Exploring the Roles of the Swi2/Snf2 Gene Family in Maize Abiotic Stress Responses
Abstract
:1. Introduction
2. Results
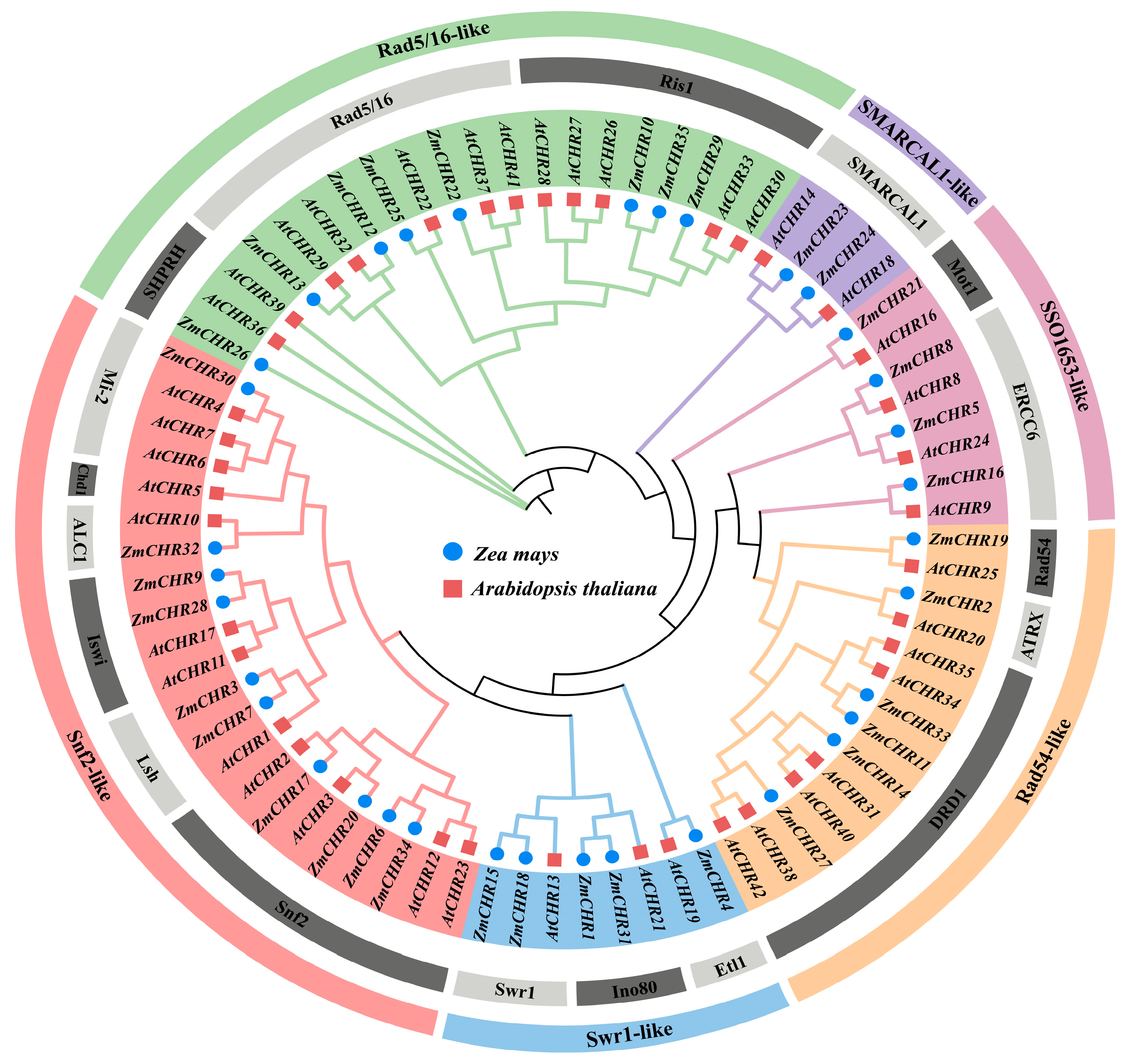
2.1. Identification of the Maize Snf2 Gene Family
2.2. Analysis of Physicochemical Properties of Maize Snf2-Family Proteins
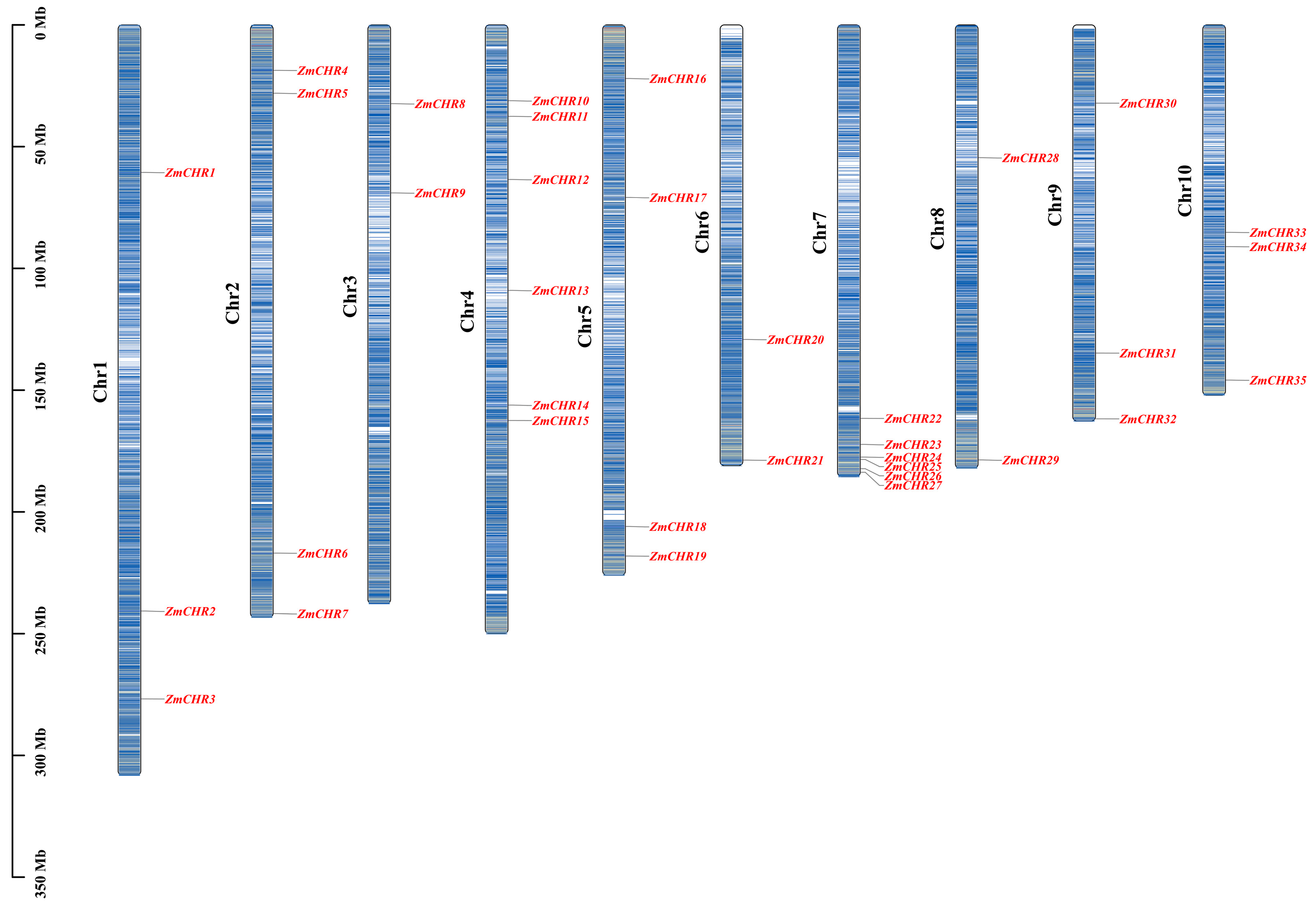
2.3. Chromosomal Distribution of Maize Snf2 Genes
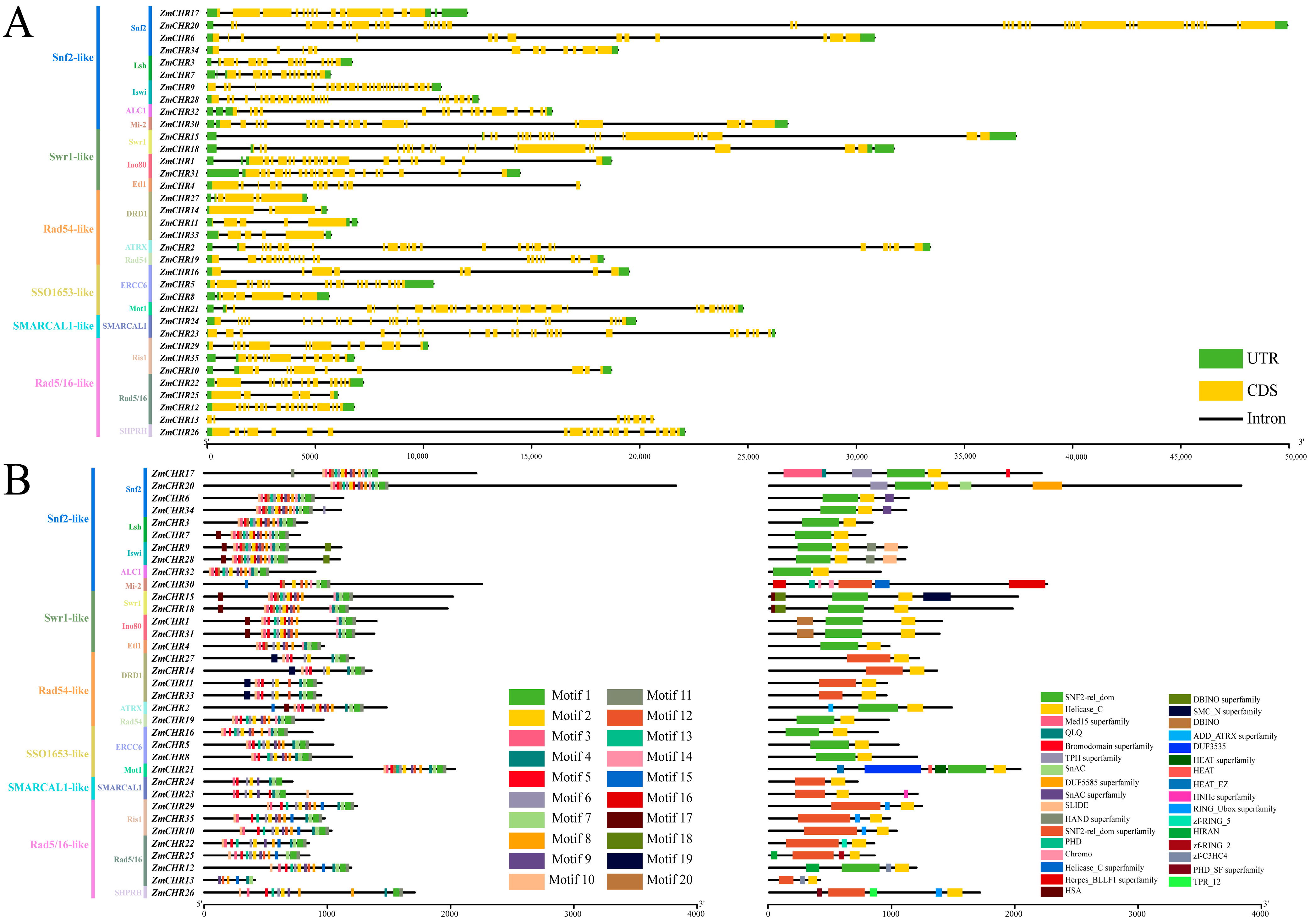
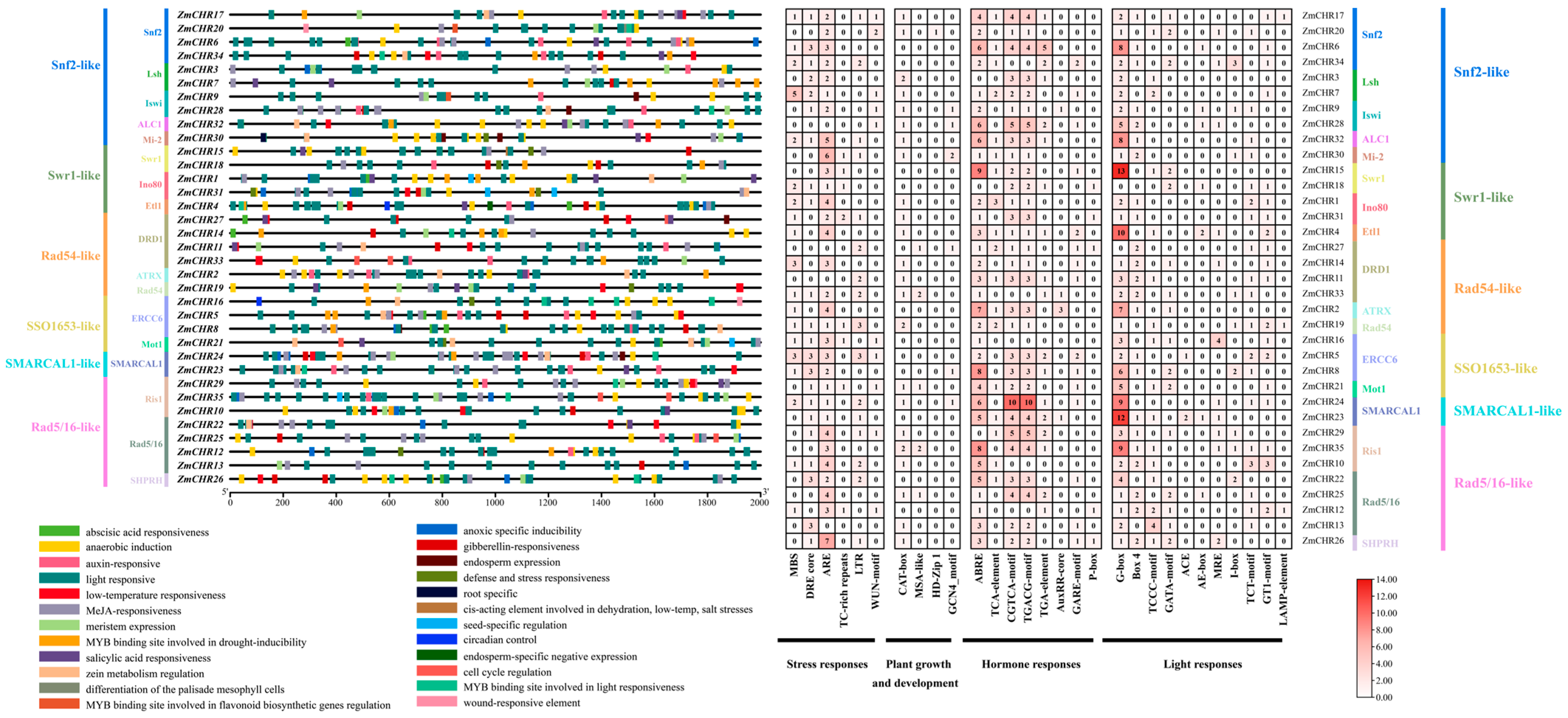
2.4. Gene Structure and Protein Domain Analysis of the Maize Snf2 Gene Family Members
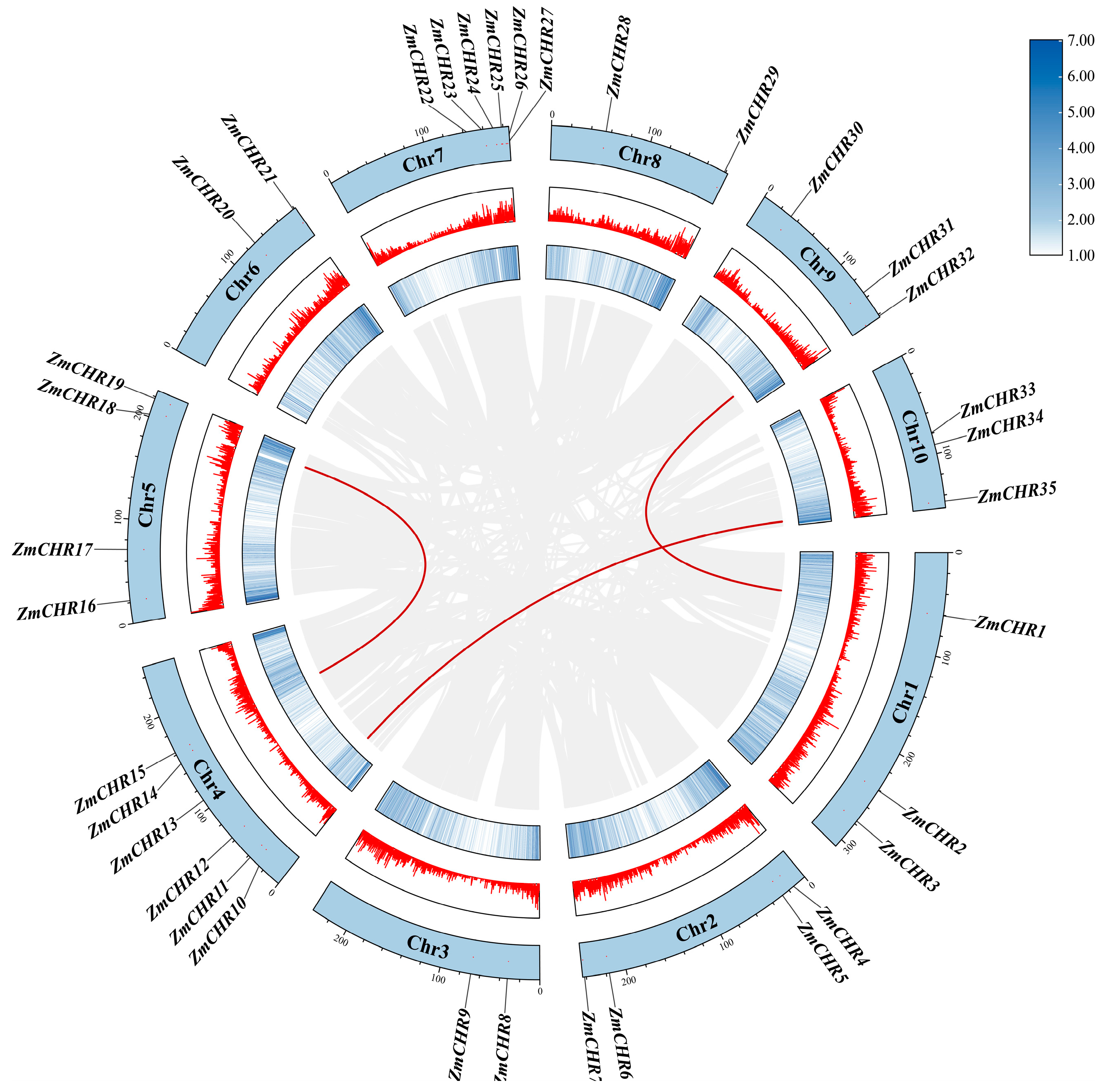
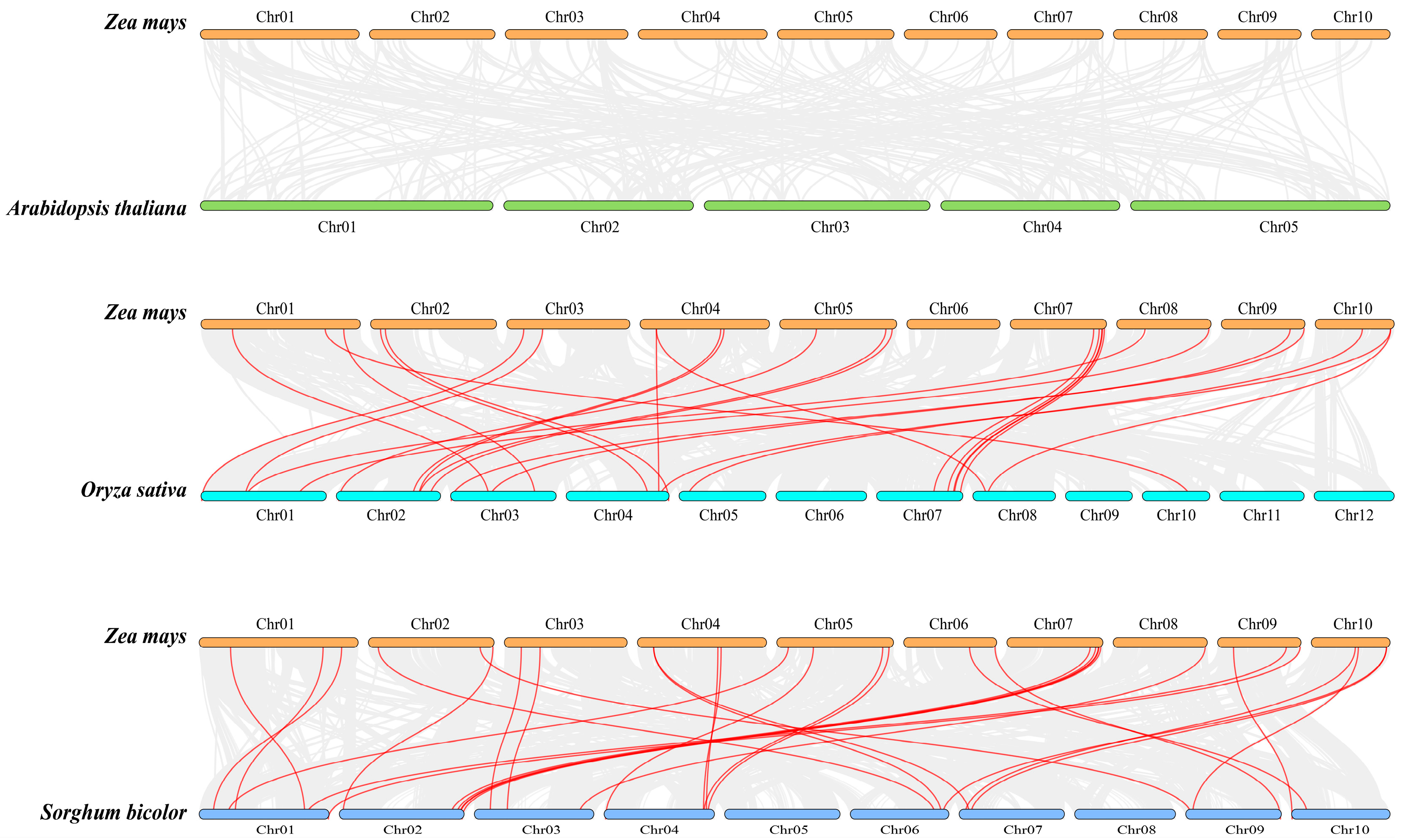
2.5. Collinearity Analysis of the Maize Snf2 Gene Family
2.6. Analysis of Cis-Regulatory Elements in the Promoters of Maize Snf2 Genes
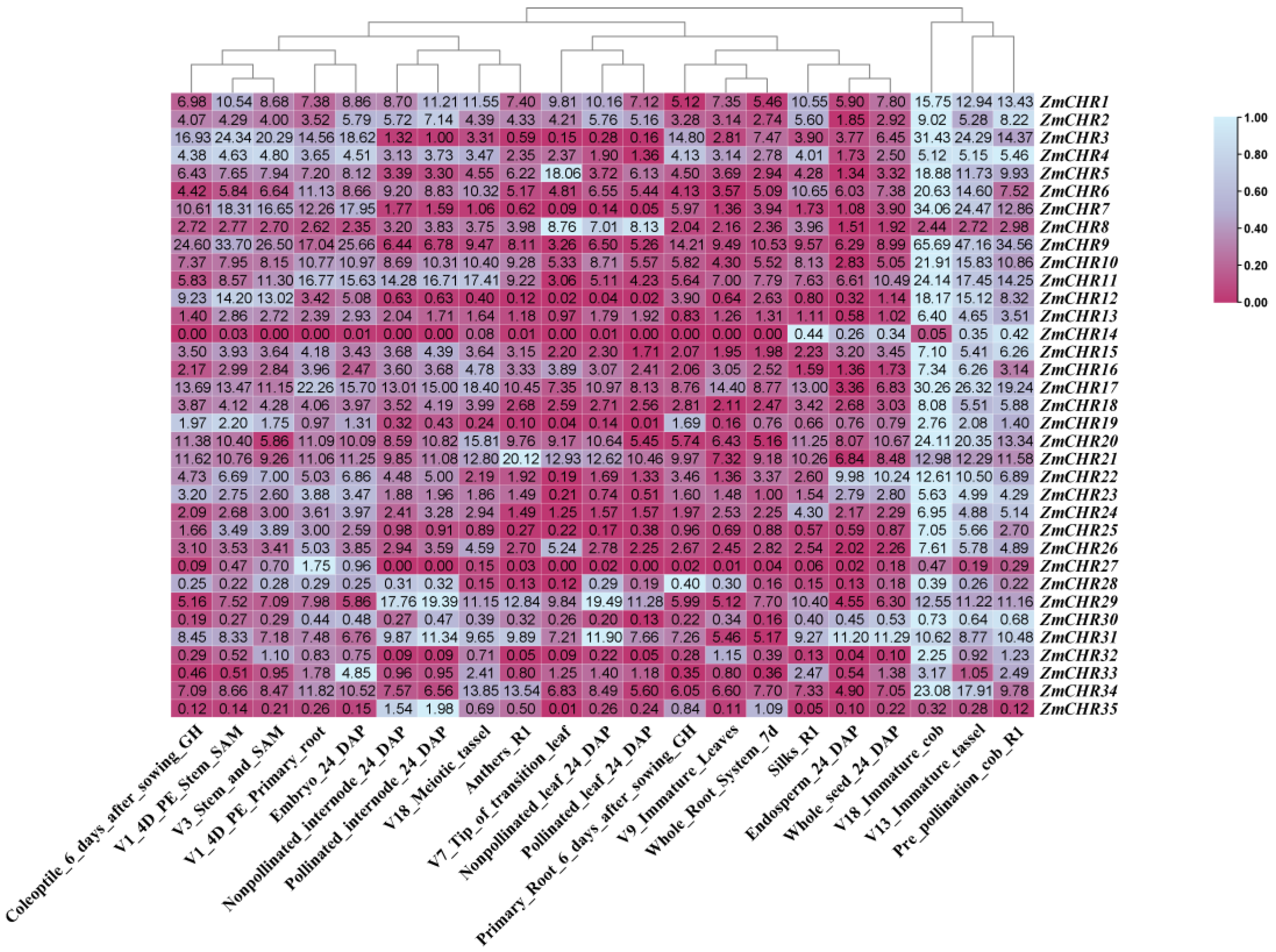
2.7. Expression Analysis of Maize Snf2 Genes in Various Tissues
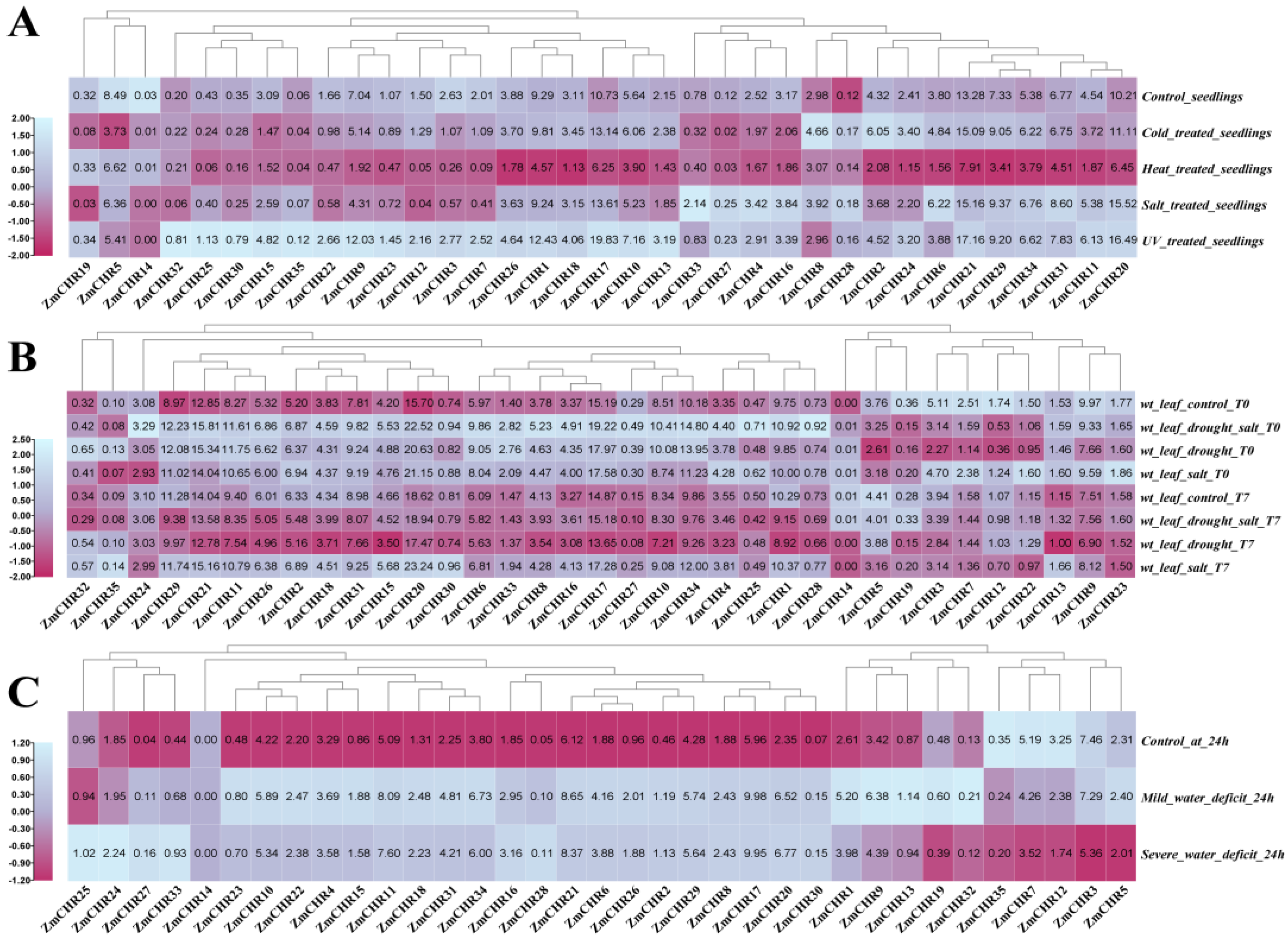
2.8. Expression Analysis of Maize Snf2 Genes under Various Stress Conditions
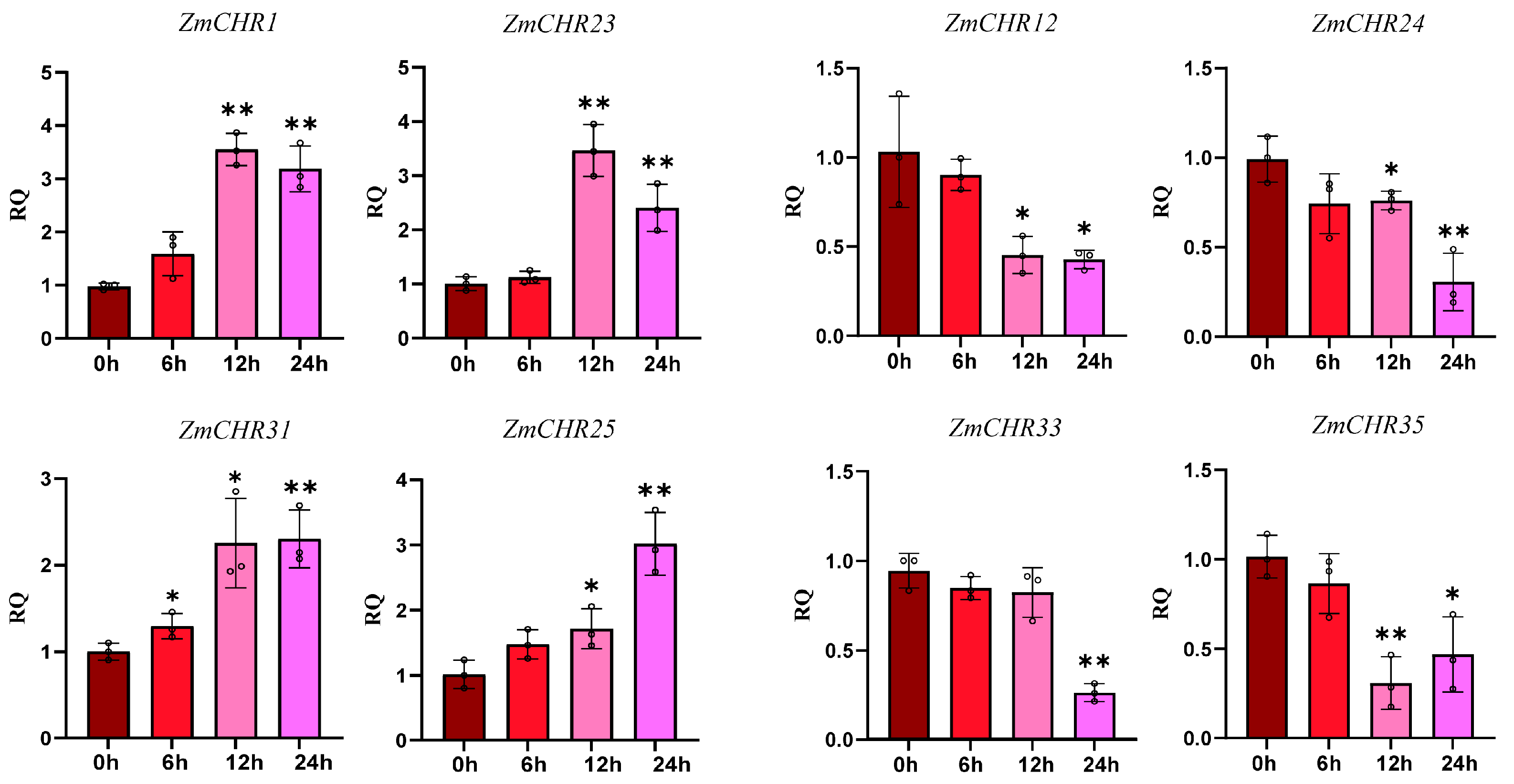
2.9. Analysis of Maize Snf2 Gene Expression under Drought Stress and qPCR Validation
3. Discussion
3.1. Functional Domains and Evolutionary Dynamics of the Maize Snf2 Gene Family
3.2. Function and Diversity: The Role of the Maize Snf2 Gene Family in Chromatin Remodeling
3.3. Differential Expression Patterns of Maize Snf2 Gene in Various Organs
3.4. Role of the Maize Snf2 Gene Family in Response to Abiotic Stress
4. Materials and Methods
4.1. Identification and Physicochemical Property Prediction of the Maize Snf2 Gene
4.2. Chromosomal Localization of the Maize Snf2 Gene
4.3. Phylogenetic, Gene Structure, and Conserved Motif Analysis of the Maize Snf2 Gene
4.4. Collinearity Analysis and Gene Duplication of the Maize Snf2 Gene
4.5. Analysis of Cis-Acting Elements in the Promoter Regions of the Maize Snf2 Gene
4.6. Expression Analysis of the Snf2 Gene Family in Maize
4.7. Plant Material and Stress Treatment Procedures
4.8. RNA Extraction and Quantitative Real-Time PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ntiamoah, E.B.; Li, D.; Appiah-Otoo, I.; Twumasi, M.A.; Yeboah, E.N. Towards a sustainable food production: Modelling the impacts of climate change on maize and soybean production in Ghana. Environ. Sci. Pollut. Res. 2022, 29, 72777–72796. [Google Scholar] [CrossRef] [PubMed]
- Shiferaw, B.; Prasanna, B.M.; Hellin, J.; Bänziger, M. Crops that feed the world 6. Past successes and future challenges to the role played by maize in global food security. Food Secur. 2011, 3, 307–327. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Liu, B.; Piao, S.; Wang, X.; Lobell, D.B.; Huang, Y.; Huang, M.T.; Yao, Y.T.; Bassu, S.; Ciais, P.; et al. Temperature increase reduces global yields of major crops in four independent estimates. Proc. Natl. Acad. Sci. USA 2017, 114, 9326–9331. [Google Scholar] [CrossRef] [PubMed]
- van Bakel, H.; Hughes, T. (Eds.) Interactions of Transcription Factors with Chromatin; Springer: Dordrecht, The Netherlands, 2011; Volume 52. [Google Scholar]
- Singh, A.; Modak, S.B.; Chaturvedi, M.M.; Purohit, J.S. SWI/SNF chromatin remodelers: Structural, functional and mechanistic implications. Cell Biochem. Biophys. 2023, 81, 167–187. [Google Scholar] [CrossRef]
- Wang, J.; Sun, Z.; Liu, H.; Yue, L.; Wang, F.; Liu, S.; Su, B.; Liu, B.; Kong, F.; Fang, C. Genome-wide identification and characterization of the soybean Snf2 gene family and expression response to rhizobia. Int. J. Mol. Sci. 2023, 24, 7250. [Google Scholar] [CrossRef]
- Ryan, D.P.; Owen-Hughes, T. Snf2-family proteins: Chromatin remodellers for any occasion. Curr. Opin. Chem. Biol. 2011, 15, 649–656. [Google Scholar] [CrossRef]
- Cairns, B.R.; Kim, Y.J.; Sayre, M.H.; Laurent, B.C.; Kornberg, R.D. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc. Natl. Acad. Sci. USA 1994, 91, 1950–1954. [Google Scholar] [CrossRef]
- Ojolo, S.P.; Cao, S.; Priyadarshani, S.; Li, W.; Yan, M.; Aslam, M.; Zhao, H.; Qin, Y. Regulation of plant growth and development: A review from a chromatin remodeling perspective. Front. Plant Sci. 2018, 9, 1232. [Google Scholar] [CrossRef]
- Arnholdt-Schmitt, B. Stress-induced cell reprogramming. A role for global genome regulation. Plant Physiol. 2004, 136, 2579–2586. [Google Scholar] [CrossRef]
- Ashapkin, V.V.; Kutueva, L.I.; Aleksandrushkina, N.I.; Vanyushin, B.F. Epigenetic mechanisms of plant adaptation to biotic and abiotic stresses. Int. J. Mol. Sci. 2020, 21, 7457. [Google Scholar] [CrossRef]
- Song, Z.T.; Liu, J.X.; Han, J.J. Chromatin remodeling factors regulate environmental stress responses in plants. J. Integr. Plant Biol. 2021, 63, 438–450. [Google Scholar] [CrossRef]
- Sarnowska, E.; Gratkowska, D.M.; Sacharowski, S.P.; Cwiek, P.; Tohge, T.; Fernie, A.R.; Siedlecki, J.A.; Koncz, C.; Sarnowski, T.J. The role of SWI/SNF chromatin remodeling complexes in hormone crosstalk. Trends Plant Sci. 2016, 21, 594–608. [Google Scholar] [CrossRef]
- Walley, J.W.; Rowe, H.C.; Xiao, Y.; Chehab, E.W.; Kliebenstein, D.J.; Wagner, D.; Dehesh, K. The chromatin remodeler SPLAYED regulates specific stress signaling pathways. PLoS Pathog. 2008, 4, e1000237. [Google Scholar] [CrossRef]
- Guo, T.; Wang, D.; Fang, J.; Zhao, J.; Yuan, S.; Xiao, L.; Li, X. Mutations in the rice OsCHR4 gene, encoding a CHD3 family chromatin remodeler, induce narrow and rolled leaves with increased cuticular wax. Int. J. Mol. Sci. 2019, 20, 2567. [Google Scholar] [CrossRef]
- Mlynárová, L.; Nap, J.P.; Bisseling, T. The SWI/SNF chromatin-remodeling gene AtCHR12 mediates temporary growth arrest in Arabidopsis thaliana upon perceiving environmental stress. Plant J. 2007, 51, 874–885. [Google Scholar] [CrossRef]
- Yu, X.; Meng, X.; Liu, Y.; Li, N.; Zhang, A.; Wang, T.-J.; Jiang, L.; Pang, J.; Zhao, X.; Qi, X.; et al. The chromatin remodeler ZmCHB101 impacts expression of osmotic stress-responsive genes in maize. Plant Mol. Biol. 2018, 97, 451–465. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, D.; Gan, T.; Liu, L.; Long, W.; Wang, Y.; Niu, M.; Li, X.; Zheng, M.; Jiang, L.; et al. CRL6, a member of the CHD protein family, is required for crown root development in rice. Plant Physiol. Biochem. 2016, 105, 185–194. [Google Scholar] [CrossRef]
- Campos-Rivero, G.; Osorio-Montalvo, P.; Sánchez-Borges, R.; Us-Camas, R.; Duarte-Aké, F.; De-La-Peña, C. Plant hormone signaling in flowering: An epigenetic point of view. J. Plant Physiol. 2017, 214, 16–27. [Google Scholar] [CrossRef]
- Flaus, A.; Martin, D.M.A.; Barton, G.J.; Owen-Hughes, T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006, 34, 2887–2905. [Google Scholar] [CrossRef]
- Knizewski, L.; Ginalski, K.; Jerzmanowski, A. Snf2 proteins in plants: Gene silencing and beyond. Trends Plant Sci. 2008, 13, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhu, N.; Wang, X.; Yi, Q.; Zhu, D.; Lai, Y.; Zhao, Y. Analysis of rice Snf2 family proteins and their potential roles in epigenetic regulation. Plant Physiol. Bioch. 2013, 70, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, S.; Yang, P.; Yang, J.; Yang, S.; Wu, K. Identification and expression analysis of Snf2 family proteins in tomato (Solanum lycopersicum). Int. J. Genom. 2019, 2019, 5080935. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Mishina, K.; Zhu, H.; Kikuchi, S.; Sassa, H.; Oono, Y.; Komatsuda, T. Genome-wide analysis of Snf2 gene family reveals potential role in regulation of spike development in barley. Int. J. Mol. Sci. 2022, 24, 457. [Google Scholar] [CrossRef] [PubMed]
- Makarevitch, I.; Waters, A.J.; West, P.T.; Stitzer, M.; Hirsch, C.N.; Ross-Ibarra, J.; Springer, N.M. Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genet. 2015, 11, e1004915. [Google Scholar]
- Forestan, C.; Aiese Cigliano, R.; Farinati, S.; Lunardon, A.; Sanseverino, W.; Varotto, S. Stress-induced and epigenetic-mediated maize transcriptome regulation study by means of transcriptome reannotation and differential expression analysis. Sci. Rep. 2016, 6, 30446. [Google Scholar] [CrossRef]
- Opitz, N.; Paschold, A.; Marcon, C.; Malik, W.A.; Lanz, C.; Piepho, H.-P.; Hochholdinger, F. Transcriptomic complexity in young maize primary roots in response to low water potentials. BMC Genom. 2014, 15, 741. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, X.; Zhou, C.; He, Z.; Shen, X. Genome-wide identification of chromatin regulators in Sorghum bicolor. 3 Biotech 2022, 12, 117. [Google Scholar] [CrossRef]
- Kwon, C.S.; Wagner, D. Unwinding chromatin for development and growth: A few genes at a time. Trends Genet. 2007, 23, 403–412. [Google Scholar] [CrossRef]
- Vincent, J.A.; Kwong, T.J.; Tsukiyama, T. ATP-dependent chromatin remodeling shapes the DNA replication landscape. Nat. Struct. Mol. Biol. 2008, 15, 477–484. [Google Scholar] [CrossRef]
- Raab, J.R.; Resnick, S.; Magnuson, T. Genome-wide transcriptional regulation mediated by biochemically distinct SWI/SNF complexes. PLoS Genet. 2015, 11, e1005748. [Google Scholar] [CrossRef] [PubMed]
- Ceballos, S.J.; Heyer, W.-D. Functions of the Snf2/Swi2 family Rad54 motor protein in homologous recombination. BBA-Gene Regul. Mech. 2011, 1809, 509–523. [Google Scholar] [CrossRef]
- Lans, H.; Marteijn, J.A.; Vermeulen, W. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenet. Chromatin 2012, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Peng, R.; Wang, J.; Yang, S. Functions of chromatin remodeling factors during plant development. J. Trop. Subtrop. Bot. 2019, 27, 565–579. [Google Scholar]
- Han, S.-K.; Wagner, D. Role of chromatin in water stress responses in plants. J. Exp. Bot. 2014, 65, 2785–2799. [Google Scholar] [CrossRef]
- Mohammadi, M.A.; Han, X.; Zhang, Z.; Xi, Y.; Boorboori, M.; Wang-Pruski, G. Phosphite application alleviates pythophthora infestans by modulation of photosynthetic and physio-biochemical metabolites in potato leaves. Pathogens 2020, 9, 170. [Google Scholar] [CrossRef]
- Fahad, M.; Liu, C.; Shen, Y.; Sajid, M.; Wu, L. enomic exploration and functional insights into the diversification of the snf2 gene family in subgenomes of Triticum aestivum. Crop Des. 2024, 3, 100050. [Google Scholar]
- Dai, X.; Shen, L. Advances and trends in omics technology development. Front. Med. 2022, 9, 911861. [Google Scholar] [CrossRef]
- Yu, X.; Jiang, L.; Wu, R.; Meng, X.; Zhang, A.; Li, N.; Xia, Q.; Qi, X.; Pang, J.; Xu, Z.-Y.; et al. The core subunit of a chromatin-remodeling complex, ZmCHB101, plays essential roles in maize growth and development. Sci. Rep. 2016, 6, 38504. [Google Scholar] [CrossRef]
- Long, J.C.; Xia, A.A.; Liu, J.H.; Jing, J.L.; Wang, Y.Z.; Qi, C.Y.; He, Y. Decrease in DNA methylation 1 (DDM1) is required for the formation of mCHH islands in maize. J. Integr. Plant Biol. 2019, 61, 749–764. [Google Scholar] [CrossRef]
- Guo, M.; Zhao, H.; He, Z.; Zhang, W.; She, Z.; Mohammadi, M.A.; Shi, C.; Yan, M.; Tian, D.; Qin, Y. Comparative expression profiling of Snf2 family genes during reproductive development and stress responses in rice. Front. Plant Sci. 2022, 13, 910663. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, Q.; Zhu, G.; Zhang, F.; Fang, X.; Ren, H.; Jiang, J.; Yang, F.; Zhang, D.; Chen, F. CHR721, interacting with OsRPA1a, is essential for both male and female reproductive development in rice. Plant Mol. Biol. 2020, 103, 473–487. [Google Scholar] [CrossRef] [PubMed]
- Jing, Y.; Guo, Q.; Lin, R. The chromatin-remodeling factor PICKLE antagonizes polycomb repression of FT to promote flowering. Plant Physiol. 2019, 181, 656–668. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chen, C.; Gao, L.; Yang, S.; Nguyen, V.; Shi, X.; Siminovitch, K.; Kohalmi, S.E.; Huang, S.; Wu, K.; et al. The Arabidopsis SWI2/SNF2 chromatin remodeler BRAHMA regulates polycomb function during vegetative development and directly activates the flowering repressor gene SVP. PLoS Genet. 2015, 11, e1004944. [Google Scholar] [CrossRef]
- Zhao, M.; Yang, S.; Chen, C.Y.; Li, C.; Shan, W.; Lu, W.; Cui, Y.; Liu, X.; Wu, K. Arabidopsis BREVIPEDICELLUS interacts with the SWI2/SNF2 chromatin remodeling ATPase BRAHMA to regulate KNAT2 and KNAT6 expression in control of inflorescence architecture. PLoS Genet. 2015, 11, e1005125. [Google Scholar] [CrossRef]
- Yang, R.; Hong, Y.; Ren, Z.; Tang, K.; Zhang, H.; Zhu, J.-K.; Zhao, C. A role for PICKLE in the regulation of cold and salt stress tolerance in Arabidopsis. Front. Plant Sci. 2019, 10, 900. [Google Scholar] [CrossRef]
- Han, S.K.; Sang, Y.; Rodrigues, A.; Wu, M.F.; Rodriguez, P.L.; Wagner, D. The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis. Plant Cell 2012, 24, 4892–4906. [Google Scholar] [CrossRef]
- Chen, Q.; Shi, X.; Ai, L.; Tian, X.; Zhang, H.; Tian, J.; Wang, Q.; Zhang, M.; Cui, S.; Yang, C.; et al. Genome-wide identification of genes encoding SWI/SNF components in soybean and the functional characterization of GmLFR1 in drought-stressed plants. Front. Plant Sci. 2023, 14, 1176376. [Google Scholar] [CrossRef]
- Yang, J.; Chang, Y.; Qin, Y.; Chen, D.; Zhu, T.; Peng, K.; Wang, H.; Tang, N.; Li, X.; Wang, Y.; et al. A lamin-like protein OsNMCP1 regulates drought resistance and root growth through chromatin accessibility modulation by interacting with a chromatin remodeller OsSWI3C in rice. New Phytol. 2020, 227, 65–83. [Google Scholar] [CrossRef]
- Casati, P.; Stapleton, A.E.; Blum, J.E.; Walbot, V. Genome-wide analysis of high-altitude maize and gene knockdown stocks implicates chromatin remodeling proteins in response to UV-B. Plant J. 2006, 46, 613–627. [Google Scholar] [CrossRef]
- Stelpflug, S.C.; Sekhon, R.S.; Vaillancourt, B.; Hirsch, C.N.; Buell, C.R.; de Leon, N.; Kaeppler, S.M. An Expanded Maize Gene Expression Atlas based on RNA Sequencing and its Use to Explore Root Development. Plant Genome 2016, 9. [Google Scholar] [CrossRef] [PubMed]
- Mo, F.; Zhang, N.; Qiu, Y.; Meng, L.; Cheng, M.; Liu, J.; Yao, L.; Lv, R.; Liu, Y.; Zhang, Y.; et al. Molecular Characterization, Gene Evolution and Expression Analysis of the F-Box Gene Family in Tomato (Solanum lycopersicum). Genes 2021, 12, 417. [Google Scholar] [CrossRef] [PubMed]
| Gene Name | Sequence ID | Length (aa) | MW (kDa) | pI | Instability Index | Aliphatic Index | GRAVY | Predicted Location(s) |
|---|---|---|---|---|---|---|---|---|
| ZmCHR1 | Zm00001eb017000_T001 | 1398 | 159.89 | 8.11 | 48.88 | 83.53 | −0.529 | Nuclear |
| ZmCHR2 | Zm00001eb046650_T001 | 1481 | 168.18 | 5.46 | 51.9 | 76.91 | −0.674 | Nuclear |
| ZmCHR3 | Zm00001eb055640_T002 | 837 | 94.16 | 5.46 | 45.07 | 92.19 | −0.417 | Nuclear |
| ZmCHR4 | Zm00001eb073870_T001 | 973 | 109.04 | 6.12 | 52.05 | 81.01 | −0.525 | Nuclear |
| ZmCHR5 | Zm00001eb076670_T001 | 1048 | 116.95 | 5.85 | 42.11 | 83.11 | −0.533 | Nuclear |
| ZmCHR6 | Zm00001eb108690_T001 | 1129 | 128.88 | 6.78 | 52.49 | 80.88 | −0.629 | Nuclear |
| ZmCHR7 | Zm00001eb117870_T001 | 779 | 88.40 | 6.21 | 44.7 | 90.8 | −0.461 | Cytoplasmic |
| ZmCHR8 | Zm00001eb127010_T001 | 1198 | 133.40 | 7.64 | 48.2 | 79.47 | −0.609 | Nuclear |
| ZmCHR9 | Zm00001eb131790_T002 | 1114 | 128.43 | 5.22 | 49.03 | 72.56 | −0.851 | Nuclear |
| ZmCHR10 | Zm00001eb171780_T003 | 1033 | 114.16 | 7.75 | 47.19 | 80.92 | −0.455 | Nuclear |
| ZmCHR11 | Zm00001eb173050_T002 | 952 | 106.75 | 6.61 | 40.88 | 86.13 | −0.418 | Nuclear |
| ZmCHR12 | Zm00001eb176980_T001 | 1193 | 132.33 | 8.52 | 48.01 | 87.04 | −0.248 | Nuclear |
| ZmCHR13 | Zm00001eb181350_T001 | 411 | 46.27 | 8.56 | 64.6 | 89.93 | −0.185 | Nuclear |
| ZmCHR14 | Zm00001eb186260_T001 | 1360 | 151.36 | 6.43 | 51.85 | 74.21 | −0.675 | Nuclear |
| ZmCHR15 | Zm00001eb187690_T003 | 2018 | 231.23 | 5.38 | 50.32 | 77.33 | −0.654 | Nuclear |
| ZmCHR16 | Zm00001eb220010_T002 | 879 | 99.64 | 8.65 | 48.91 | 81.95 | −0.501 | Nuclear |
| ZmCHR17 | Zm00001eb229760_T001 | 2208 | 246.56 | 8.85 | 56.41 | 66.3 | −0.815 | Nuclear |
| ZmCHR18 | Zm00001eb251100_T003 | 1975 | 226.47 | 5.29 | 52.17 | 76.95 | −0.653 | Nuclear |
| ZmCHR19 | Zm00001eb255780_T002 | 968 | 107.38 | 6.42 | 49.17 | 81.27 | −0.46 | Nuclear |
| ZmCHR20 | Zm00001eb280740_T001 | 3828 | 414.73 | 5.07 | 52.19 | 71.07 | −0.647 | Nuclear |
| ZmCHR21 | Zm00001eb297280_T001 | 2036 | 225.90 | 5.87 | 44.01 | 97.49 | −0.188 | Nuclear |
| ZmCHR22 | Zm00001eb322510_T001 | 850 | 95.87 | 8.63 | 51.51 | 76.42 | −0.48 | Nuclear |
| ZmCHR23 | Zm00001eb326710_T001 | 1202 | 135.36 | 8.39 | 50.3 | 88.78 | −0.38 | Nuclear |
| ZmCHR24 | Zm00001eb328680_T001 | 717 | 78.84 | 9.66 | 50.48 | 94.3 | −0.153 | Mitochondrial |
| ZmCHR25 | Zm00001eb329060_T003 | 854 | 95.16 | 8.68 | 41.69 | 90.91 | −0.265 | Nuclear |
| ZmCHR26 | Zm00001eb330720_T003 | 1709 | 193.32 | 6.68 | 48.47 | 84.96 | −0.375 | Nuclear |
| ZmCHR27 | Zm00001eb331510_T001 | 1214 | 137.93 | 8.05 | 43.49 | 82.06 | −0.404 | Nuclear |
| ZmCHR28 | Zm00001eb341650_T002 | 1102 | 127.34 | 5.25 | 48.09 | 72.99 | −0.853 | Nuclear |
| ZmCHR29 | Zm00001eb369880_T003 | 1239 | 138.32 | 5.15 | 52.28 | 72.32 | −0.513 | Nuclear |
| ZmCHR30 | Zm00001eb379660_T002 | 2254 | 249.35 | 6.04 | 53.3 | 84.63 | −0.444 | Nuclear |
| ZmCHR31 | Zm00001eb394610_T002 | 1380 | 157.94 | 8.79 | 46.64 | 82.78 | −0.553 | Cytoplasmic |
| ZmCHR32 | Zm00001eb404510_T002 | 904 | 101.65 | 6.55 | 46.13 | 94.21 | −0.214 | Nuclear |
| ZmCHR33 | Zm00001eb416780_T001 | 950 | 106.54 | 6.61 | 43.06 | 84.57 | −0.442 | Nuclear |
| ZmCHR34 | Zm00001eb417900_T002 | 1110 | 126.98 | 6.04 | 52.86 | 81.82 | −0.624 | Nuclear |
| ZmCHR35 | Zm00001eb431470_T001 | 979 | 109.84 | 6.29 | 51.09 | 78.77 | −0.464 | Nuclear |
| ZmCHR1 | Zm00001eb017000_T001 | 1398 | 159.89 | 8.11 | 48.88 | 83.53 | −0.529 | Nuclear |
| ZmCHR2 | Zm00001eb046650_T001 | 1481 | 168.18 | 5.46 | 51.9 | 76.91 | −0.674 | Nuclear |
| ZmCHR3 | Zm00001eb055640_T002 | 837 | 94.16 | 5.46 | 45.07 | 92.19 | −0.417 | Nuclear |
| ZmCHR4 | Zm00001eb073870_T001 | 973 | 109.04 | 6.12 | 52.05 | 81.01 | −0.525 | Nuclear |
| ZmCHR5 | Zm00001eb076670_T001 | 1048 | 116.95 | 5.85 | 42.11 | 83.11 | −0.533 | Nuclear |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, J.; Wang, Q.; Qian, B.; Liu, Q.; Wang, Z.; Liu, Y.; Chen, Z.; Wu, W.; Zhang, C.; Yin, Y. Exploring the Roles of the Swi2/Snf2 Gene Family in Maize Abiotic Stress Responses. Int. J. Mol. Sci. 2024, 25, 9686. https://doi.org/10.3390/ijms25179686
Han J, Wang Q, Qian B, Liu Q, Wang Z, Liu Y, Chen Z, Wu W, Zhang C, Yin Y. Exploring the Roles of the Swi2/Snf2 Gene Family in Maize Abiotic Stress Responses. International Journal of Molecular Sciences. 2024; 25(17):9686. https://doi.org/10.3390/ijms25179686
Chicago/Turabian StyleHan, Jiarui, Qi Wang, Buxuan Qian, Qing Liu, Ziyu Wang, Yang Liu, Ziqi Chen, Weilin Wu, Chuang Zhang, and Yuejia Yin. 2024. "Exploring the Roles of the Swi2/Snf2 Gene Family in Maize Abiotic Stress Responses" International Journal of Molecular Sciences 25, no. 17: 9686. https://doi.org/10.3390/ijms25179686




