R2R3 MYB Transcription Factor GhMYB201 Promotes Cotton Fiber Elongation via Cell Wall Loosening and Very-Long-Chain Fatty Acid Synthesis
Abstract
1. Introduction
2. Results
2.1. GhMYB201 Is a Transcriptional Activator Preferentially Expressed in Elongating Fibers
2.2. GhMYB201 Knockout Negatively Affected Fiber Elongation
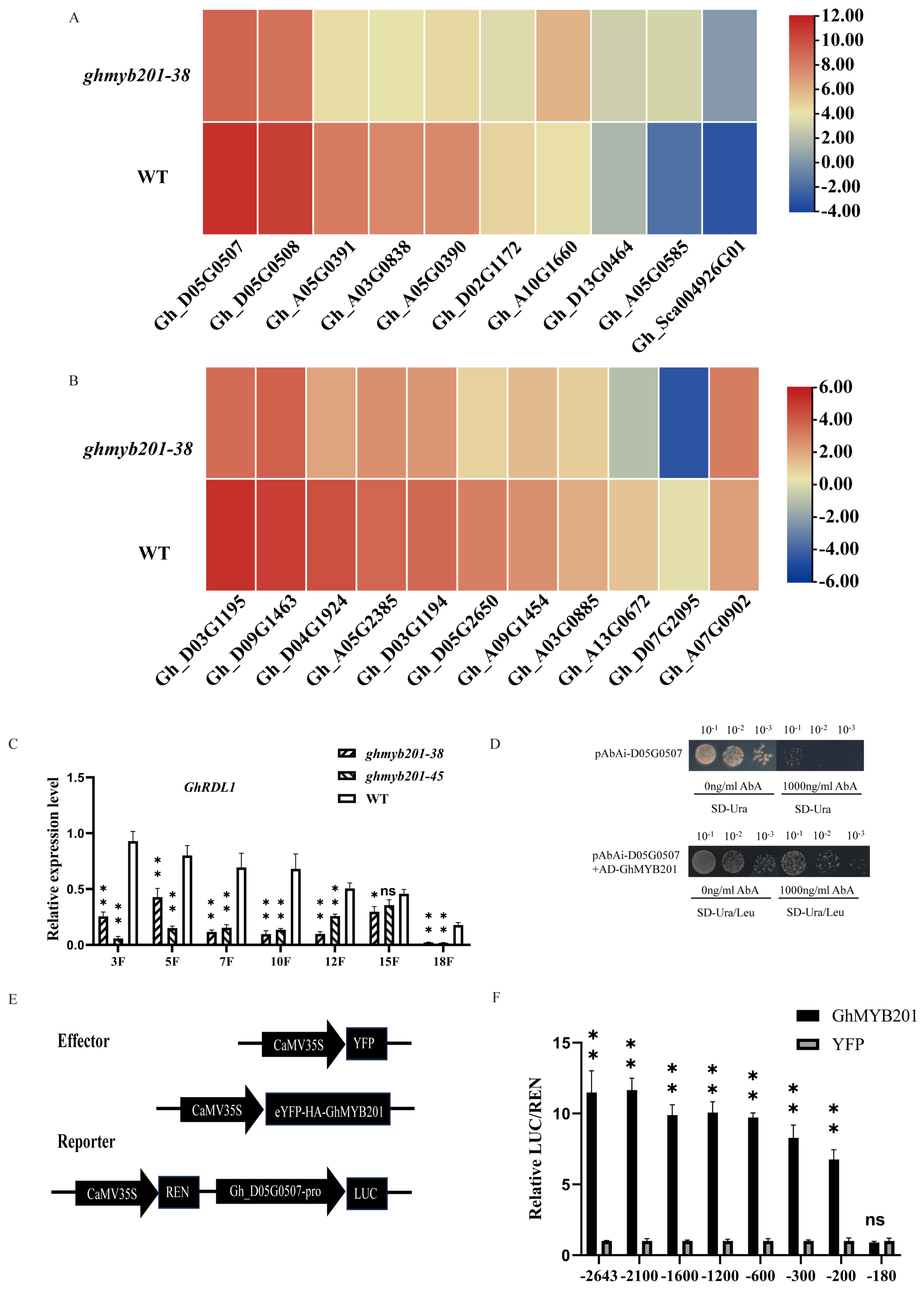
2.3. GhMYB201 Transcriptionally Activates Cell Wall Loosening-Related Genes
2.4. GhMYB201 Activates the Expression of GhKCSs and Changes the VLCFA Contents
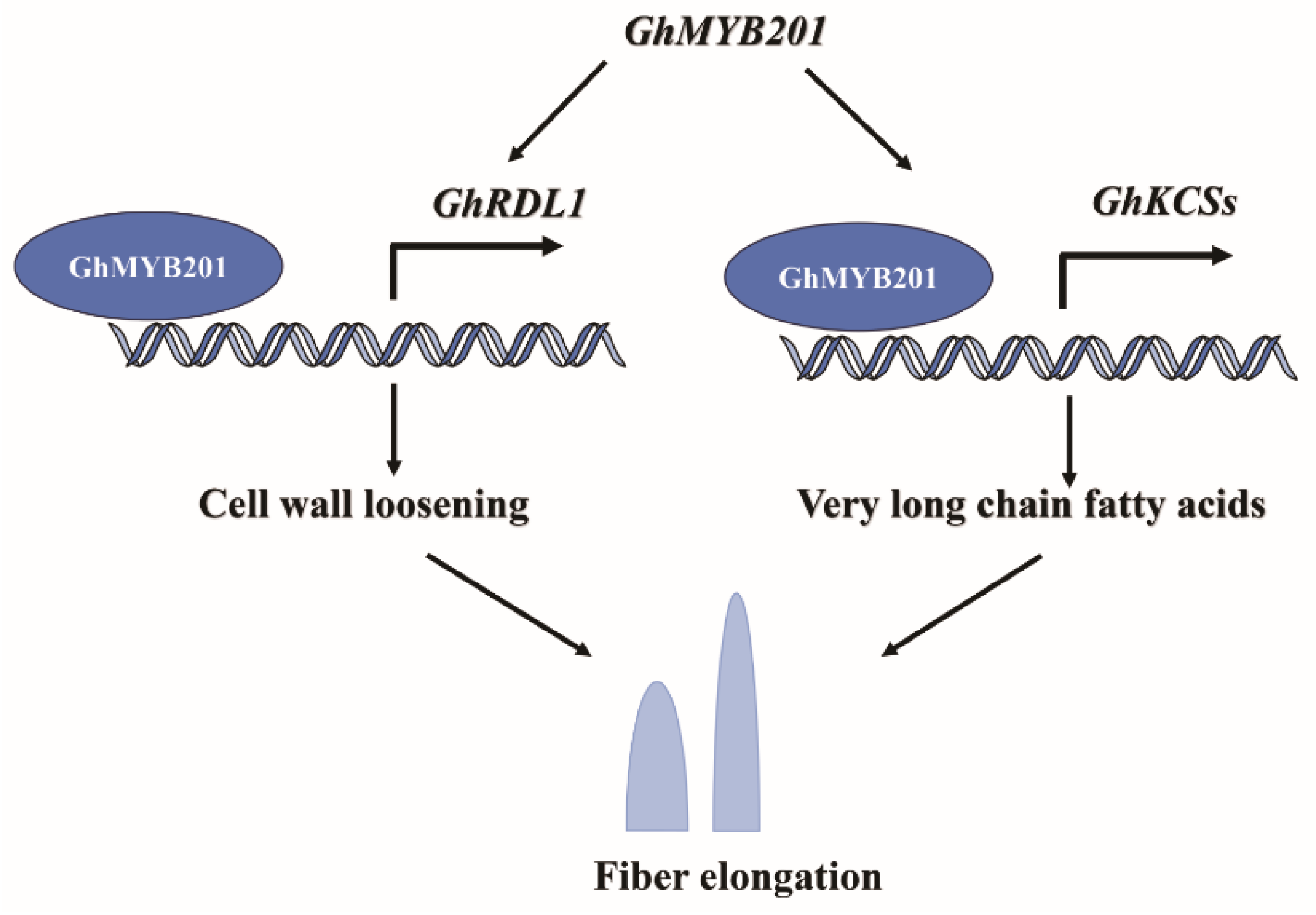
3. Discussion
4. Materials and Methods
4.1. Identification and Cloning of GhMYB201s
4.2. Total RNA Isolation, qRT-PCR Analysis, and Transcriptome Analysis
4.3. Generation of Knockout Cottons
4.4. Observation of Cotton Fiber Phenotype
4.5. Arabidopsis Growth and Transformation
4.6. Transactivation Activity Assay in Yeast and Yeast One-Hybrid Assay
4.7. Transient Assays in Nicotiana Benthamiana
4.8. Fatty Acid Extractions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Haigler, C.H.; Betancur, L.; Stiff, M.R.; Tuttle, J.R. Cotton fiber: A powerful single-cell model for cell wall and cellulose research. Front. Plant Sci. 2012, 3, 104. [Google Scholar] [CrossRef]
- Lee, J.J.; Woodward, A.W.; Chen, Z.J. Gene expression changes and early events in cotton fibre development. Ann. Bot. 2007, 100, 1391–1401. [Google Scholar] [CrossRef]
- Qin, Y.M.; Zhu, Y.X. How cotton fibers elongate: A tale of linear cell-growth mode. Curr. Opin. Plant Biol. 2011, 14, 106–111. [Google Scholar] [CrossRef]
- Wang, N.N.; Li, Y.; Chen, Y.H.; Lu, R.; Zhou, L.; Wang, Y.; Zheng, Y.; Li, X.B. Phosphorylation of WRKY16 by MPK3-1 is essential for its transcriptional activity during fiber initiation and elongation in cotton (Gossypium hirsutum). Plant Cell 2021, 33, 2736–2752. [Google Scholar] [CrossRef]
- Cao, J.F.; Zhao, B.; Huang, C.C.; Chen, Z.W.; Zhao, T.; Liu, H.R.; Hu, G.J.; Shangguan, X.X.; Shan, C.M.; Wang, L.J.; et al. The miR319-Targeted GhTCP4 Promotes the Transition from Cell Elongation to Wall Thickening in Cotton Fiber. Mol. Plant 2020, 13, 1063–1077. [Google Scholar] [CrossRef]
- Ding, X.; Li, X.; Wang, L.; Zeng, J.; Huang, L.; Xiong, L.; Song, S.; Zhao, J.; Hou, L.; Wang, F.; et al. Sucrose enhanced reactive oxygen species generation promotes cotton fibre initiation and secondary cell wall deposition. Plant Biotechnol. J. 2021, 19, 1092–1094. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Chen, Z.; Yang, Z.; Wang, M.; Jin, S.; Wang, G.; Zhang, L.; Wang, L.; Li, J.; Saeed, S.; et al. A comprehensive overview of cotton genomics, biotechnology and molecular biological studies. Sci. China Life Sci. 2023, 66, 2214–2256. [Google Scholar] [CrossRef]
- Wang, N.N.; Ni, P.; Wei, Y.L.; Hu, R.; Li, Y.; Li, X.B.; Zheng, Y. Phosphatidic acid interacts with an HD-ZIP transcription factor GhHOX4 to influence its function in fiber elongation of cotton (Gossypium hirsutum). Plant J. 2024, 118, 423–436. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Zhu, L.; Zhou, X.; Fu, X.; Zhang, Y.; Zhao, P.; Jiang, B.; Wang, H.; Xiao, G. Gibberellic acid promotes single-celled fiber elongation through the activation of two signaling cascades in cotton. Dev. Cell 2024, 59, 723–739 e4. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; He, S.P.; Xu, S.W.; Li, L.; Zheng, Y.; Li, X.B. The transcription factor ERF108 interacts with AUXIN RESPONSE FACTORs to mediate cotton fiber secondary cell wall biosynthesis. Plant Cell 2023, 35, 4133–4154. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Gou, J.Y.; Li, F.G.; Shangguan, X.X.; Zhao, B.; Yang, C.Q.; Wang, L.J.; Yuan, S.; Liu, C.J.; Chen, X.Y. A cotton BURP domain protein interacts with alpha-expansin and their co-expression promotes plant growth and fruit production. Mol. Plant 2013, 6, 945–958. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.M.; Hu, C.Y.; Pang, Y.; Kastaniotis, A.J.; Hiltunen, J.K.; Zhu, Y.X. Saturated very-long-chain fatty acids promote cotton fiber and Arabidopsis cell elongation by activating ethylene biosynthesis. Plant Cell 2007, 19, 3692–3704. [Google Scholar] [CrossRef]
- Tian, Z.; Zhang, Y.; Zhu, L.; Jiang, B.; Wang, H.; Gao, R.; Friml, J.; Xiao, G. Strigolactones act downstream of gibberellins to regulate fiber cell elongation and cell wall thickness in cotton (Gossypium hirsutum). Plant Cell 2022, 34, 4816–4839. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Liu, Z.; Ge, X.; Lu, L.; Qin, W.; Qanmber, G.; Liu, L.; Wang, Z.; Li, F. Brassinosteroids regulate cotton fiber elongation by modulating very-long-chain fatty acid biosynthesis. Plant Cell 2023, 35, 2114–2131. [Google Scholar] [CrossRef] [PubMed]
- Shan, C.M.; Shangguan, X.X.; Zhao, B.; Zhang, X.F.; Chao, L.M.; Yang, C.Q.; Wang, L.J.; Zhu, H.Y.; Zeng, Y.D.; Guo, W.Z.; et al. Control of cotton fibre elongation by a homeodomain transcription factor GhHOX3. Nat. Commun. 2014, 5, 5519. [Google Scholar] [CrossRef]
- Zhao, B.; Cao, J.F.; Hu, G.J.; Chen, Z.W.; Wang, L.Y.; Shangguan, X.X.; Wang, L.J.; Mao, Y.B.; Zhang, T.Z.; Wendel, J.F.; et al. Core cis-element variation confers subgenome-biased expression of a transcription factor that functions in cotton fiber elongation. New Phytol. 2018, 218, 1061–1075. [Google Scholar] [CrossRef]
- Zeng, J.; Yao, D.; Luo, M.; Ding, L.; Wang, Y.; Yan, X.; Ye, S.E.; Wang, C.; Wu, Y.; Zhang, J.; et al. Fiber-specific increase of carotenoid content promotes cotton fiber elongation by increasing abscisic acid and ethylene biosynthesis. Crop J. 2023, 11, 774–784. [Google Scholar] [CrossRef]
- Machado, A.; Wu, Y.; Yang, Y.; Llewellyn, D.J.; Dennis, E.S. The MYB transcription factor GhMYB25 regulates early fibre and trichome development. Plant J. 2009, 59, 52–62. [Google Scholar] [CrossRef]
- Pu, L.; Li, Q.; Fan, X.; Yang, W.; Xue, Y. The R2R3 MYB transcription factor GhMYB109 is required for cotton fiber development. Genetics 2008, 180, 811–820. [Google Scholar] [CrossRef]
- Sun, W.; Gao, Z.; Wang, J.; Huang, Y.; Chen, Y.; Li, J.; Lv, M.; Wang, J.; Luo, M.; Zuo, K. Cotton fiber elongation requires the transcription factor GhMYB212 to regulate sucrose transportation into expanding fibers. New Phytol. 2019, 222, 864–881. [Google Scholar] [CrossRef]
- Xu, F.; Li, G.; He, S.; Zeng, Z.; Wang, Q.; Zhang, H.; Yan, X.; Hu, Y.; Tian, H.; Luo, M. Sphingolipid inhibitor response gene GhMYB86 controls fiber elongation by regulating microtubule arrangement. J. Integr. Plant Biol. 2024. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.C.; Liu, H.L.; Xu, Y.Y.; Zhao, J.R.; Guo, Y.W.; Long, L.; Gao, W.; Song, C.P. Heterogeneous expression of the cotton R2R3-MYB transcription factor GbMYB60 increases salt sensitivity in transgenic. Plant Cell Tissue Org. 2018, 133, 15–25. [Google Scholar] [CrossRef]
- Walford, S.A.; Wu, Y.; Llewellyn, D.J.; Dennis, E.S. GhMYB25-like: A key factor in early cotton fibre development. Plant J. 2011, 65, 785–797. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.; Lian, B.; Hao, P.; Fu, X.; Zhang, M.; Lu, J.; Ma, L.; Yu, S.; Wei, H.; Wang, H. GhMYB30-GhMUR3 affects fiber elongation and secondary wall thickening in cotton. Plant J. 2024, 117, 694–712. [Google Scholar] [CrossRef]
- Cominelli, E.; Galbiati, M.; Vavasseur, A.; Conti, L.; Sala, T.; Vuylsteke, M.; Leonhardt, N.; Dellaporta, S.L.; Tonelli, C. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr. Biol. 2005, 15, 1196–1200. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.E.; Kwon, Y.; Kim, J.H.; Noh, H.; Hong, S.W.; Lee, H. A dual role for MYB60 in stomatal regulation and root growth of Arabidopsis thaliana under drought stress. Plant Mol. Biol. 2011, 77, 91–103. [Google Scholar] [CrossRef]
- Wang, C.; Lv, Y.; Xu, W.; Zhang, T.; Guo, W. Aberrant phenotype and transcriptome expression during fiber cell wall thickening caused by the mutation of the Im gene in immature fiber (im) mutant in Gossypium hirsutum L. BMC Genom. 2014, 15, 94. [Google Scholar] [CrossRef]
- Park, J.S.; Kim, J.B.; Cho, K.J.; Cheon, C.I.; Sung, M.K.; Choung, M.G.; Roh, K.H. Arabidopsis R2R3-MYB transcription factor AtMYB60 functions as a transcriptional repressor of anthocyanin biosynthesis in lettuce (Lactuca sativa). Plant Cell Rep. 2008, 27, 985–994. [Google Scholar] [CrossRef]
- Kim, H.J.; Triplett, B.A. Cotton Fiber Growth in Planta and in Vitro. Models for Plant Cell Elongation and Cell Wall Biogenesis. Plant Physiol. 2001, 127, 1361–1366. [Google Scholar] [CrossRef]
- Lee, J.; Burns, T.H.; Light, G.; Sun, Y.; Fokar, M.; Kasukabe, Y.; Fujisawa, K.; Maekawa, Y.; Allen, R.D. Xyloglucan endotransglycosylase/hydrolase genes in cotton and their role in fiber elongation. Planta 2010, 232, 1191–1205. [Google Scholar] [CrossRef]
- Huang, G.; Wu, Z.; Percy, R.G.; Bai, M.; Li, Y.; Frelichowski, J.E.; Hu, J.; Wang, K.; Yu, J.Z.; Zhu, Y. Genome sequence of Gossypium herbaceum and genome updates of Gossypium arboreum and Gossypium hirsutum provide insights into cotton A-genome evolution. Nat. Genet. 2020, 52, 516–524. [Google Scholar] [CrossRef]
- Fang, L.; Wang, Q.; Hu, Y.; Jia, Y.; Chen, J.; Liu, B.; Zhang, Z.; Guan, X.; Chen, S.; Zhou, B.; et al. Genomic analyses in cotton identify signatures of selection and loci associated with fiber quality and yield traits. Nat. Genet. 2017, 49, 1089–1098. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Aibar, S.; Fontanillo, C.; Droste, C.; De Las Rivas, J. Functional Gene Networks: R/Bioc package to generate and analyse gene networks derived from functional enrichment and clustering. Bioinformatics 2015, 31, 1686–1688. [Google Scholar] [CrossRef] [PubMed]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Lu, L.; Liu, H.Y.; Li, S.; Xing, F.; Chen, L.L. CRISPR-P: A web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant 2014, 7, 1494–1496. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, J.; Sun, L.; Ma, Y.; Xu, J.; Liang, S.; Deng, J.; Tan, J.; Zhang, Q.; Tu, L.; et al. High efficient multisites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system. Plant Biotechnol. J. 2018, 16, 137–150. [Google Scholar] [CrossRef]
- Luo, M.; Xiao, Y.; Li, X.; Lu, X.; Deng, W.; Li, D.; Hou, L.; Hu, M.; Li, Y.; Pei, Y. GhDET2, a steroid 5alpha-reductase, plays an important role in cotton fiber cell initiation and elongation. Plant J. 2007, 51, 419–430. [Google Scholar] [CrossRef]
- Yan, Q.; Wang, Y.; Li, Q.; Zhang, Z.; Ding, H.; Zhang, Y.; Liu, H.; Luo, M.; Liu, D.; Song, W.; et al. Up-regulation of GhTT2-3A in cotton fibres during secondary wall thickening results in brown fibres with improved quality. Plant Biotechnol. J. 2018, 16, 1735–1747. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Welti, R.; Li, W.; Li, M.; Sang, Y.; Biesiada, H.; Zhou, H.E.; Rajashekar, C.B.; Williams, T.D.; Wang, X. Profiling membrane lipids in plant stress responses. Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. J. Biol. Chem. 2002, 277, 31994–32002. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Li, L.; Li, M.; Lam, S.M.; Wang, G.; Wu, Y.; Zhang, H.; Niu, C.; Zhang, X.; Liu, X.; et al. Microbiota Depletion Impairs Thermogenesis of Brown Adipose Tissue and Browning of White Adipose Tissue. Cell Rep. 2019, 26, 2720–2737.e5. [Google Scholar] [CrossRef] [PubMed]



| Average Length of Upper Quartile Fibers (mm) | Length Uniformity (%) | Fiber Strength (cN/tex) | Micronaire Value | Fiber Elongation (%) | |
|---|---|---|---|---|---|
| WT | 29.5 ± 0.2 | 84.65 ± 0.95 | 31.6 ± 0.80 | 5.15 ± 0.05 | 6.40 ± 0.40 |
| ghmyb201-38 | 21.25 ± 0.35 ** | 79.5 ± 0.20 * | 23.45 ± 1.15 * | 7.1 ± 0.10 ** | 6.10 ± 0.20 ns |
| ghmyb201-45 | 22.25 ± 0.35 ** | 81.45 ± 0.05 ns | 23.5 ± 0.30 * | 7.25 ± 0.25 * | 6.40 ± 0.10 ns |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suo, Q.; Fang, N.; Zeng, J.; Yan, F.; Zhu, X.; Wang, Y.; Yu, W.; Chen, J.; Liang, A.; Li, Y.; et al. R2R3 MYB Transcription Factor GhMYB201 Promotes Cotton Fiber Elongation via Cell Wall Loosening and Very-Long-Chain Fatty Acid Synthesis. Int. J. Mol. Sci. 2024, 25, 9559. https://doi.org/10.3390/ijms25179559
Suo Q, Fang N, Zeng J, Yan F, Zhu X, Wang Y, Yu W, Chen J, Liang A, Li Y, et al. R2R3 MYB Transcription Factor GhMYB201 Promotes Cotton Fiber Elongation via Cell Wall Loosening and Very-Long-Chain Fatty Acid Synthesis. International Journal of Molecular Sciences. 2024; 25(17):9559. https://doi.org/10.3390/ijms25179559
Chicago/Turabian StyleSuo, Qingwei, Nianjuan Fang, Jianyan Zeng, Fulin Yan, Xi Zhu, Yi Wang, Wanting Yu, Junmin Chen, Aimin Liang, Yaohua Li, and et al. 2024. "R2R3 MYB Transcription Factor GhMYB201 Promotes Cotton Fiber Elongation via Cell Wall Loosening and Very-Long-Chain Fatty Acid Synthesis" International Journal of Molecular Sciences 25, no. 17: 9559. https://doi.org/10.3390/ijms25179559
APA StyleSuo, Q., Fang, N., Zeng, J., Yan, F., Zhu, X., Wang, Y., Yu, W., Chen, J., Liang, A., Li, Y., Kong, J., & Xiao, Y. (2024). R2R3 MYB Transcription Factor GhMYB201 Promotes Cotton Fiber Elongation via Cell Wall Loosening and Very-Long-Chain Fatty Acid Synthesis. International Journal of Molecular Sciences, 25(17), 9559. https://doi.org/10.3390/ijms25179559






