcriptome Screening and Identification of Chemosensory Proteins in the Double-Spine European Spruce Bark Beetle, Ips duplicatus (Coleoptera: Scolytinae)" International Journal of Molecular Sciences 25, no. 17: 9513. https://doi.org/10.3390/ijms25179513
APA StyleJohny, J., Große-Wilde, E., Kalinová, B., & Roy, A. (2024). Antennal Transcriptome Screening and Identification of Chemosensory Proteins in the Double-Spine European Spruce Bark Beetle, Ips duplicatus (Coleoptera: Scolytinae). International Journal of Molecular Sciences, 25(17), 9513. https://doi.org/10.3390/ijms25179513






