Transcriptomic Alterations in Spliceosome Components in Advanced Heart Failure: Status of Cardiac-Specific Alternative Splicing Factors
Abstract
:1. Introduction
2. Results
2.1. Clinical Characteristics of Patients
2.2. mRNA Expression of Spliceosome Components in Advanced Heart Failure Patients
2.2.1. Alterations in E, A, B and C Complexes
2.2.2. Cardiac-Specific AS Factors
3. Discussion
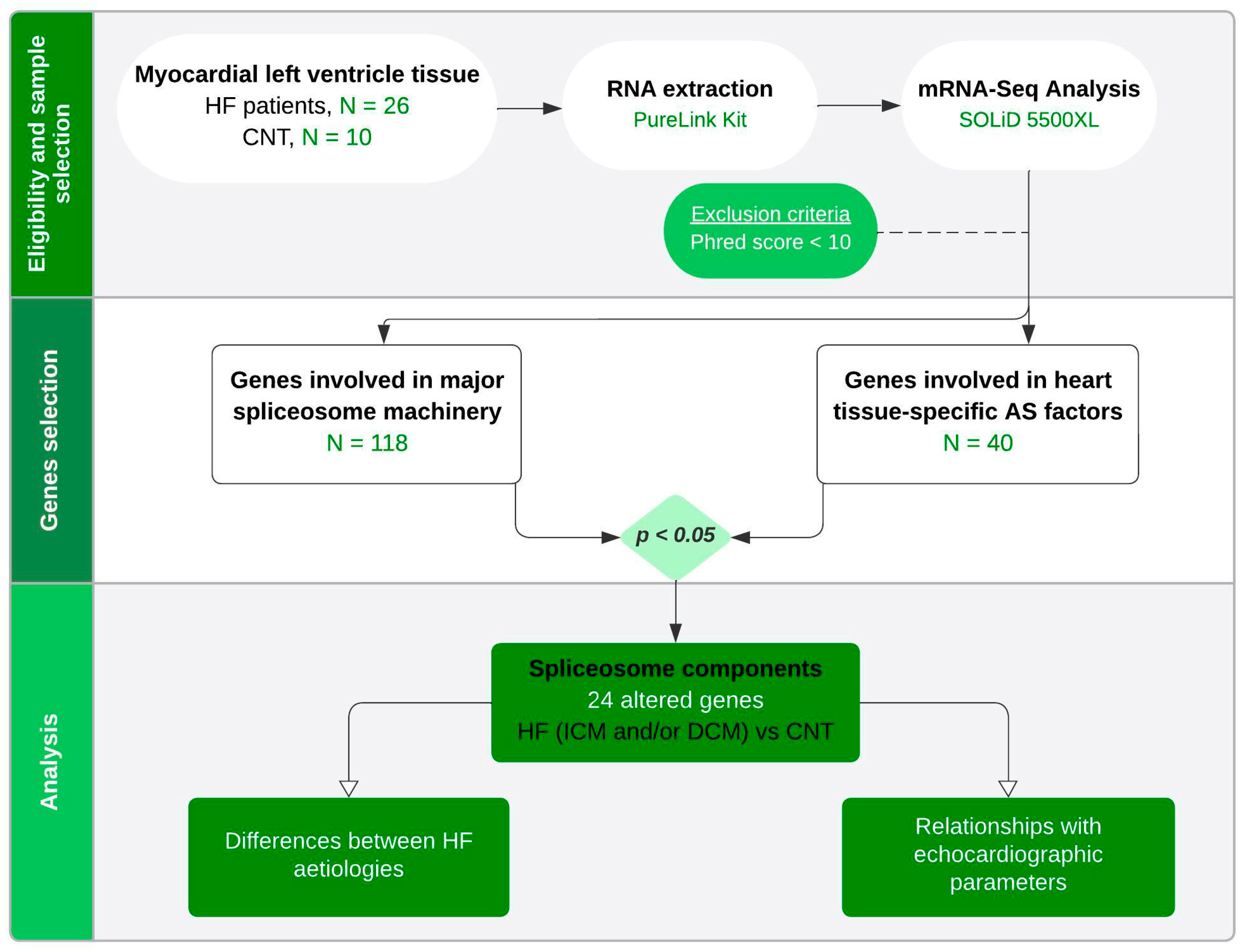
4. Materials and Methods
4.1. Tissue Sample Collection
4.2. RNA Extraction and Integrity
4.3. mRNA-Seq Analysis
4.4. Statistical Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bozkurt, B.; Coats, A.J.S.; Tsutsui, H.; Abdelhamid, C.M.; Adamopoulos, S.; Albert, N.; Anker, S.D.; Atherton, J.; Bohm, M.; Butler, J.; et al. Universal definition and classification of heart failure: A report of the Heart Failure Society of America, Heart Failure Association of the European Society of Cardiology, Japanese Heart Failure Society and Writing Committee of the Universal Definition of Heart Failure: Endorsed by the Canadian Heart Failure Society, Heart Failure Association of India, Cardiac Society of Australia and New Zealand, and Chinese Heart Failure Association. Eur. J. Heart Fail. 2021, 23, 352–380. [Google Scholar]
- Bozkurt, B.; Colvin, M.; Cook, J.; Cooper, L.T.; Deswal, A.; Fonarow, G.C.; Francis, G.S.; Lenihan, D.; Lewis, E.F.; McNamara, D.M.; et al. Current Diagnostic and Treatment Strategies for Specific Dilated Cardiomyopathies: A Scientific Statement From the American Heart Association. Circulation 2016, 134, e579–e646. [Google Scholar] [CrossRef]
- Baldasseroni, S.; Opasich, C.; Gorini, M.; Lucci, D.; Marchionni, N.; Marini, M.; Campana, C.; Perini, G.; Deorsola, A.; Masotti, G.; et al. Left bundle-branch block is associated with increased 1-year sudden and total mortality rate in 5517 outpatients with congestive heart failure: A report from the Italian network on congestive heart failure. Am. Heart J. 2002, 143, 398–405. [Google Scholar] [CrossRef]
- Herrer, I.; Rosello-Lleti, E.; Rivera, M.; Molina-Navarro, M.M.; Tarazon, E.; Ortega, A.; Martinez-Dolz, L.; Trivino, J.C.; Lago, F.; Gonzalez-Juanatey, J.R.; et al. RNA-sequencing analysis reveals new alterations in cardiomyocyte cytoskeletal genes in patients with heart failure. Lab. Investig. 2014, 94, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Tarazon, E.; Rosello-Lleti, E.; Rivera, M.; Ortega, A.; Molina-Navarro, M.M.; Trivino, J.C.; Lago, F.; Gonzalez-Juanatey, J.R.; Orosa, P.; Montero, J.A.; et al. RNA sequencing analysis and atrial natriuretic peptide production in patients with dilated and ischemic cardiomyopathy. PLoS ONE 2014, 9, e90157. [Google Scholar] [CrossRef] [PubMed]
- Gimenez-Escamilla, I.; Benedicto, C.; Perez-Carrillo, L.; Delgado-Arija, M.; Gonzalez-Torrent, I.; Vilchez, R.; Martinez-Dolz, L.; Portoles, M.; Tarazon, E.; Rosello-Lleti, E. Alterations in Mitochondrial Oxidative Phosphorylation System: Relationship of Complex V and Cardiac Dysfunction in Human Heart Failure. Antioxidants 2024, 13, 285. [Google Scholar] [CrossRef]
- Rosello-Lleti, E.; Alonso, J.; Cortes, R.; Almenar, L.; Martinez-Dolz, L.; Sanchez-Lazaro, I.; Lago, F.; Azorin, I.; Juanatey, J.R.; Portoles, M.; et al. Cardiac protein changes in ischaemic and dilated cardiomyopathy: A proteomic study of human left ventricular tissue. J. Cell. Mol. Med. 2012, 16, 2471–2486. [Google Scholar] [CrossRef] [PubMed]
- Lara-Pezzi, E.; Dopazo, A.; Manzanares, M. Understanding cardiovascular disease: A journey through the genome (and what we found there). Dis. Models Mech. 2012, 5, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Chen, L. Alternative splicing: Human disease and quantitative analysis from high-throughput sequencing. Comput. Struct. Biotechnol. J. 2021, 19, 183–195. [Google Scholar] [CrossRef]
- Wahl, M.C.; Luhrmann, R. SnapShot: Spliceosome Dynamics I. Cell 2015, 161, 1474. [Google Scholar] [CrossRef]
- Cao, J.; Wei, Z.; Nie, Y.; Chen, H.Z. Therapeutic potential of alternative splicing in cardiovascular diseases. EBioMedicine 2024, 101, 104995. [Google Scholar] [CrossRef] [PubMed]
- Badr, E.; ElHefnawi, M.; Heath, L.S. Computational Identification of Tissue-Specific Splicing Regulatory Elements in Human Genes from RNA-Seq Data. PLoS ONE 2016, 11, e0166978. [Google Scholar] [CrossRef]
- Tao, Y.; Zhang, Q.; Wang, H.; Yang, X.; Mu, H. Alternative splicing and related RNA binding proteins in human health and disease. Signal Transduct. Target. Ther. 2024, 9, 26. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Patel, P.N.; Gorham, J.M.; McDonough, B.; DePalma, S.R.; Adler, E.E.; Lam, L.; MacRae, C.A.; Mohiuddin, S.M.; Fatkin, D.; et al. Identification of pathogenic gene mutations in LMNA and MYBPC3 that alter RNA splicing. Proc. Natl. Acad. Sci. USA 2017, 114, 7689–7694. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Schafer, S.; Greaser, M.L.; Radke, M.H.; Liss, M.; Govindarajan, T.; Maatz, H.; Schulz, H.; Li, S.; Parrish, A.M.; et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat. Med. 2012, 18, 766–773. [Google Scholar] [CrossRef]
- Nutter, C.A.; Jaworski, E.A.; Verma, S.K.; Deshmukh, V.; Wang, Q.; Botvinnik, O.B.; Lozano, M.J.; Abass, I.J.; Ijaz, T.; Brasier, A.R.; et al. Dysregulation of RBFOX2 Is an Early Event in Cardiac Pathogenesis of Diabetes. Cell Rep. 2016, 15, 2200–2213. [Google Scholar] [CrossRef]
- Varga, Z.V.; Pipicz, M.; Baan, J.A.; Baranyai, T.; Koncsos, G.; Leszek, P.; Kusmierczyk, M.; Sanchez-Cabo, F.; Garcia-Pavia, P.; Brenner, G.J.; et al. Alternative Splicing of NOX4 in the Failing Human Heart. Front. Physiol. 2017, 8, 935. [Google Scholar] [CrossRef]
- Cvitkovic, I.; Jurica, M.S. Spliceosome database: A tool for tracking components of the spliceosome. Nucleic Acids Res. 2013, 41, D132–D141. [Google Scholar] [CrossRef]
- Seal, R.L.; Braschi, B.; Gray, K.; Jones, T.E.M.; Tweedie, S.; Haim-Vilmovsky, L.; Bruford, E.A. Genenames.org: The HGNC resources in 2023. Nucleic Acids Res. 2023, 51, D1003–D1009. [Google Scholar] [CrossRef]
- van den Hoogenhof, M.M.; Pinto, Y.M.; Creemers, E.E. RNA Splicing: Regulation and Dysregulation in the Heart. Circ. Res. 2016, 118, 454–468. [Google Scholar] [CrossRef]
- Ponikowski, P.; Voors, A.A.; Anker, S.D.; Bueno, H.; Cleland, J.G.; Coats, A.J.; Falk, V.; Gonzalez-Juanatey, J.R.; Harjola, V.P.; Jankowska, E.A.; et al. 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: The Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC). Developed with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur. J. Heart Fail 2016, 18, 891–975. [Google Scholar]
- Rogalska, M.E.; Vivori, C.; Valcarcel, J. Regulation of pre-mRNA splicing: Roles in physiology and disease, and therapeutic prospects. Nat. Rev. Genet. 2023, 24, 251–269. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, S.; Zhang, L.; Issaian, A.; Hill, R.C.; Espinosa, S.; Shi, S.; Cui, Y.; Kappel, K.; Das, R.; et al. A unified mechanism for intron and exon definition and back-splicing. Nature 2019, 573, 375–380. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Bae, B.; Cuddleston, W.H.; Miura, P. Coordination of alternative splicing and alternative polyadenylation revealed by targeted long read sequencing. Nat. Commun. 2023, 14, 5506. [Google Scholar] [CrossRef]
- Qiu, F.; Fu, Y.; Lu, C.; Feng, Y.; Wang, Q.; Huo, Z.; Jia, X.; Chen, C.; Chen, S.; Xu, A. Small Nuclear Ribonucleoprotein Polypeptide A-Mediated Alternative Polyadenylation of STAT5B during Th1 Cell Differentiation. J. Immunol. 2017, 199, 3106–3115. [Google Scholar] [CrossRef]
- Bansal, S.S.; Ismahil, M.A.; Goel, M.; Patel, B.; Hamid, T.; Rokosh, G.; Prabhu, S.D. Activated T Lymphocytes are Essential Drivers of Pathological Remodeling in Ischemic Heart Failure. Circ. Heart Fail. 2017, 10, e003688. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Li, M.; Huo, Z.; Chen, L.; Liu, S.; Deng, K.; Lu, X.; Chen, S.; Fu, Y.; Xu, A. U1 snRNP proteins promote proximal alternative polyadenylation sites by directly interacting with 3′ end processing core factors. J. Mol. Cell Biol. 2022, 14, mjac054. [Google Scholar] [CrossRef]
- Ribera, J.; Portoles, I.; Cordoba-Jover, B.; Rodriguez-Vita, J.; Casals, G.; Gonzalez-de la Presa, B.; Graupera, M.; Solsona-Vilarrasa, E.; Garcia-Ruiz, C.; Fernandez-Checa, J.C.; et al. The loss of DHX15 impairs endothelial energy metabolism, lymphatic drainage and tumor metastasis in mice. Commun. Biol. 2021, 4, 1192. [Google Scholar] [CrossRef]
- Maul-Newby, H.M.; Amorello, A.N.; Sharma, T.; Kim, J.H.; Modena, M.S.; Prichard, B.E.; Jurica, M.S. A model for DHX15 mediated disassembly of A-complex spliceosomes. RNA 2022, 28, 583–595. [Google Scholar] [CrossRef]
- Zhou, Y.; Alimohamadi, S.; Wang, L.; Liu, Z.; Wall, J.B.; Yin, C.; Liu, J.; Qian, L. A Loss of Function Screen of Epigenetic Modifiers and Splicing Factors during Early Stage of Cardiac Reprogramming. Stem Cells Int. 2018, 2018, 3814747. [Google Scholar] [CrossRef]
- Laflamme, M.A.; Murry, C.E. Heart regeneration. Nature 2011, 473, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, R.; Georgiou, M.; Yang, C.; Szymanska, K.; Lahat, A.; Vasconcelos, E.J.R.; Ji, Y.; Moya Molina, M.; Collin, J.; Queen, R.; et al. PRPF8-mediated dysregulation of hBrr2 helicase disrupts human spliceosome kinetics and 5 -splice-site selection causing tissue-specific defects. Nat. Commun. 2024, 15, 3138. [Google Scholar] [CrossRef]
- Gonzalez-Rodriguez, P.; Klionsky, D.J.; Joseph, B. Autophagy regulation by RNA alternative splicing and implications in human diseases. Nat. Commun. 2022, 13, 2735. [Google Scholar] [CrossRef]
- Xu, G.; Li, T.; Chen, J.; Li, C.; Zhao, H.; Yao, C.; Dong, H.; Wen, K.; Wang, K.; Zhao, J.; et al. Autosomal dominant retinitis pigmentosa-associated gene PRPF8 is essential for hypoxia-induced mitophagy through regulating ULK1 mRNA splicing. Autophagy 2018, 14, 1818–1830. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.; Toan, S.; Li, R.; Zhou, H. Therapeutic strategies in ischemic cardiomyopathy: Focus on mitochondrial quality surveillance. EBioMedicine 2022, 84, 104260. [Google Scholar] [CrossRef]
- Ye, J.; Ying, J.; Chen, H.; Wu, Z.; Huang, C.; Zhang, C.; Chen, Z.; Chen, H. PPIH acts as a potential predictive biomarker for patients with common solid tumors. BMC Cancer 2024, 24, 681. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, S.; Qin, J.; Zhang, X.; Lu, G.; Liu, H.; Guo, H.; Wu, L.; Shender, V.O.; Shao, C.; et al. Splicing factor BUD31 promotes ovarian cancer progression through sustaining the expression of anti-apoptotic BCL2L12. Nat. Commun. 2022, 13, 6246. [Google Scholar] [CrossRef]
- Sales-Lee, J.; Perry, D.S.; Bowser, B.A.; Diedrich, J.K.; Rao, B.; Beusch, I.; Yates, J.R., 3rd; Roy, S.W.; Madhani, H.D. Coupling of spliceosome complexity to intron diversity. Curr. Biol. 2021, 31, 4898–4910.e4. [Google Scholar] [CrossRef]
- Hamann, F.; Enders, M.; Ficner, R. Structural basis for RNA translocation by DEAH-box ATPases. Nucleic Acids Res. 2019, 47, 4349–4362. [Google Scholar] [CrossRef]
- Dreyer, J.; Schleicher, M.; Tappe, A.; Schilling, K.; Kuner, T.; Kusumawidijaja, G.; Muller-Esterl, W.; Oess, S.; Kuner, R. Nitric oxide synthase (NOS)-interacting protein interacts with neuronal NOS and regulates its distribution and activity. J. Neurosci. 2004, 24, 10454–10465. [Google Scholar] [CrossRef]
- Rosello-Lleti, E.; Carnicer, R.; Tarazon, E.; Ortega, A.; Gil-Cayuela, C.; Lago, F.; Gonzalez-Juanatey, J.R.; Portoles, M.; Rivera, M. Human Ischemic Cardiomyopathy Shows Cardiac Nos1 Translocation and its Increased Levels are Related to Left Ventricular Performance. Sci. Rep. 2016, 6, 24060. [Google Scholar] [CrossRef] [PubMed]
- Coltri, P.; Effenberger, K.; Chalkley, R.J.; Burlingame, A.L.; Jurica, M.S. Breaking up the C complex spliceosome shows stable association of proteins with the lariat intron intermediate. PLoS ONE 2011, 6, e19061. [Google Scholar] [CrossRef] [PubMed]
- Brauch, K.M.; Karst, M.L.; Herron, K.J.; de Andrade, M.; Pellikka, P.A.; Rodeheffer, R.J.; Michels, V.V.; Olson, T.M. Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J. Am. Coll. Cardiol. 2009, 54, 930–941. [Google Scholar] [CrossRef] [PubMed]
- Montanes-Agudo, P.; Pinto, Y.M.; Creemers, E.E. Splicing factors in the heart: Uncovering shared and unique targets. J. Mol. Cell. Cardiol. 2023, 179, 72–79. [Google Scholar] [CrossRef]
- Montaville, P.; Dai, Y.; Cheung, C.Y.; Giller, K.; Becker, S.; Michalak, M.; Webb, S.E.; Miller, A.L.; Krebs, J. Nuclear translocation of the calcium-binding protein ALG-2 induced by the RNA-binding protein RBM22. Biochim. Biophys. Acta 2006, 1763, 1335–1343. [Google Scholar] [CrossRef]
- Janowicz, A.; Michalak, M.; Krebs, J. Stress induced subcellular distribution of ALG-2, RBM22 and hSlu7. Biochim. Biophys. Acta 2011, 1813, 1045–1049. [Google Scholar] [CrossRef]
- Guo, J.; Liu, H.B.; Sun, C.; Yan, X.Q.; Hu, J.; Yu, J.; Yuan, Y.; Du, Z.M. MicroRNA-155 Promotes Myocardial Infarction-Induced Apoptosis by Targeting RNA-Binding Protein QKI. Oxid. Med. Cell Longev. 2019, 2019, 4579806. [Google Scholar] [CrossRef]
- Chen, X.; Liu, Y.; Xu, C.; Ba, L.; Liu, Z.; Li, X.; Huang, J.; Simpson, E.; Gao, H.; Cao, D.; et al. QKI is a critical pre-mRNA alternative splicing regulator of cardiac myofibrillogenesis and contractile function. Nat. Commun. 2021, 12, 89. [Google Scholar] [CrossRef]
- Montanes-Agudo, P.; Aufiero, S.; Schepers, E.N.; van der Made, I.; Cocera-Ortega, L.; Ernault, A.C.; Richard, S.; Kuster, D.W.D.; Christoffels, V.M.; Pinto, Y.M.; et al. The RNA-binding protein QKI governs a muscle-specific alternative splicing program that shapes the contractile function of cardiomyocytes. Cardiovasc. Res. 2023, 119, 1161–1174. [Google Scholar] [CrossRef]
- Zhang, Y.; Si, Y.; Ma, N.; Mei, J. The RNA-binding protein PCBP2 inhibits Ang II-induced hypertrophy of cardiomyocytes though promoting GPR56 mRNA degeneration. Biochem. Biophys. Res. Commun. 2015, 464, 679–684. [Google Scholar] [CrossRef]
- Ortiz-Sanchez, P.; Villalba-Orero, M.; Lopez-Olaneta, M.M.; Larrasa-Alonso, J.; Sanchez-Cabo, F.; Marti-Gomez, C.; Camafeita, E.; Gomez-Salinero, J.M.; Ramos-Hernandez, L.; Nielsen, P.J.; et al. Loss of SRSF3 in Cardiomyocytes Leads to Decapping of Contraction-Related mRNAs and Severe Systolic Dysfunction. Circ. Res. 2019, 125, 170–183. [Google Scholar] [CrossRef] [PubMed]
- Akerberg, A.A.; Trembley, M.; Butty, V.; Schwertner, A.; Zhao, L.; Beerens, M.; Liu, X.; Mahamdeh, M.; Yuan, S.; Boyer, L.; et al. RBPMS2 Is a Myocardial-Enriched Splicing Regulator Required for Cardiac Function. Circ. Res. 2022, 131, 980–1000. [Google Scholar] [CrossRef]
- Ye, J.; Beetz, N.; O’Keeffe, S.; Tapia, J.C.; Macpherson, L.; Chen, W.V.; Bassel-Duby, R.; Olson, E.N.; Maniatis, T. hnRNP U protein is required for normal pre-mRNA splicing and postnatal heart development and function. Proc. Natl. Acad. Sci. USA 2015, 112, E3020–E3029. [Google Scholar] [CrossRef] [PubMed]
- Aslan, G.S.; Jae, N.; Manavski, Y.; Fouani, Y.; Shumliakivska, M.; Kettenhausen, L.; Kirchhof, L.; Gunther, S.; Fischer, A.; Luxan, G.; et al. Malat1 deficiency prevents neonatal heart regeneration by inducing cardiomyocyte binucleation. JCI Insight 2023, 8, e162124. [Google Scholar] [CrossRef]
- Soubise, B.; Jiang, Y.; Douet-Guilbert, N.; Troadec, M.B. RBM22, a Key Player of Pre-mRNA Splicing and Gene Expression Regulation, Is Altered in Cancer. Cancers 2022, 14, 643. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Ren, S.; Lee, J.H.; Qiu, J.; Chapski, D.J.; Rau, C.D.; Zhou, Y.; Abdellatif, M.; Nakano, A.; Vondriska, T.M.; et al. RBFox1-mediated RNA splicing regulates cardiac hypertrophy and heart failure. J. Clin. Investig. 2016, 126, 195–206. [Google Scholar] [CrossRef]
- Love, S.L.; Emerson, J.D.; Koide, K.; Hoskins, A.A. Pre-mRNA splicing-associated diseases and therapies. RNA Biol. 2023, 20, 525–538. [Google Scholar] [CrossRef]
- Zong, F.Y.; Fu, X.; Wei, W.J.; Luo, Y.G.; Heiner, M.; Cao, L.J.; Fang, Z.; Fang, R.; Lu, D.; Ji, H.; et al. The RNA-binding protein QKI suppresses cancer-associated aberrant splicing. PLoS Genet. 2014, 10, e1004289. [Google Scholar] [CrossRef] [PubMed]
- Griffin, C.; Saint-Jeannet, J.P. Spliceosomopathies: Diseases and mechanisms. Dev. Dyn. 2020, 249, 1038–1046. [Google Scholar] [CrossRef]
- Martin, S.S.; Aday, A.W.; Almarzooq, Z.I.; Anderson, C.A.M.; Arora, P.; Avery, C.L.; Baker-Smith, C.M.; Barone Gibbs, B.; Beaton, A.Z.; Boehme, A.K.; et al. 2024 Heart Disease and Stroke Statistics: A Report of US and Global Data From the American Heart Association. Circulation 2024, 149, e347–e913. [Google Scholar]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Macrae, D.J. The Council for International Organizations and Medical Sciences (CIOMS) guidelines on ethics of clinical trials. Proc. Am. Thorac. Soc. 2007, 4, 176–178, discussion 178–179. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef]


| mRNA-Seq | ||
|---|---|---|
| ICM (n = 13) | DCM (n = 13) | |
| Gender male (%) | 100 | 92 |
| Age (years) | 54 ± 8 | 51 ± 11 |
| NYHA class | III–IV | III–IV |
| BMI (kg/m2) | 27 ± 4 | 27 ± 5 |
| Haemoglobin (g/dL) | 14 ± 3 | 13 ± 3 |
| Haematocrit (%) | 41 ± 6 | 39 ± 7 |
| Total cholesterol (mg/dL) | 162 ± 41 | 147 ± 37 |
| NT-proBNP (pg/mL) | 3684 ± 2285 | 4071 ± 3584 |
| Prior hypertension (%) | 33 | 17 |
| Prior smoking (%) | 92 * | 50 |
| Diabetes mellitus (%) | 42 | 17 |
| Echocardiographic study | ||
| LVEF (%) | 25 ± 5 * | 17 ± 8 |
| LVESD (mm) | 57 ± 8 *** | 74 ± 10 |
| LVEDD (mm) | 65 ± 8 *** | 81 ± 8 |
| LVMI (g/cm2) | 139 ± 26 *** | 245 ± 64 |
| Duration of disease (months) | 45 ± 40 | 75 ± 68 |
| Treatment (%) | ||
| Renin–angiotensin system inhibitors | 71 | 100 |
| B-blokers | 57 | 73 |
| Aldosterone antagonists | 100 | 91 |
| Digoxin | 43 | 64 |
| Diuretics | 71 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giménez-Escamilla, I.; Pérez-Carrillo, L.; González-Torrent, I.; Delgado-Arija, M.; Benedicto, C.; Portolés, M.; Tarazón, E.; Roselló-Lletí, E. Transcriptomic Alterations in Spliceosome Components in Advanced Heart Failure: Status of Cardiac-Specific Alternative Splicing Factors. Int. J. Mol. Sci. 2024, 25, 9590. https://doi.org/10.3390/ijms25179590
Giménez-Escamilla I, Pérez-Carrillo L, González-Torrent I, Delgado-Arija M, Benedicto C, Portolés M, Tarazón E, Roselló-Lletí E. Transcriptomic Alterations in Spliceosome Components in Advanced Heart Failure: Status of Cardiac-Specific Alternative Splicing Factors. International Journal of Molecular Sciences. 2024; 25(17):9590. https://doi.org/10.3390/ijms25179590
Chicago/Turabian StyleGiménez-Escamilla, Isaac, Lorena Pérez-Carrillo, Irene González-Torrent, Marta Delgado-Arija, Carlota Benedicto, Manuel Portolés, Estefanía Tarazón, and Esther Roselló-Lletí. 2024. "Transcriptomic Alterations in Spliceosome Components in Advanced Heart Failure: Status of Cardiac-Specific Alternative Splicing Factors" International Journal of Molecular Sciences 25, no. 17: 9590. https://doi.org/10.3390/ijms25179590
APA StyleGiménez-Escamilla, I., Pérez-Carrillo, L., González-Torrent, I., Delgado-Arija, M., Benedicto, C., Portolés, M., Tarazón, E., & Roselló-Lletí, E. (2024). Transcriptomic Alterations in Spliceosome Components in Advanced Heart Failure: Status of Cardiac-Specific Alternative Splicing Factors. International Journal of Molecular Sciences, 25(17), 9590. https://doi.org/10.3390/ijms25179590






