Potential Involvements of Cilia-Centrosomal Genes in Primary Congenital Glaucoma
Abstract
1. Introduction
2. Results
2.1. Identification of Rare and Common Variants in CEP164 and INPP5E
2.2. Haplotype Analysis of CEP164 and INPP5E Genes
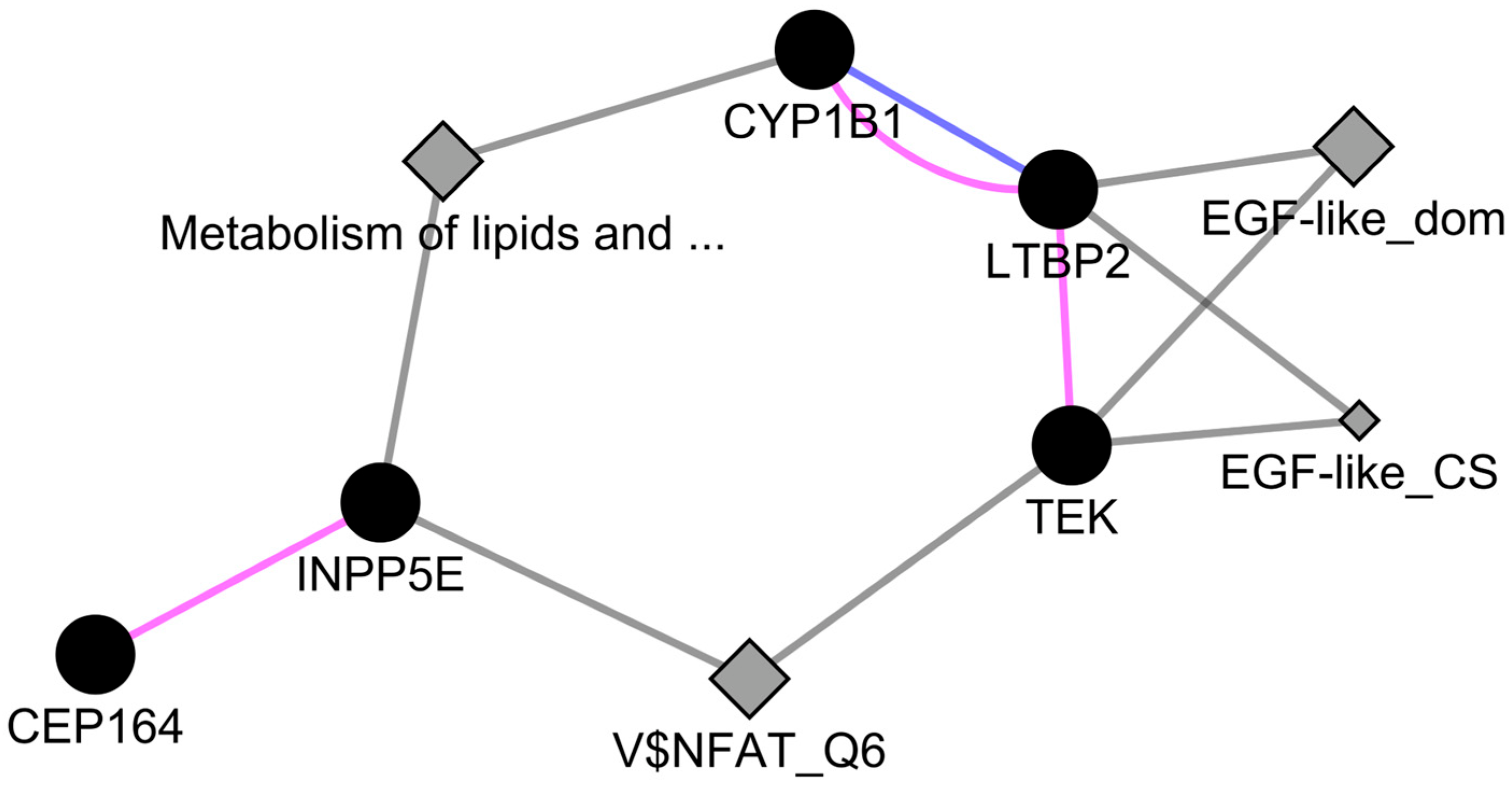
2.3. Genetic and Physical Interactions between CEP164 and CYP1B1
2.4. Genotype–Phenotype Correlation
3. Discussion
4. Materials and Methods
4.1. Study Approval
4.2. Enrolment of Cases and Controls
4.3. Targeted Sequencing, Cell Culture, and Pull-Down Assay
4.4. Allele and Haplotype Analysis
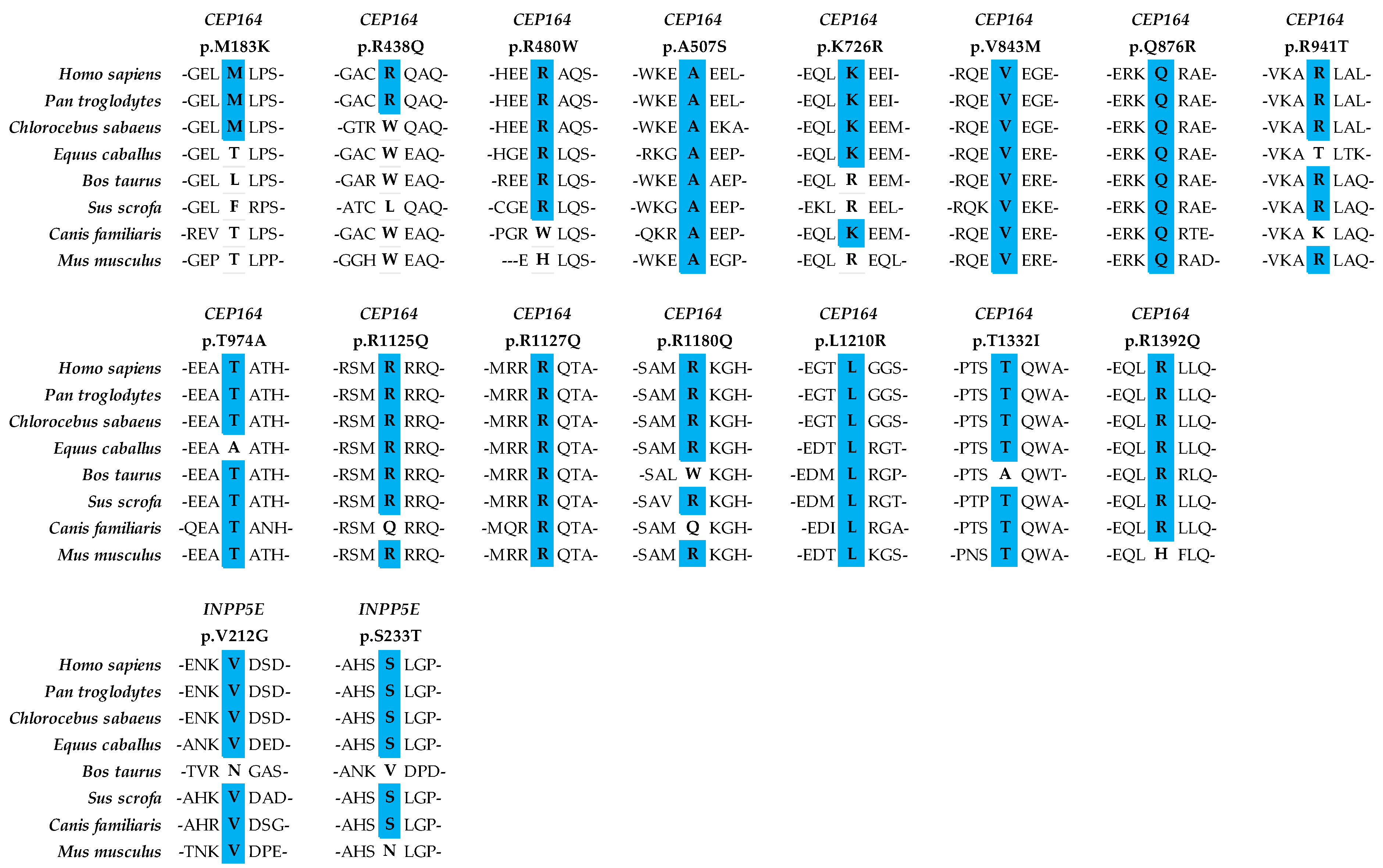
4.5. Conservation of Amino Acids
4.6. Network Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- de Luise, V.P.; Anderson, D.R. Primary infantile glaucoma (congenital glaucoma). Surv. Ophthalmol. 1983, 28, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Karaconji, T.; Zagora, S.; Grigg, J.R. Approach to childhood glaucoma: A review. Clin. Exp. Ophthalmol. 2022, 50, 232–246. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Iwata, T. Exploring the genetic landscape of childhood glaucoma. Children 2024, 11, 454. [Google Scholar] [CrossRef] [PubMed]
- Stoilov, I.; Akarsu, A.N.; Sarfarazi, M. Identification of three different truncating mutations in cytochrome P4501B1 (CYP1B1) as the principal cause of primary congenital glaucoma (Buphthalmos) in families linked to the GLC3A locus on chromosome 2p21. Hum. Mol. Genet. 1997, 6, 641–647. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhou, Y.; Du, L.; Wei, M.; Chen, X. Overview of Cytochrome P450 1B1 gene mutations in patients with primary congenital glaucoma. Exp. Eye Res. 2011, 93, 572–579. [Google Scholar] [CrossRef]
- Chouiter, L.; Nadifi, S. Analysis of CYP1B1 Gene Mutations in Patients with Primary Congenital Glaucoma. J. Pediatr. Genet. 2017, 6, 205–214. [Google Scholar] [CrossRef]
- Ali, M.; McKibbin, M.; Booth, A.; Parry, D.A.; Jain, P.; Riazuddin, S.A.; Hejtmancik, J.F.; Khan, S.N.; Firasat, S.; Shires, M.; et al. Null mutations in LTBP2 cause primary congenital glaucoma. Am. J. Hum. Genet. 2009, 84, 664–671. [Google Scholar] [CrossRef]
- Narooie-Nejad, M.; Paylakhi, S.H.; Shojaee, S.; Fazlali, Z.; Rezaei Kanavi, M.; Nilforushan, N.; Yazdani, S.; Babrzadeh, F.; Suri, F.; Ronaghi, M.; et al. Loss of function mutations in the gene encoding latent transforming growth factor beta binding protein 2, LTBP2, cause primary congenital glaucoma. Hum. Mol. Genet. 2009, 18, 3969–3977. [Google Scholar] [CrossRef]
- Souma, T.; Tompson, S.W.; Thomson, B.R.; Siggs, O.M.; Kizhatil, K.; Yamaguchi, S.; Feng, L.; Limviphuvadh, V.; Whisenhunt, K.N.; Maurer-Stroh, S.; et al. Angiopoietin receptor TEK mutations underlie primary congenital glaucoma with variable expressivity. J. Clin. Invest. 2016, 126, 2575–2587. [Google Scholar] [CrossRef]
- Kabra, M.; Zhang, W.; Rathi, S.; Mandal, A.K.; Senthil, S.; Pyatla, G.; Ramappa, M.; Banerjee, S.; Shekhar, K.; Marmamula, S.; et al. Angiopoietin receptor TEK interacts with CYP1B1 in primary congenital glaucoma. Hum. Genet. 2017, 136, 941–949. [Google Scholar] [CrossRef]
- Ling, C.; Zhang, D.; Zhang, J.; Sun, H.; Du, Q.; Li, X. Updates on the molecular genetics of primary congenital glaucoma (Review). Exp. Ther. Med. 2020, 20, 968–977. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Gharahkhani, P.; Hamel, A.R.; Ong, J.S.; Rentería, M.E.; Mehta, P.; Dong, X.; Pasutto, F.; Hammond, C.; Young, T.L.; et al. Large-scale multitrait genome-wide association analyses identify hundreds of glaucoma risk loci. Nat. Genet. 2023, 55, 1116–1125. [Google Scholar] [CrossRef] [PubMed]
- Kaur, K.; Reddy, A.B.; Mukhopadhyay, A.; Mandal, A.K.; Hasnain, S.E.; Ray, K.; Thomas, R.; Balasubramanian, D.; Chakrabarti, S. Myocilin gene implicated in primary congenital glaucoma. Clin. Genet. 2005, 67, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, S.; Kaur, K.; Rao, K.N.; Mandal, A.K.; Kaur, I.; Parikh, R.S.; Thomas, R. The transcription factor gene FOXC1 exhibits a limited role in primary congenital glaucoma. Invest. Ophthalmol. Vis. Sci. 2009, 50, 75–83. [Google Scholar] [CrossRef]
- Makhoul, N.J.; Wehbi, Z.; El Hadi, D.; Noureddine, B.; Boustany, R.M.; Al-Haddad, C. Whole-exome screening for primary congenital glaucoma in Lebanon. Ophthalmic Genet. 2023, 44, 234–245. [Google Scholar] [CrossRef]
- Yadav, M.; Yadav, A.; Bhardwaj, A.; Dhull, C.S.; Sachdeva, S.; Yadav, R.; Tanwar, M. A rare optineurin mutation in an Indian family with coexistence of JOAG and PCG. Indian J. Ophthalmol. 2023, 71, 3016–3023. [Google Scholar] [CrossRef]
- Fu, H.; Siggs, O.M.; Knight, L.S.; Staffieri, S.E.; Ruddle, J.B.; Birsner, A.E.; Collantes, E.R.; Craig, J.E.; Wiggs, J.L.; D’Amato, R.J. Thrombospondin 1 missense alleles induce extracellular matrix protein aggregation and TM dysfunction in congenital glaucoma. J. Clin. Invest. 2022, 132, e156967. [Google Scholar] [CrossRef]
- Morales-Cámara, S.; Alexandre-Moreno, S.; Bonet-Fernández, J.M.; Atienzar-Aroca, R.; Aroca-Aguilar, J.D.; Ferre-Fernández, J.J.; Méndez, C.D.; Morales, L.; Fernández-Sánchez, L.; Cuenca, N.; et al. Role of GUCA1C in Primary Congenital Glaucoma and in the Retina: Functional Evaluation in Zebrafish. Genes 2020, 11, 550. [Google Scholar] [CrossRef]
- Gupta, V.; Somarajan, B.I.; Kaur, G.; Gupta, S.; Singh, R.; Pradhan, D.; Singh, H.; Kaur, P.; Sharma, A.; Chawla, B.; et al. Exome sequencing identifies procollagen-lysine 2-oxoglutarate 5-dioxygenase 2 mutations in primary congenital and juvenile glaucoma. Indian J. Ophthalmol. 2021, 69, 2710–2716. [Google Scholar] [CrossRef]
- Mauri, L.; Uebe, S.; Sticht, H.; Vossmerbaeumer, U.; Weisschuh, N.; Manfredini, E.; Maselli, E.; Patrosso, M.; Weinreb, R.N.; Penco, S.; et al. Expanding the clinical spectrum of COL1A1 mutations in different forms of glaucoma. Orphanet. J. Rare Dis. 2016, 11, 108. [Google Scholar] [CrossRef]
- Medina-Trillo, C.; Aroca-Aguilar, J.D.; Ferre-Fernandez, J.J.; Alexandre-Moreno, S.; Morales, L.; Mendez-Hernandez, C.D.; Garcia-Feijoo, J.; Escribano, J. Role of FOXC2 and PITX2 rare variants associated with mild functional alterations as modifier factors in congenital glaucoma. PLoS ONE 2019, 14, e0211029. [Google Scholar] [CrossRef] [PubMed]
- Hadrami, M.; Bonnet, C.; Zeitz, C.; Veten, F.; Biya, M.; Hamed, C.T.; Condroyer, C.; Wang, P.; Sidi, M.M.; Cheikh, S.; et al. Mutation profile of glaucoma candidate genes in Mauritanian families with primary congenital glaucoma. Mol. Vis. 2019, 25, 373–381. [Google Scholar] [PubMed]
- Bonet-Fernández, J.M.; Aroca-Aguilar, J.D.; Corton, M.; Ramírez, A.I.; Alexandre-Moreno, S.; García-Antón, M.T.; Salazar, J.J.; Ferre-Fernández, J.J.; Atienzar-Aroca, R.; Villaverde, C.; et al. CPAMD8 loss-of-function underlies non-dominant congenital glaucoma with variable anterior segment dysgenesis and abnormal extracellular matrix. Hum. Genet. 2020, 139, 1209–1231. [Google Scholar] [CrossRef] [PubMed]
- Young, T.L.; Whisenhunt, K.N.; Jin, J.; LaMartina, S.M.; Martin, S.M.; Souma, T.; Limviphuvadh, V.; Suri, F.; Souzeau, E.; Zhang, X.; et al. SVEP1 as a Genetic Modifier of TEK-Related Primary Congenital Glaucoma. Invest. Ophthalmol. Vis. Sci. 2020, 61, 6. [Google Scholar] [CrossRef]
- Labelle-Dumais, C.; Pyatla, G.; Paylakhi, S.; Tolman, N.G.; Hameed, S.; Seymens, Y.; Dang, E.; Mandal, A.K.; Senthil, S.; Khanna, R.C.; et al. Loss of PRSS56 function leads to ocular angle defects and increased susceptibility to high intraocular pressure. Dis. Model. Mech. 2020, 13, dmm042853. [Google Scholar] [CrossRef]
- Shivanna, M.; Anand, M.; Chakrabarti, S.; Khanna, H. Ocular Ciliopathies: Genetic and Mechanistic Insights into Developing Therapies. Curr. Med. Chem. 2019, 26, 3120–3131. [Google Scholar] [CrossRef]
- Seo, S.; Sonn, S.K.; Kweon, H.Y.; Jin, J.; Kume, T.; Ko, J.Y.; Park, J.H.; Oh, G.T. Primary Cilium in Neural Crest Cells Crucial for Anterior Segment Development and Corneal Avascularity. Invest. Ophthalmol. Vis. Sci. 2024, 65, 30. [Google Scholar] [CrossRef]
- Portal, C.; Rompolas, P.; Lwigale, P.; Iomini, C. Primary cilia deficiency in neural crest cells models anterior segment dysgenesis in mouse. Elife 2019, 8, e52423. [Google Scholar] [CrossRef]
- Ueda, J.; Wentz–Hunter, K.K.; Cheng, E.L.; Fukuchi, T.; Abe, H.; Yue, B.Y. Ultrastructural localization of myocilin in human trabecular meshwork cells and tissues. J. Histochem. Cytochem. 2000, 48, 1321–1330. [Google Scholar] [CrossRef]
- Noda, S.; Mashima, Y.; Obazawa, M.; Kubota, R.; Oguchi, Y.; Kudoh, J.; Minoshima, S.; Shimizu, N. Myocilin expression in the astrocytes of the optic nerve head. Biochem. Biophys. Res. Commun. 2000, 276, 1129–1135. [Google Scholar] [CrossRef]
- Luo, N.; Conwell, M.D.; Chen, X.; Kettenhofen, C.I.; Westlake, C.J.; Cantor, L.B.; Wells, C.D.; Weinreb, R.N.; Corson, T.W.; Spandau, D.F.; et al. Primary cilia signaling mediates intraocular pressure sensation. Proc. Natl. Acad. Sci. USA 2014, 111, 12871–12876. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Zhou, J. The Primary Cilium as a Therapeutic Target in Ocular Diseases. Front. Pharmacol. 2020, 11, 977. [Google Scholar] [CrossRef] [PubMed]
- Reed, M.; Takemaru, K.I.; Ying, G.; Frederick, J.M.; Baehr, W. Deletion of CEP164 in mouse photoreceptors post-ciliogenesis interrupts ciliary intraflagellar transport (IFT). PLoS Genet. 2022, 18, e1010154. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Xie, T.; Nawy, S.; Shen, Y. The development and the genetic diseases of the ciliary body. Cell Insight 2024, 3, 100162. [Google Scholar] [CrossRef]
- Teilmann, S.C.; Christensen, S.T. Localization of the angiopoietin receptors Tie-1 and Tie-2 on the primary cilia in the female reproductive organs. Cell Biol. Int. 2005, 29, 340–346. [Google Scholar] [CrossRef]
- Yoon, M.J.; Cho, C.H.; Lee, C.S.; Jang, I.H.; Ryu, S.H.; Koh, G.Y. Localization of Tie2 and phospholipase D in endothelial caveolae is involved in angiopoietin-1-induced MEK/ERK phosphorylation and migration in endothelial cells. Biochem. Biophys. Res. Commun. 2003, 308, 101–105. [Google Scholar] [CrossRef]
- Graser, S.; Stierhof, Y.-D.; Lavoie, S.B.; Gassner, O.S.; Lamla, S.; Le Clech, M.; Nigg, E.A. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 2007, 179, 321–330. [Google Scholar] [CrossRef]
- Sivasubramaniam, S.; Sun, X.; Pan, Y.R.; Wang, S.; Eva, Y.H.L. Cep164 is a mediator protein required for the maintenance of genomic stability through modulation of MDC1, RPA, and CHK1. Genes Dev. 2008, 22, 587–600. [Google Scholar] [CrossRef]
- Luo, N.; Lu, J.; Sun, Y. Evidence of a role of inositol polyphosphate 5-phosphatase INPP5E in cilia formation in zebrafish. Vision Res. 2012, 75, 98–107. [Google Scholar] [CrossRef]
- Zhang, R.; Tang, J.; Li, T.; Zhou, J.; Pan, W. INPP5E and Coordination of Signaling Networks in Cilia. Front. Mol. Biosci. 2022, 9, 885592. [Google Scholar] [CrossRef]
- Plotnikova, O.V.; Seo, S.; Cottle, D.L.; Conduit, S.; Hakim, S.; Dyson, J.M.; Mitchell, C.A.; Smyth, I.M. INPP5E interacts with AURKA, linking phosphoinositide signaling to primary cilium stability. J. Cell Sci. 2015, 128, 364–372. [Google Scholar] [PubMed]
- Yue, H.; Li, S.; Qin, J.; Gao, T.; Lyu, J.; Liu, Y.; Wang, X.; Guan, Z.; Zhu, Z.; Niu, B.; et al. Down-Regulation of Inpp5e Associated With Abnormal Ciliogenesis During Embryonic Neurodevelopment Under Inositol Deficiency. Front. Neurol. 2021, 12, 579998. [Google Scholar] [CrossRef] [PubMed]
- Humbert, M.C.; Weihbrecht, K.; Searby, C.C.; Li, Y.; Pope, R.M.; Sheffield, V.C.; Seo, S. ARL13B, PDE6D, and CEP164 form a functional network for INPP5E ciliary targeting. Proc. Natl. Acad. Sci. USA 2012, 109, 19691–19696. [Google Scholar] [CrossRef] [PubMed]
- Maria, M.; Lamers, I.J.; Schmidts, M.; Ajmal, M.; Jaffar, S.; Ullah, E.; Mustafa, B.; Ahmad, S.; Nazmutdinova, K.; Hoskins, B.; et al. Genetic and clinical characterization of Pakistani families with Bardet-Biedl syndrome extends the genetic and phenotypic spectrum. Sci. Rep. 2016, 6, 34764. [Google Scholar] [CrossRef]
- Vilboux, T.; Doherty, D.A.; Glass, I.A.; Parisi, M.A.; Phelps, I.G.; Cullinane, A.R.; Zein, W.; Brooks, B.P.; Heller, T.; Soldatos, A.; et al. Molecular genetic findings and clinical correlations in 100 patients with Joubert syndrome and related disorders prospectively evaluated at a single center. Genet. Med. 2017, 19, 875–882. [Google Scholar] [CrossRef]
- Jacoby, M.; Cox, J.J.; Gayral, S.; Hampshire, D.J.; Ayub, M.; Blockmans, M.; Pernot, E.; Kisseleva, M.V.; Compere, P.; Schiffmann, S.N.; et al. INPP5E mutations cause primary cilium signaling defects, ciliary instability and ciliopathies in human and mouse. Nat. Genet. 2009, 41, 1027–1031. [Google Scholar] [CrossRef]
- Travaglini, L.; Brancati, F.; Silhavy, J.; Iannicelli, M.; Nickerson, E.; Elkhartoufi, N.; Scott, E.; Spencer, E.; Gabriel, S.; Thomas, S.; et al. Phenotypic spectrum and prevalence of INPP5E mutations in Joubert syndrome and related disorders. Eur. J. Hum. Genet. 2013, 21, 1074–1078. [Google Scholar] [CrossRef]
- Slaats, G.G.; Ghosh, A.K.; Falke, L.L.; Le Corre, S.; Shaltiel, I.A.; van de Hoek, G.; Klasson, T.D.; Stokman, M.F.; Logister, I.; Verhaar, M.C. Nephronophthisis-associated CEP164 regulates cell cycle progression, apoptosis and epithelial-to-mesenchymal transition. PLoS Genet. 2014, 10, e1004594. [Google Scholar] [CrossRef]
- Yue, H.; Zhu, X.; Li, S.; Wang, F.; Wang, X.; Guan, Z.; Zhu, Z.; Niu, B.; Zhang, T.; Guo, J.; et al. Relationship Between INPP5E Gene Expression and Embryonic Neural Development in a Mouse Model of Neural Tube Defect. Med. Sci. Monit. 2018, 24, 2053–2059. [Google Scholar] [CrossRef]
- Chaki, M.; Airik, R.; Ghosh, A.K.; Giles, R.H.; Chen, R.; Slaats, G.G.; Wang, H.; Hurd, T.W.; Zhou, W.; Cluckey, A.; et al. Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell 2012, 150, 533–548. [Google Scholar] [CrossRef]
- Devlin, L.A.; Ramsbottom, S.A.; Overman, L.M.; Lisgo, S.N.; Clowry, G.; Molinari, E.; Powell, L.; Miles, C.G.; Sayer, J.A. Embryonic and foetal expression patterns of the ciliopathy gene CEP164. PLoS ONE 2020, 15, e0221914. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-J.; Suh, W.; Park, S.C.; Kim, C.Y.; Park, K.H.; Kook, M.S.; Kim, Y.Y.; Kim, C.-S.; Park, C.K.; Ki, C.-S.; et al. Mutation spectrum of CYP1B1 and MYOC genes in Korean patients with primary congenital glaucoma. Mol. Vis. 2011, 17, 2093–2101. [Google Scholar] [PubMed]
- Chen, Y.; Jiang, D.; Yu, L.; Katz, B.; Zhang, K.; Wan, B.; Sun, X. CYP1B1 and MYOC mutations in 116 Chinese patients with primary congenital glaucoma. Arch. Ophthalmol. 2008, 126, 1443–1447. [Google Scholar] [CrossRef] [PubMed]
- Geyer, O.; Wolf, A.; Levinger, E.; Harari-Shacham, A.; Walton, D.S.; Shochat, C.; Korem, S.; Bercovich, D. Genotype/phenotype correlation in primary congenital glaucoma patients from different ethnic groups of the Israeli population. Am. J. Ophthalmol. 2011, 151, 263–271. [Google Scholar] [CrossRef]
- Suh, W.; Kee, C. A clinical and molecular genetics study of primary congenital glaucoma in South Korea. Br. J. Ophthalmol. 2012, 96, 1372–1377. [Google Scholar] [CrossRef]
- Hilal, L.; Boutayeb, S.; Serrou, A.; Refass-Buret, L.; Shisseh, H.; Bencherifa, F.; El Mzibri, M.; Benazzouz, B.; Berraho, A. Screening of CYP1B1 and MYOC in Moroccan families with primary congenital glaucoma: Three novel mutations in CYP1B1. Mol. Vis. 2010, 16, 1215–1226. [Google Scholar]
- Fassad, M.R.; Amin, A.K.; Morsy, H.A.; Issa, N.M.; Bayoumi, N.H.; El Shafei, S.A.; Kholeif, S.F. CYP1B1 and myocilin gene mutations in Egyptian patients with primary congenital glaucoma. Egypt J. Med. Hum. Genet. 2017, 18, 219–224. [Google Scholar] [CrossRef][Green Version]
- Akbas, A.C.; Erdem, E.; Bozdogan, S.T.; Harbiyeli, I.I.; Yagmur, M. CYP1B1 and MYOC Gene Analysis of Patients with Primary Congenital Glaucoma in the Cukurova Region of Turkey. J. Pediatr. Genet. 2023. [Google Scholar] [CrossRef]
- Vincent, A.L.; Billingsley, G.; Buys, Y.; Levin, A.V.; Priston, M.; Trope, G.; Williams-Lyn, D.; Héon, E. Digenic inheritance of early-onset glaucoma: CYP1B1, a potential modifier gene. Am. J. Hum. Genet. 2002, 70, 448–460. [Google Scholar] [CrossRef]
- Stenson, P.D.; Ball, E.V.; Mort, M.; Phillips, A.D.; Shiel, J.A.; Thomas, N.S.; Abeysinghe, S.; Krawczak, M.; Cooper, D.N. Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 2003, 21, 577–581. [Google Scholar] [CrossRef]
- Naik, A.; Sihota, R.; Mahalingam, K.; Angmo, D.; Dada, T.; Kumar, A.; Kumar, A.; Gupta, A. Evaluation of visual field changes with retinal nerve fiber layer thickness in primary congenital glaucoma. Indian J. Ophthalmol. 2022, 70, 3556–3561. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Falcón, S.; Glaser, T.; Go, M.S.; Kelly, M.P.; Chen, X.; Freedman, S.F.; El-Dairi, M. Retinal injury identified by overhead-mounted optical coherence tomography in two young children with infantile-onset glaucoma. J. AAPOS 2023, 27, 28.e21–28.e26. [Google Scholar] [CrossRef] [PubMed]
- Sen, J.; Harpavat, S.; Peters, M.A.; Cepko, C.L. Retinoic acid regulates the expression of dorsoventral topographic guidance molecules in the chick retina. Development 2005, 132, 5147–5159. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chakrabarti, S.; Kaur, K.; Kaur, I.; Mandal, A.K.; Parikh, R.S.; Thomas, R.; Majumder, P.P. Globally, CYP1B1 Mutations in Primary Congenital Glaucoma Are Strongly Structured by Geographic and Haplotype Backgrounds. Invest. Opthalmol. Vis. Sci. 2006, 47, 43–47. [Google Scholar] [CrossRef]
- Gibbs, R.A.; Boerwinkle, E.; Doddapaneni, H.; Han, Y.; Korchina, V.; Kovar, C.; Lee, S.; Muzny, D.; Reid, J.G.; Zhu, Y.; et al. A global references for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar]
- Chen, S.; Francioli, L.C.; Goodrich, J.K.; Collins, R.L.; Kanai, M.; Wang, Q.; Alföldi, J.; Watts, N.A.; Vittal, C.; Gauthier, L.D.; et al. A genomic mutational constraint map using variation in 76,156 human genomes. Nature 2024, 625, 92–100. [Google Scholar] [CrossRef]
- All of Us Research Program I; Denny, J.C.; Rutter, J.L.; Goldstein, D.B.; Philippakis, A.; Smoller, J.W.; Jenkins, G.; Dishman, E. The “All of Us” Research Program. N. Engl. J. Med. 2019, 381, 668–676. [Google Scholar] [CrossRef]
- IIoannidis, N.M.; Rothstein, J.H.; Pejaver, V.; Middha, S.; McDonnell, S.K.; Baheti, S.; Musolf, A.; Li, Q.; Holzinger, E.; Karyadi, D.; et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am. J. Hum. Genet. 2016, 99, 877–885. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.D.M.J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
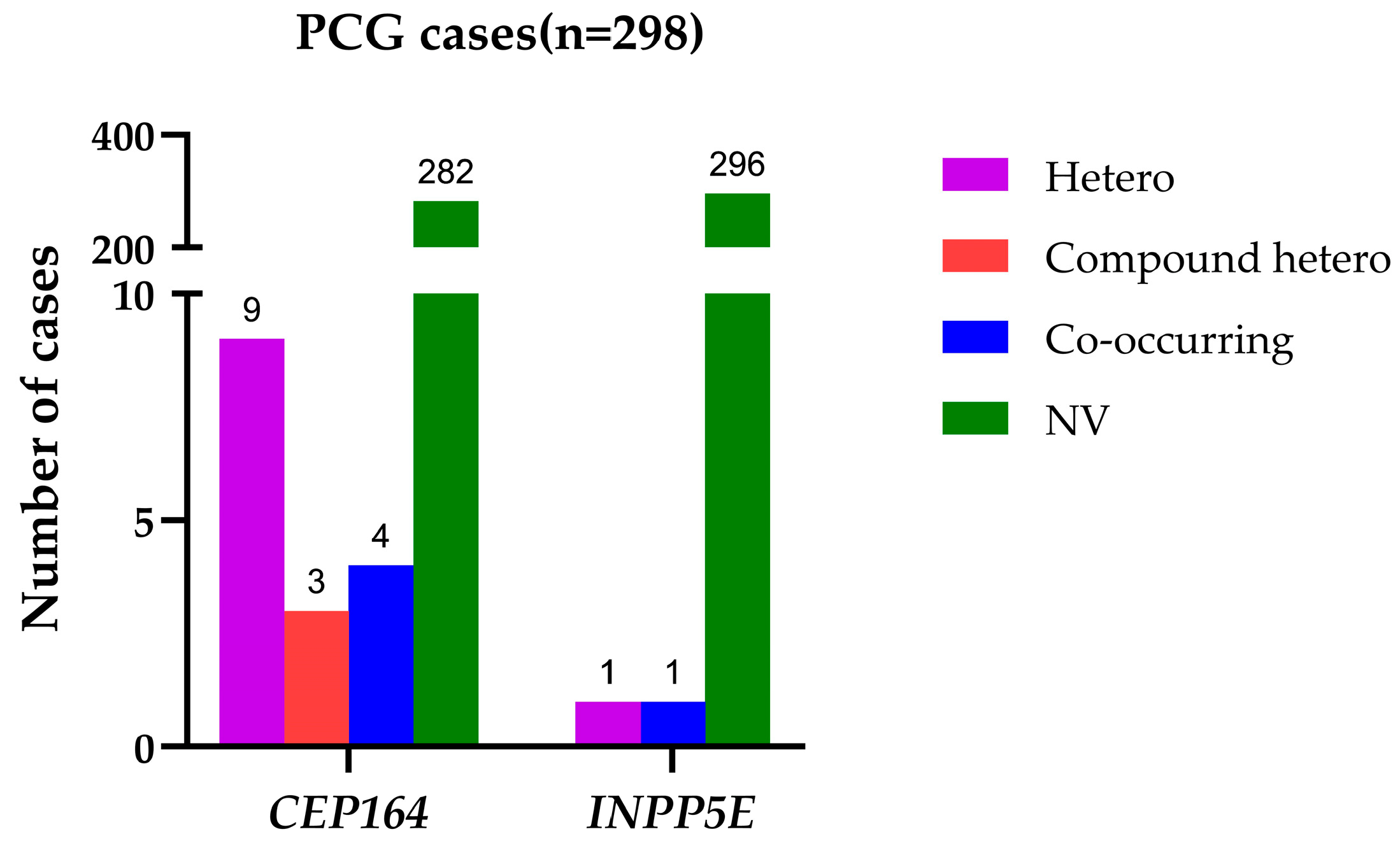

| Chromosomal Position (GRCh38) | Amino Acid Change | dbSNP ID | REVEL Score | Cases MAF (n = 298) | Controls MAF (n = 1757) | p Value | Odds Ratio (95% CI) | 1000 Genomes (n = 2504); gnomAD v4.1 (n = 807,162); All of Us (n = 245,400) |
|---|---|---|---|---|---|---|---|---|
| A | ||||||||
| 11: 117361989T > A | p.M183K | rs144206271 | 0.18 | 0.0016 | 0.0003 | 0.15 | 5.91 (0.37–94.74) | 0.0008; 0.00063; 0.0006 |
| 11: 117375787G > A | p.R438Q | rs137987733 | 0.05 | 0.0016 | 0.0003 | 0.15 | 5.91 (0.37–94.74) | NA; 0.00018; 0.0001 |
| 11: 117381729C > T | p.R480W | rs112209873 | 0.04 | 0.0016 | 0 | - | - | 0.001; 0.00019; 0.00068 |
| 11: 117381810G > T | p.A507S | NA | 0.08 | 0.0016 | 0 | - | - | NA |
| 11: 117391109A > G | p.K726R | rs2044597036 | 0.02 | 0.0016 | 0 | - | - | NA |
| 11: 117393037G > A | p.V843M | rs566117718 | 0.09 | 0.0016 | 0.0008 | 0.55 | 1.96 (0.20–18.98) | 0.0012; 0.00005; 0.000016 |
| 11: 117394360A > G | p.Q876R | rs752659513 | 0.13 | 0.0033 | 0.0003 | 0.01 | 11.86 (1.07–131.20) | NA; 0.000044; 0.000006 |
| 11: 117394981G > C | p.R941T | rs749310077 | 0.03 | 0.0016 | 0 | - | - | NA; 0.0000006; NA |
| 11: 117395553A > G | p.T974A | rs56699807 | 0.04 | 0.0033 | 0.0003 | 0.01 | 11.86 (1.07–131.20) | 0.0002; 0.0003; 0.0003 |
| 11: 117397186G > A | p.R1125Q | rs767918200 | 0.03 | 0.0016 | 0.0008 | 0.55 | 1.96 (0.20–18.98) | NA; 0.000015; 0.000002 |
| 11: 117397192G > A | p.R1127Q | rs753895198 | 0.16 | 0.0016 | 0 | - | - | NA; 0.000009; 0.000016 |
| 11: 117407962G > A | p.R1180Q | rs568896676 | 0.02 | 0.0050 | 0.0006 | 0.004 | 8.92 (1.48–53.61) | NA; 0.00001; 0.000012 |
| 11: 117408909T > G | p.L1210R | rs767571570 | 0.17 | 0.0016 | 0 | - | - | NA; 0.000003; NA |
| 11: 117409864C > T | p.T1332I | rs760788324 | 0.03 | 0.0016 | 0 | - | - | NA; 0.000006; NA |
| 11: 117411806G > A | p.R1392Q | rs772989312 | 0.07 | 0.0016 | 0.0011 | 0.72 | 1.47 (0.16–13.24) | NA; 0.000012; 0.000016 |
| B | ||||||||
| 9: 136438722C > G | p.S233T | rs568767788 | 0.35 | 0.0016 | 0.0003 | 0.15 | 5.91 (0.37–94.74) | 0.00039; 0.000026; 0.000006 |
| 9: 136438785A > C | p.V212G | rs533861933 | 0.54 | 0.0016 | 0.0017 | 0.98 | 0.98 (0.11–8.18) | 0.00019; 0.0000068; 0.00001 |
| Genes | Haplotypes | Overall Haplotype Frequency | Frequency in Cases | Frequency in Controls | Chi Square | Uncorrected p Value | Corrected p Value * |
|---|---|---|---|---|---|---|---|
| CEP164 | C-C | 0.747 | 0.792 | 0.739 | 7.464 | 0.006 | 0.056 |
| CEP164 | C-G | 0.198 | 0.171 | 0.203 | 3.179 | 0.075 | 0.414 |
| CEP164 | G-C | 0.055 | 0.037 | 0.058 | 4.383 | 0.036 | 0.242 |
| INPP5E | C-G-G | 0.844 | 0.834 | 0.846 | 0.613 | 0.434 | 0.954 |
| INPP5E | A-A-G | 0.084 | 0.131 | 0.076 | 20.037 | 7.6 × 10−6 | 0.0005 |
| INPP5E | A-A-A | 0.055 | 0.032 | 0.059 | 7.234 | 0.007 | 0.055 |
| Genotype Combinations in PCG Cases (Number of Cases) | Intraocular Pressure (mmHg) | Cup-to-Disc Ratio | Corneal Diameter (mm) | Visual Acuity (logMAR) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| At Presen-tation | After 3 Months | After 1 Year | At Presen-tation | After 3 Months | After 1 Year | At Presen-tation | After 3 Months | After 1 Year | At Presen-tation | After 3 Months | After 1 Year | |
| Cases with unique heterozygous CEP164 alleles only (n = 9) | 25.75 ± 6.63 | 12.86 ± 3.8 | 11.63 ± 1.69 | 0.48 ± 0.13 | 0.54 ± 0.22 | 0.46 ± 0.2 | 13.06 ± 0.98 | 12.71 ± 1.19 | 13.1 ± 0.74 | NA | NA | 1.19 ± 0.47 |
| Cases with compound heterozygous CEP164 alleles (n = 3) | 28 ± 6.93 | 15.33 ± 5.03 | 10.33 ± 2.52 | NA | 0.47 ± 0.23 | 0.5 ± 0.26 | 13.67 ± 0.58 | 13.33 ± 0.58 | 13.67 ± 0.29 | NA | NA | |
| Cases with co-occurring CEP164 alleles along with heterozygous alleles of other genes (n = 4) | 24.25 ± 6.65 | NA | 17.67 ± 5.86 | 0.83 ± 0.06 | NA | 0.63 ± 0.31 | 15 ± 2.29 | NA | NA | NA | NA | 2.34 ± 0.75 |
| * p value (Unique heterozygous CEP164 alleles versus co-occurring CEP164 and alleles of other genes) | 0.72 | NA | 0.019 | 0.004 | NA | 0.284 | 0.056 | NA | NA | NA | NA | 0.053 |
| * p value (Compound heterozygous CEP164 alleles versus co-occurring CEP164 and alleles of other genes) | 0.5 | NA | 0.117 | NA | NA | 0.598 | 0.384 | NA | NA | NA | NA | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pyatla, G.; Kabra, M.; Mandal, A.K.; Zhang, W.; Mishra, A.; Bera, S.; Rathi, S.; Patnaik, S.; Anthony, A.A.; Dixit, R.; et al. Potential Involvements of Cilia-Centrosomal Genes in Primary Congenital Glaucoma. Int. J. Mol. Sci. 2024, 25, 10028. https://doi.org/10.3390/ijms251810028
Pyatla G, Kabra M, Mandal AK, Zhang W, Mishra A, Bera S, Rathi S, Patnaik S, Anthony AA, Dixit R, et al. Potential Involvements of Cilia-Centrosomal Genes in Primary Congenital Glaucoma. International Journal of Molecular Sciences. 2024; 25(18):10028. https://doi.org/10.3390/ijms251810028
Chicago/Turabian StylePyatla, Goutham, Meha Kabra, Anil K. Mandal, Wei Zhang, Ashish Mishra, Samir Bera, Sonika Rathi, Satish Patnaik, Alice A. Anthony, Ritu Dixit, and et al. 2024. "Potential Involvements of Cilia-Centrosomal Genes in Primary Congenital Glaucoma" International Journal of Molecular Sciences 25, no. 18: 10028. https://doi.org/10.3390/ijms251810028
APA StylePyatla, G., Kabra, M., Mandal, A. K., Zhang, W., Mishra, A., Bera, S., Rathi, S., Patnaik, S., Anthony, A. A., Dixit, R., Banerjee, S., Shekhar, K., Marmamula, S., Kaur, I., Khanna, R. C., & Chakrabarti, S. (2024). Potential Involvements of Cilia-Centrosomal Genes in Primary Congenital Glaucoma. International Journal of Molecular Sciences, 25(18), 10028. https://doi.org/10.3390/ijms251810028






