Modification of the Trizol Method for the Extraction of RNA from Prorocentrum triestinum ACIZ_LEM2
Abstract
:1. Introduction
2. Experimental Design
2.1. Materials
- Conical tubes (50 mL) (Greiner Bio-One, Kremsmünster, Upper Austria, Austria, catalog number: 227261).
- 1.5 mL and 2 mL microtubes (MercK, Darmstadt, Germany).
- BOECO 2000 mL balloon flask (MercK, Darmstadt, Germany).
- Analog Timer Timer 24 Hours Adapter (ESHOPANGIE, Santiago, Chile).
- Micropipette tips (0.5–20 µL, 100–200 µL, 1000 µL) (Gilson™, Middleton, WI, USA).
- Parafilm (Greenwich, CT, USA 06836)
- Nitrile Gloves (Winkler, Santiago, Chile).
- Invitrogen™ TRIzol™ Reagent (Invitrogen, Waltham, CA, USA, Cat. No.: 15596018)
- Chloroform (Winkler, Lampa, Santiago, Chile, Cat. No.: CL-0595).
- Ethanol 96% EMSURE® Reag. Ph Eur (MercK, Darmstadt, Germany, Cat. No.: 159010).
- Iso-PROPYL Alcohol (Winkler, Lampa, Santiago, Chile).
- Glycogen, RNA grade (Thermo Fisher Scientific, Waltham, CA, USA, Cat. No.: R0561).
- OmniPur® Water, DEPC Treated, Autoclaved, Nuclease-Free—CAS 7732-18-5—Calbiochem (Merck, Darmstadt, Germany).
- Face or screen protector.
- Commercial Bleach, 6% Sodium Hypochlorite (Clorox®, Oakland, CA, USA).
- TAE Buffer, 40X Molecular Biology Grade (Promega, Madison, WI, USA, Cat. No.: V4271).
- Lafken Agarose (Fermelo Biotec, Santiago, Chile, Cat. No.: FER00AL200G).
- PAGE GelRed Nucleic Acid Gel Stain 0.1 mL (Biotium, Fremont, CA 94538, USA, Cat. No.: 41008-T).
- 6X Blue/Orange Dye (Promega, Madison, WI, USA, Cat. No.: G1881).
- Nuclease-free water (Sigma-Aldrich, Burlington, VT, USA).
2.2. Equipment
- Mini V/PCR vertical laminar flow cabinet (il Telstar, SLU, Barcelona, Spain).
- KIMBLE® PELLET PESTLE® Cordless Motor (Merck, Darmstadt, Germany).
- ChemiDoc MP Imaging System (BIO-RAD, Hercules, CA, USA).Labnet AccuBlock D1100 Digital Dry Bath (TEquipment.NET, Long Branch, NJ, USA).Centrifuges M-240/M-240R (Boeco Germany, Hamburg, Germany).
- Thermo Sorvall ST 8R centrifuge (Thermo Scientific™, Waltham, MA, USA).
- Horizontal electrophoresis chamber (Select Bioproducts) (Bio Rad, Hercules, CA, USA).
- Power source (BioRad, Hercules, CA, USA).
- BAE224C precision digital analytical balance (Revel) (Velab, Puebla, Mexico).
- BOECO Vacuum Pump R-300. (Boeco Germany, Hamburg, Germany).
- Operon Ultra-Low Temperature Freezer −86 °C. (Thermo Fisher Scientific, Waltham, MA, USA).
- Agilent 4150 (Agilent, Santa Clara, CA, USA).
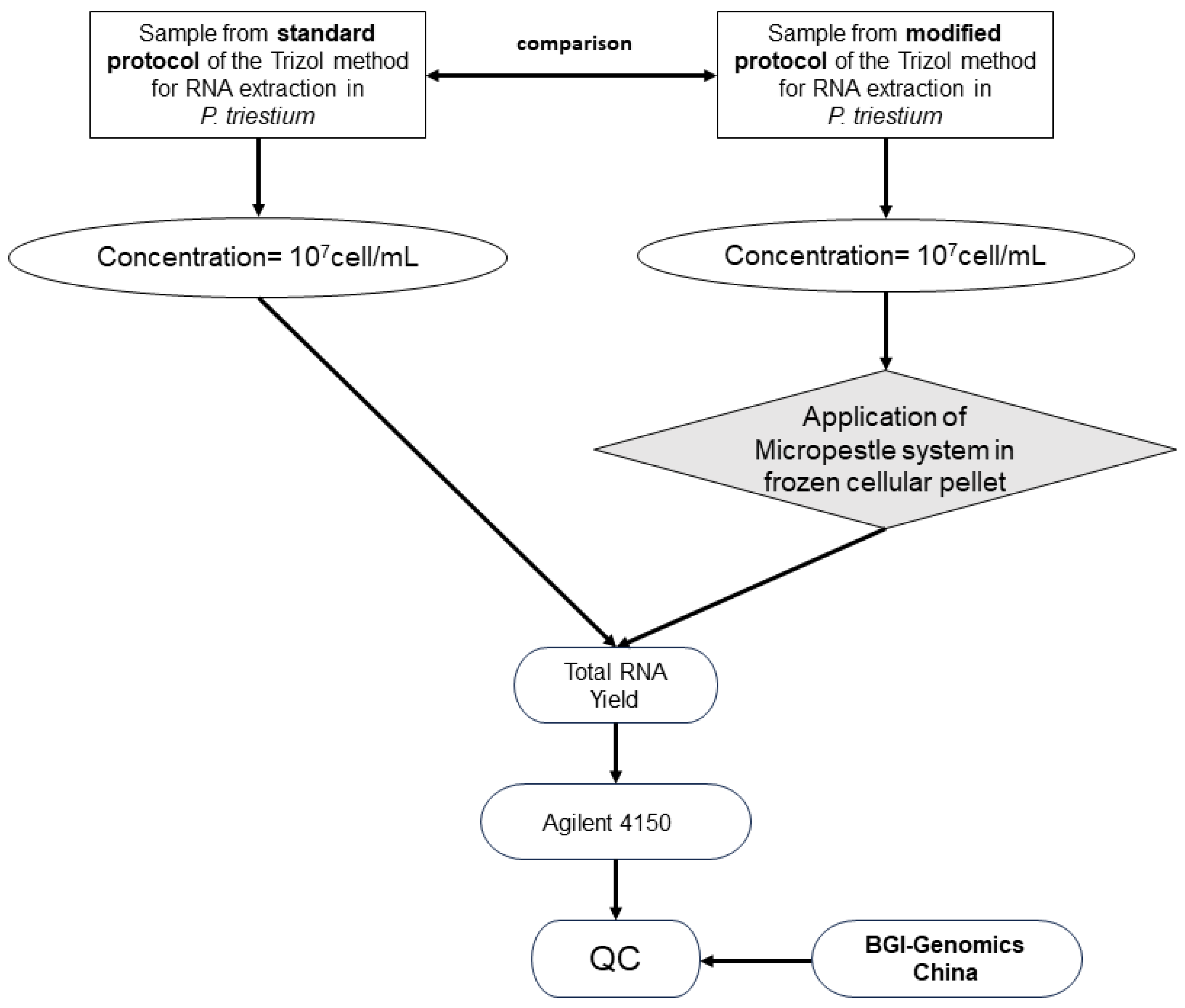
3. Procedure
3.1. Test Description
- Resuspend in 750 µL of Trizol Reagent (Invitrogen, Carlsbad, CA, USA) for every 250 µL of cell pellets (obtained from 400 mL of culture with a total of 107 cells/mL) and then store at −80 °C.
- 2.
- Thaw the samples at room temperature and centrifuge at 12,000 rpm for 1 min [14].
- 3.
- Transfer the supernatant to a new Eppendorf tube,
- 4.
- Homogenize using a Micropestle System (KIMBLE® PELLET PESTLE® Cordless Motor) until thawed [14].
- 5.
- 6.
- Add 200 µL of chloroform then centrifuge the samples at 12,000 rpm for 15 min at 4 °C.
- 7.
- Recover the supernatant and transfer to new tubes, then add 1 µL of molecular glycogen and 500 µL of isopropanol (Winkler), incubating at 4 °C for 10 min, then centrifuging at 12,000 rpm for 10 min at 4 °C.
- 8.
- Perform a wash with 1 mL of 75% molecular grade ethanol.
- 9.
- Centrifuge the samples at 7500 rpm for 5 min at 4 °C, observing a white granule of RNA, then discard the supernatant and dry at a temperature of 60 °C for 10 to 15 min using a Labnet AccuBlock D1100 Digital Dry Bath (TEquipment.NET, Long Branch, NJ, USA).
- 10.
- Add 50 µL of nuclease-free water (Sigma-Aldrich) to solubilize the RNA granule at 60 °C for 10 to 15 min.

3.2. Methodologies for Analyzing RNA Quality
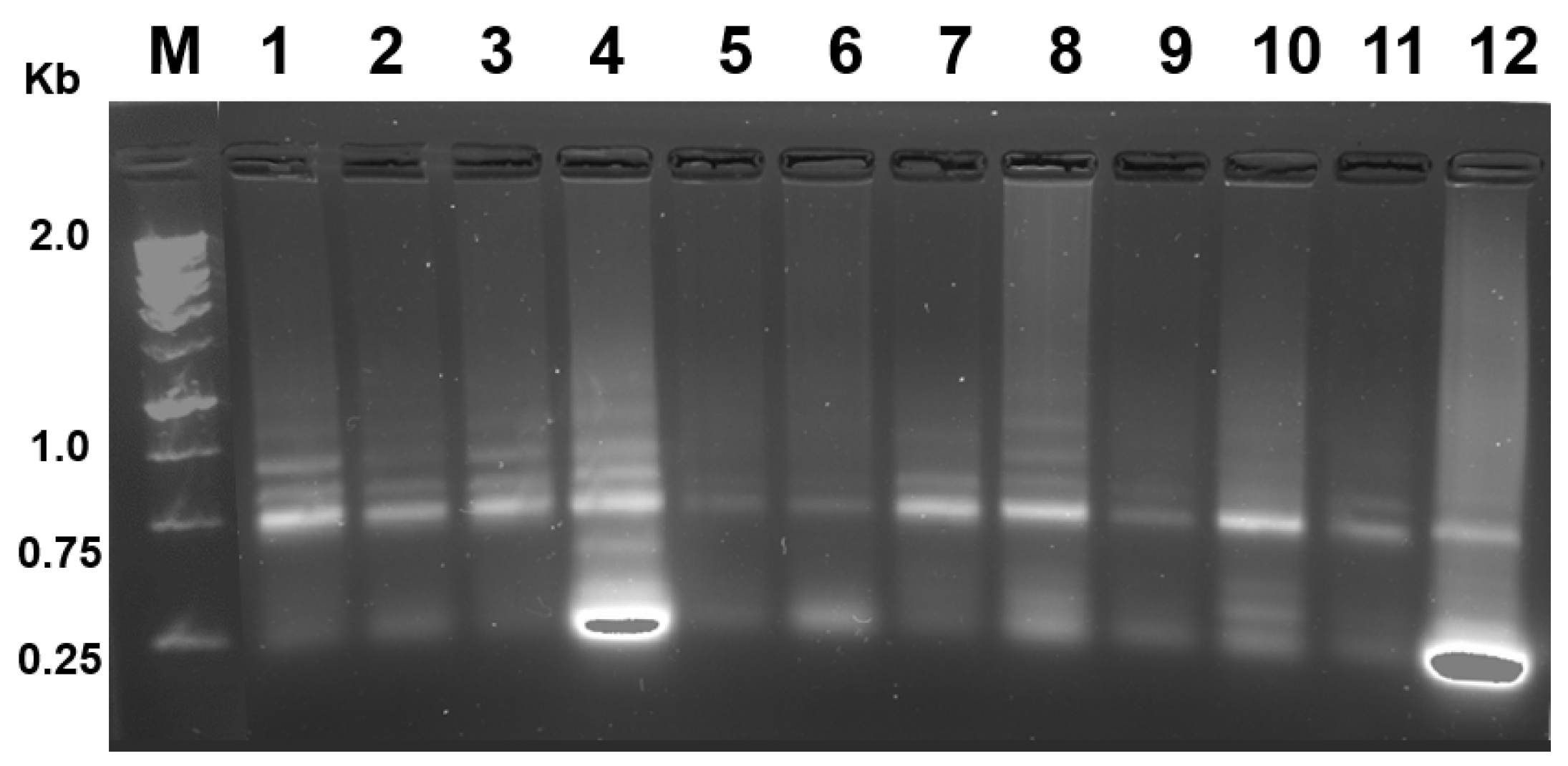
4. Results
Yield and Quality of Total RNA Isolated from P. triestinum
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lorons, D.; Rodrigues, K.F.; Sidik, M.J.; Chin, G.J.W.L. RNA Isolation from Environmental Samples of a Harmful Algal Bloom for Metatranscriptome Next-Generation Sequencing. Pertanika J. Sci. Technol. 2022, 30, 2707–2725. [Google Scholar] [CrossRef]
- Meng, F.-Q.; Song, J.-T.; Zhou, J.; Cai, Z.-H. Transcriptomic Profile and Sexual Reproduction-Relevant Genes of Alexandrium minutum in Response to Nutritional Deficiency. Front. Microbiol. 2019, 10, 2629. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Ou, L.; Dai, X.; Cui, L.; Dong, Y.; Wang, P.; Li, D.; Lu, D. An overview of Prorocentrum donghaiense blooms in China: Species identification, occurrences, ecological consequences, and factors regulating prevalence. Harmful Algae 2022, 114, 102207. [Google Scholar] [CrossRef] [PubMed]
- Akbar, M.A.; Ahmad, A.; Usup, G.; Bunawan, H. RNA-Seq as an Emerging Tool for Marine Dinoflagellate Transcriptome Analysis: Process and Challenges. Processes 2018, 6, 5. [Google Scholar] [CrossRef]
- Chang, S.; Puryear, J.; Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 1993, 11, 113–116. [Google Scholar] [CrossRef]
- Mundt, F.; Heinrich, S.; Hanelt, D. RNA isolation from taxonomically diverse photosynthetic protists. Limnol. Oceanogr. Methods 2018, 17, 190–199. [Google Scholar] [CrossRef]
- Harlow, L.D.; Koutoulis, A.; Hallegraeff, G.M. A novel, simplified technique for preservation and rapid isolation of total RNA from the toxic dinoflagellate Alexandrium catenella (Dinophyceae). Phycologia 2006, 45, 311–318. [Google Scholar] [CrossRef]
- Akbar, M.A.; Yusof, N.Y.M.; Sahrani, F.K.; Usup, G.; Ahmad, A.; Baharum, S.N.; Muhammad, N.A.N.; Bunawan, H. Insights into Alexandrium minutum Nutrient Acquisition, Metabolism and Saxitoxin Biosynthesis through Comprehensive Transcriptome Survey. Biology 2021, 10, 826. [Google Scholar] [CrossRef] [PubMed]
- Carnicer, O.; Hu, Y.-Y.; Ebenezer, V.; Irwin, A.J.; Finkel, Z.V. Genomic architecture constrains macromolecular allocation in dinoflagellates. Protist 2023, 174, 125992. [Google Scholar] [CrossRef] [PubMed]
- Santiago-Vazquez, L.Z.; Ranzer, L.K.; Kerr, R.G. Comparison of two total RNA extraction protocols using the marine gorgonian coral Pseudopterogorgia elisabethae and its symbiont Symbiodinium sp. Electron. J. Biotechnol. 2006, 9, 598–603. [Google Scholar] [CrossRef]
- Mylne, J.S.; Mason, M.G.; Botella, J.R. Rapid isolation of high-quality RNA from symbiotic dinoflagellates. Phycologia 1998, 37, 307–309. [Google Scholar] [CrossRef]
- Rosic, N.N.; Hoegh-Guldberg, O. A method for extracting a high-quality RNA from Symbiodinium sp. J. Appl. Phycol. 2009, 22, 139–146. [Google Scholar] [CrossRef]
- Guillard, R.R.; Ryther, J.H. Studies of marine planktonic diatoms: I. Cyclotella nana hustedt, and detonula Confervacea (cleve) Gran. Can. J. Microbiol. 1962, 8, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lin, S. Complex Gene Structure of The Form Ii Rubisco In The Dinoflagellate Prorocentrum Minimum (Dinophyceae). J. Phycol. 2003, 39, 1160–1171. [Google Scholar] [CrossRef]
- Schiwitza, S.; Arndt, H.; Nitsche, F. First description of an euryoecious acanthoecid choanoflagellate species, Enibas tolerabilis gen. et sp. nov. from a salar in the Chilean Andes based on morphological and transcriptomic data. Eur. J. Protistol. 2018, 67, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Strand, C.; Enell, J.; Hedenfalk, I.; Fernö, M. RNA quality in frozen breast cancer samples and the influence on gene expression analysis—A comparison of three evaluation methods using microcapillary electrophoresis traces. BMC Mol. Biol. 2007, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, A.; Mueller, O.; Stocker, S.; Salowsky, R.; Leiber, M.; Gassmann, M.; Lightfoot, S.; Menzel, W.; Granzow, M.; Ragg, T. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 2006, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Babovskaya, A.A.; Trifonova, E.A.; Serebrova, V.N.; Svarovskaya, M.G.; Zarubin, A.A.; Zhilyakova, O.; Gabidulina, T.; Poltanova, A.A.; Rychkova, L.; Stepanov, V.A. Protocol of Transcriptome Analysis of Decidual Placenta Cells. Mol. Biol. 2022, 56, 276–282. [Google Scholar] [CrossRef]
- Futschik, M.E.; Kemmner, W.; Schäfer, R.; Sers, C. Chapter 7—The human transcriptome: Implications for the understanding of human disease. In Molecular Pathology; Coleman, W.B., Tsongalis, G.J., Eds.; Academic Press: Cambridge, MA, USA, 2009; pp. 123–149. Available online: https://www.sciencedirect.com/science/article/abs/pii/B978012374419700007X (accessed on 22 May 2024).
- Agilent Technologies. Sample Quality Control in Next-Generation Sequencing Workflows, 2020. Agilent Technologies Web Site. Available online: https://www.chem-agilent.com/appnote/pdf/application_ngs-sample-qc-automated-electrophoresis-5994-2142en-agilent.pdf (accessed on 22 May 2024).
- Aranda, P.S.; LaJoie, D.M.; Jorcyk, C.L. Bleach gel: A simple agarose gel for analyzing RNA quality. Electrophoresis 2012, 33, 366–369. [Google Scholar] [CrossRef]
- Ávalos, V.; Cameron, H.; Barría, S.; Riquelme, C.; Espinoza, O.; Guzmán, L.; Yarimizu, K.; Okazaki, M.; Nagai, S. Dinoflagellate toxins recorded during an extensive coastal bloom in northern Chile. In Proceedings of the 18th International Conference on Harmful Algae News, Nantes, France, 21–26 October 2018; Reguera, B., Bresnan, E., Eds.; Department of Biology, University of Copenhagen: Copenhagen, Denmark, 2019; pp. 14–15. [Google Scholar]
- Wang, D.-Z.; Dong, H.-P.; Li, C.; Xie, Z.-X.; Lin, L.; Hong, H.-S. Identification and Characterization of Cell Wall Proteins of a Toxic Dinoflagellate Alexandrium catenella Using 2-D DIGE and MALDI TOF-TOF Mass Spectrometry. Evid.-Based Complement. Altern. Med. 2011, 2011, 984080. [Google Scholar] [CrossRef] [PubMed]
- Fleige, S.; Pfaffl, M.W. RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Aspects Med. 2006, 27, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Monstein, H.J.; Nylander, A.G.; Chen, D. RNA extraction from gastrointestinal tract and pancreas by a modified Chomczynski and Sacchi method. Biotechniques 1995, 19, 340–344. [Google Scholar] [PubMed]

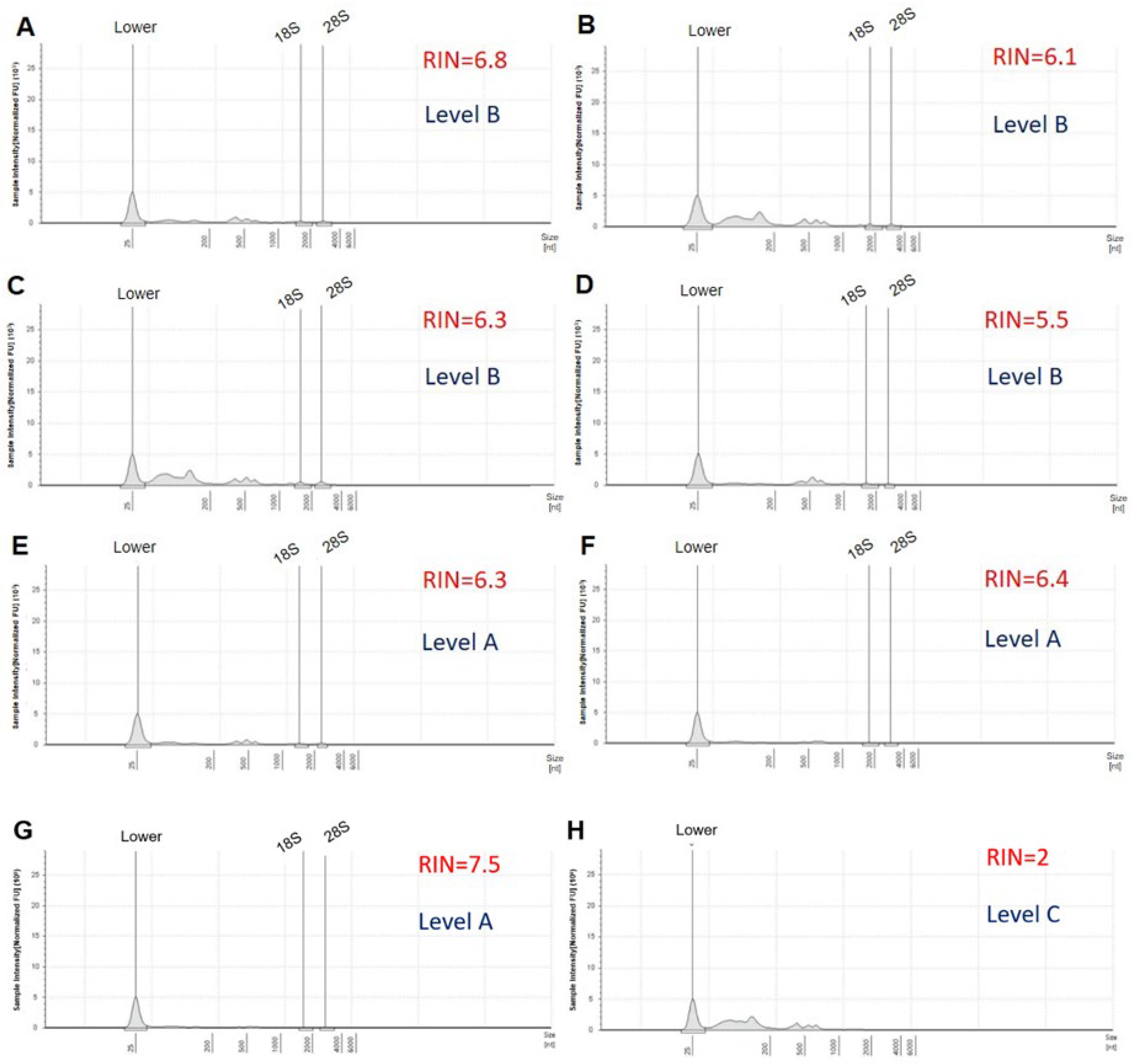

| Samples | (C) ng/µL | (V) µL | (M) µg | RIN | 28S/18S | Level | Remark |
|---|---|---|---|---|---|---|---|
| 1 | 127 | 33 | 4.19 | 6.8 | 0.9 | B | 5S peak is high |
| 2 | 187 | 89 | 16.64 | 6.1 | 0.8 | B | 5S peak is high |
| 3 | 310 | 89 | 27.59 | 6.3 | 1 | B | 5S peak is high |
| 4 | 203 | 35 | 7.11 | 5.5 | 0.6 | B | RIN < 6; 28S/18S < 0.8 |
| 5 | 195 | 30 | 5.85 | 6.3 | 1 | A | |
| 6 | 165 | 31 | 5.12 | 6.4 | 0.8 | A | |
| 7 | 311 | 84 | 26.12 | 7.5 | 1.1 | A | |
| 8 | 183.07 | 35 | 6.41 | 5.6 | 0.7 | B | RIN < 6; 28S/18S < 0.8 |
| 9 | 331.14 | 60 | 19.87 | 5.7 | 0.9 | B | RIN < 6; 5S peak is high |
| 10 | 420.00 | 95 | 39.90 | 7.8 | 0.6 | B | 28S/18S < 0.8; 5S peak is high |
| 11 | 296.78 | 65 | 19.29 | 5 | 0.8 | B | RIN < 6; 28S/18S < 0.8; 5S peak is high |
| 12 | 94.22 | 33 | 3.39 | 5.6 | 0.7 | B | RIN < 6; 28S/18S < 0.8; 5S peak is high |
| 13 | 221 | 32 | 7.07 | 2 | 0 | C | RIN < 6; 28S/18S < 0.8 |
| 14 | 545.98 | 32 | 17.47 | 1 | 0 | C | RIN < 6; 28S/18S < 0.8; 5S peak is high |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huarachi-Olivera, R.; Teresa Mata, M.; Ardiles-Candia, A.; Escobar-Méndez, V.; Gatica-Cortes, C.; Ahumada, M.; Orrego, J.; Vidal-Veuthey, B.; Cárdenas, J.P.; González, L.; et al. Modification of the Trizol Method for the Extraction of RNA from Prorocentrum triestinum ACIZ_LEM2. Int. J. Mol. Sci. 2024, 25, 9642. https://doi.org/10.3390/ijms25179642
Huarachi-Olivera R, Teresa Mata M, Ardiles-Candia A, Escobar-Méndez V, Gatica-Cortes C, Ahumada M, Orrego J, Vidal-Veuthey B, Cárdenas JP, González L, et al. Modification of the Trizol Method for the Extraction of RNA from Prorocentrum triestinum ACIZ_LEM2. International Journal of Molecular Sciences. 2024; 25(17):9642. https://doi.org/10.3390/ijms25179642
Chicago/Turabian StyleHuarachi-Olivera, Ronald, María Teresa Mata, Alonso Ardiles-Candia, Valentina Escobar-Méndez, Carlos Gatica-Cortes, Matías Ahumada, José Orrego, Boris Vidal-Veuthey, Juan P. Cárdenas, Leonel González, and et al. 2024. "Modification of the Trizol Method for the Extraction of RNA from Prorocentrum triestinum ACIZ_LEM2" International Journal of Molecular Sciences 25, no. 17: 9642. https://doi.org/10.3390/ijms25179642
APA StyleHuarachi-Olivera, R., Teresa Mata, M., Ardiles-Candia, A., Escobar-Méndez, V., Gatica-Cortes, C., Ahumada, M., Orrego, J., Vidal-Veuthey, B., Cárdenas, J. P., González, L., & Riquelme, C. (2024). Modification of the Trizol Method for the Extraction of RNA from Prorocentrum triestinum ACIZ_LEM2. International Journal of Molecular Sciences, 25(17), 9642. https://doi.org/10.3390/ijms25179642






