Differential Cytoophidium Assembly between Saccharomyces cerevisiae and Schizosaccharomyces pombe
Abstract
:1. Introduction
2. Results
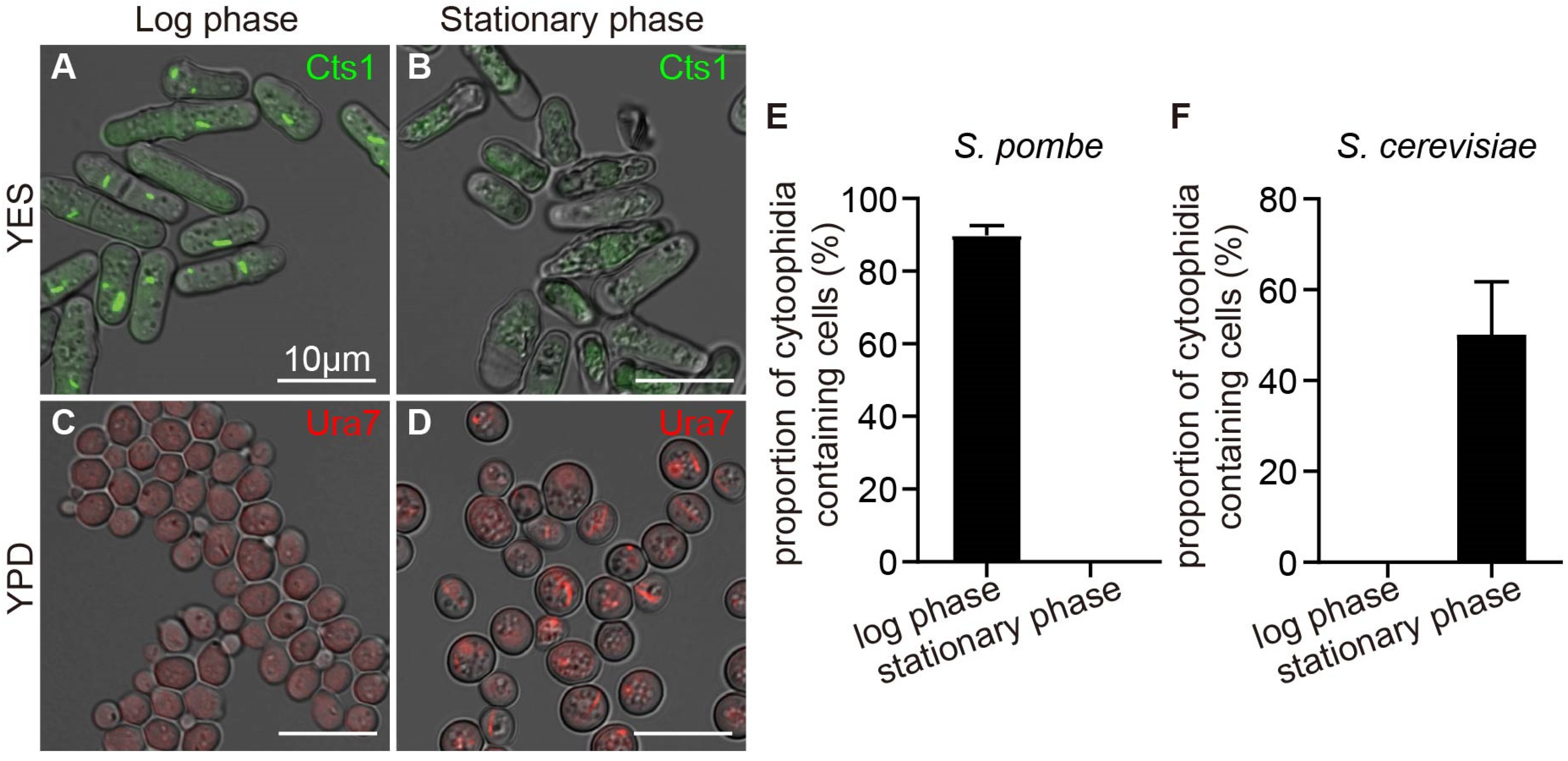
2.1. Comparison of Cytoophidia between S. cerevisiae and S. pombe
2.2. Co-Cultivation of Fission Yeast and Budding Yeast
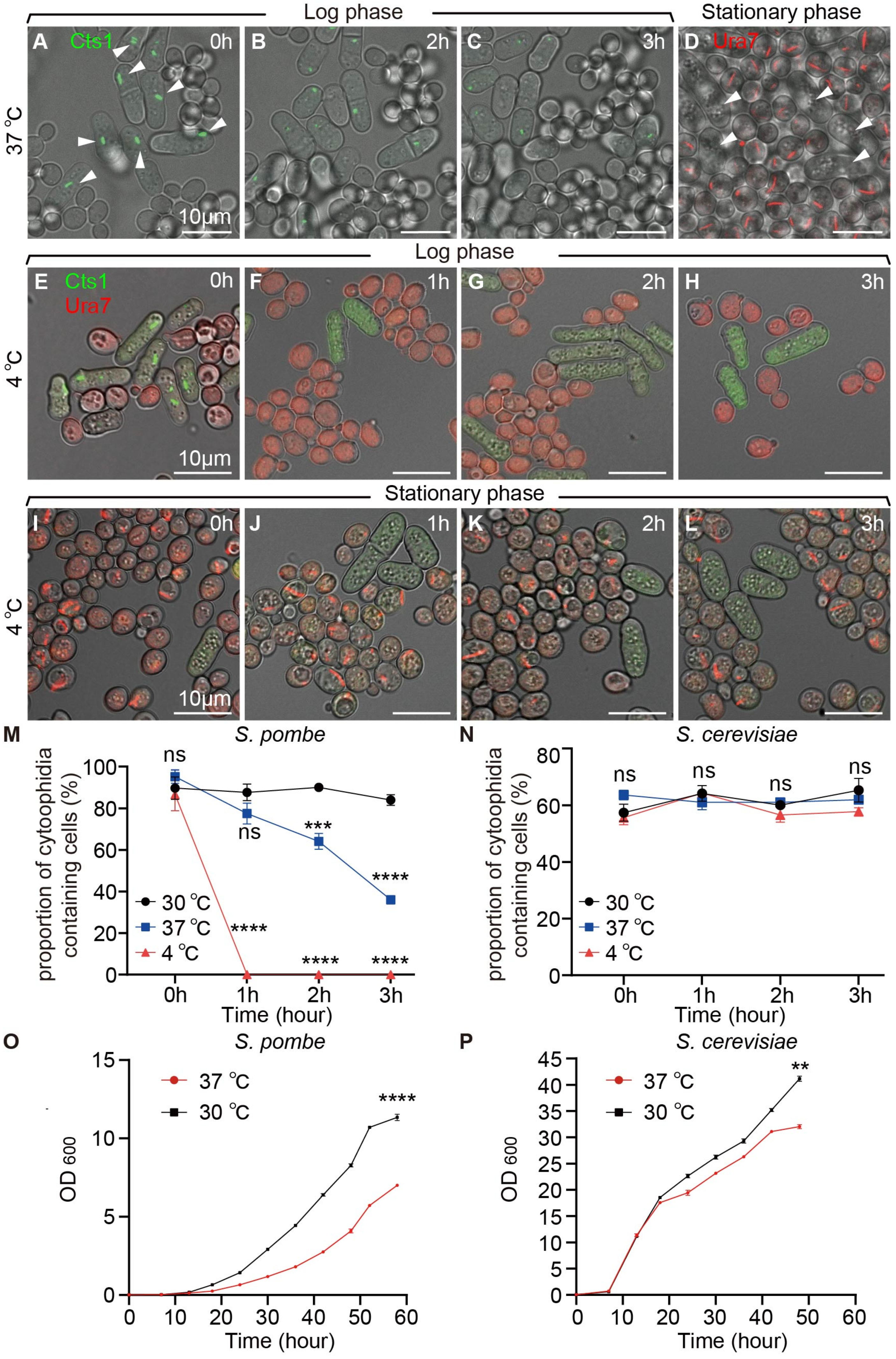
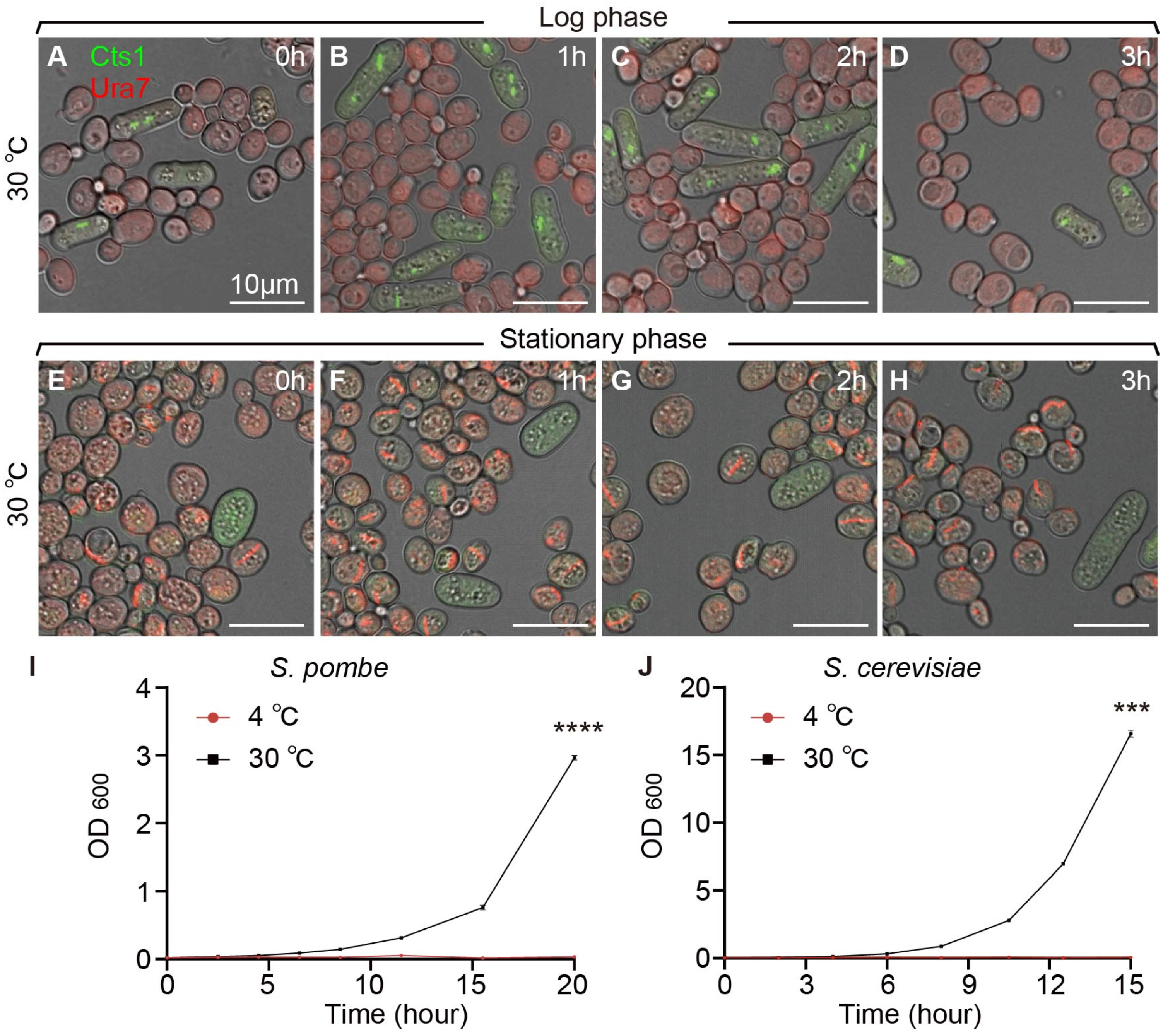
2.3. Temperature Affects Cytoophidium Assembly in Fission Yeast but Not in Budding Yeast
2.4. pH Values Influence Cytoophidium Assembly Differentially in Budding Yeast and Fission Yeast
2.5. Cytoophidia of Both Yeast Species Are Influenced by Yeast-Saturated Cultured Medium
2.6. Overexpression of CTPS Protein in Both Yeast Species Can Affect Cytoophidium Assembly
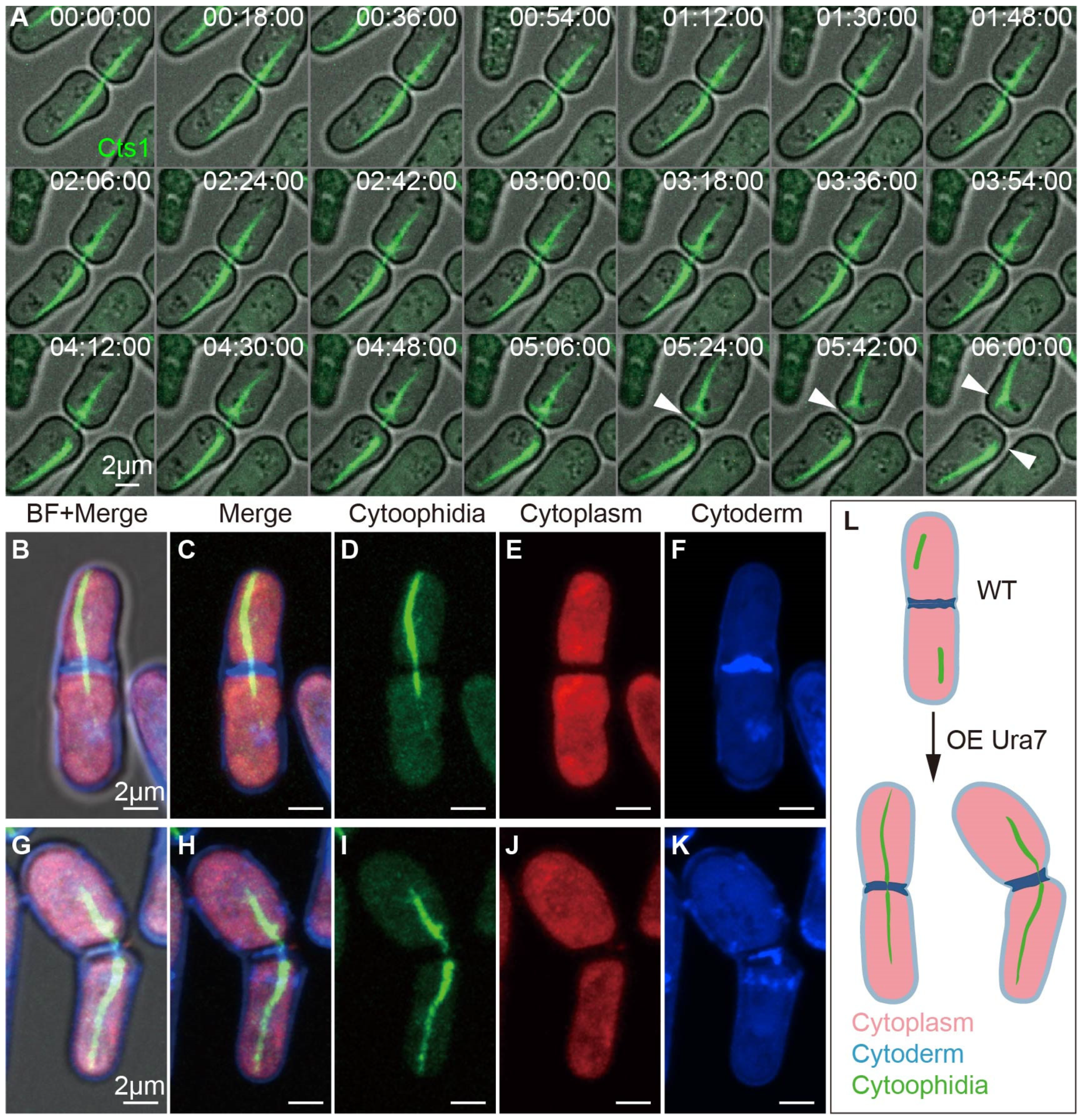
2.7. Interspecies Overexpression of Ura7 Generates Giant Cytoophidia crossing Cell Membrane
3. Discussion
3.1. Comparison of Cytoophidia in S. cerevisiae and S. pombe under External Environmental Alterations
3.2. Comparison of Cytoophidia in S. cerevisiae and S. pombe under Intracellular CTPS Alterations
3.3. Connection between Cytoophidia and Cell Growth
4. Materials and Methods
4.1. Yeast Strain and Culture Medium
4.2. Plasmid Construction
4.3. Growth Assays
4.4. Cell Fixation, Image Acquisition, and Live-Cell Imaging
4.5. Quantification and Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chang, Y.-F.; Carman, G.M. Protein phase separation: A new phase in cell biology. Prog. Lipid Res. 1956, 47, 333–339. [Google Scholar]
- Liu, J.-L. The Cytoophidium and Its Kind: Filamentation and Compartmentation of Metabolic Enzymes. Annu. Rev. Cell Dev. Biol. 2016, 32, 349–372. [Google Scholar]
- Liu, J.L. Intracellular compartmentation of CTP synthase in Drosophila. J. Genet. Genom. 2010, 37, 281–296. [Google Scholar]
- Ingerson-Mahar, M.; Briegel, A.; Werner, J.N.; Jensen, G.J.; Gitai, Z. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nat. Cell Biol. 2010, 12, 739–746. [Google Scholar]
- Noree, C.; Sato, B.K.; Broyer, R.M.; Wilhelm, J.E. Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. J. Cell Biol. 2010, 190, 541–551. [Google Scholar]
- Chen, K.; Zhang, J.; Tastan, Ö.Y.; Deussen, Z.A.; Siswick, M.Y.Y.; Liu, J.L. Glutamine analogs promote cytoophidium assembly in human and Drosophila cells. J. Genet. Genom. 2011, 38, 391–402. [Google Scholar]
- Chang, C.-C.; Jeng, Y.M.; Peng, M.; Keppeke, G.D.; Sung, L.Y.; Liu, J.L. CTP synthase forms the cytoophidium in human hepatocellular carcinoma. Exp. Cell Res. 2017, 361, 292–299. [Google Scholar]
- Zhang, J.; Hulme, L.; Liu, J.L. Asymmetric inheritance of cytoophidia in Schizosaccharomyces pombe. Biol. Open 2014, 3, 1092–1097. [Google Scholar]
- Daumann, M.; Hickl, D.; Zimmer, D.; DeTar, R.A.; Kunz, H.H.; Möhlmann, T. Characterization of filament-forming CTP synthases from Arabidopsis thaliana. Plant J. 2018, 96, 316–328. [Google Scholar]
- Zhou, S.; Xiang, H.; Liu, J.L. CTP synthase forms cytoophidia in archaea. J. Genet. Genom. 2020, 47, 213–223. [Google Scholar]
- Carcamo, W.C.; Satoh, M.; Kasahara, H.; Terada, N.; Hamazaki, T.; Chan, J.Y.F.; Yao, B.; Tamayo, S.; Covini, G.; von Mühlen, C.A.; et al. Induction of Cytoplasmic Rods and Rings Structures by Inhibition of the CTP and GTP Synthetic Pathway in Mammalian Cells. PLoS ONE 2011, 6, e29690. [Google Scholar]
- Peng, M.; Chang, C.C.; Liu, J.L.; Sung, L.Y. CTPS and IMPDH form cytoophidia in developmental thymocytes. Exp. Cell Res. 2021, 405, 112662. [Google Scholar] [PubMed]
- Zhou, X.; Guo, C.J.; Hu, H.H.; Zhong, J.; Sun, Q.; Liu, D.; Zhou, S.; Chang, C.C.; Liu, J.L. Drosophila CTP synthase can form distinct substrate- and product-bound filaments. J. Genet. Genom. 2019, 46, 537–545. [Google Scholar]
- Zhou, X.; Guo, C.J.; Chang, C.C.; Zhong, J.; Hu, H.H.; Lu, G.M.; Liu, J.L. Structural basis for ligand binding modes of CTP synthase. Proc. Natl. Acad. Sci. USA 2021, 118, e2026621118. [Google Scholar] [PubMed]
- Lynch, E.M.; DiMattia, M.A.; Albanese, S.; van Zundert, G.C.; Hansen, J.M.; Quispe, J.D.; Kennedy, M.A.; Verras, A.; Borrelli, K.; Toms, A.V.; et al. Structural basis for isoform-specific inhibition of human CTPS1. Proc. Natl. Acad. Sci. USA 2021, 118, e2107968118. [Google Scholar]
- Hansen, J.M.; Horowitz, A.; Lynch, E.M.; Farrell, D.P.; Quispe, J.; DiMaio, F.; Kollman, J.M. Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation. eLife 2021, 10, e73368. [Google Scholar]
- Barry, R.M.; Bitbol, A.F.; Lorestani, A.; Charles, E.J.; Habrian, C.H.; Hansen, J.M.; Li, H.J.; Baldwin, E.P.; Wingreen, N.S.; Kollman, J.M.; et al. Large-scale filament formation inhibits the activity of CTP synthetase. eLife 2014, 3, e03638. [Google Scholar]
- Guo, C.J.; Liu, J.L. Cytoophidia and filaments: You must unlearn what you have learned. Biochem. Soc. Trans. 2023, 51, 1245–12566. [Google Scholar]
- Zhang, J.; Liu, J.L. Temperature-sensitive cytoophidium assembly in Schizosaccharomyces pombe. J. Genet. Genom. 2019, 46, 423–432. [Google Scholar]
- Chang, C.C.; Keppeke, G.D.; Antos, C.L.; Peng, M.; Andrade, L.E.C.; Sung, L.Y.; Liu, J.L. CTPS forms the cytoophidium in zebrafish. Exp. Cell Res. 2021, 405, 112684. [Google Scholar]
- Andreadis, C.; Hulme, L.; Wensley, K.; Liu, J.L. The TOR pathway modulates cytoophidium formation in Schizosaccharomyces pombe. J. Biol. Chem. 2019, 294, 14686–14703. [Google Scholar] [PubMed]
- Sun, Z.; Liu, J. mTOR-S6K1 pathway mediates cytoophidium assembly. J. Genet. Genom. 2019, 46, 65–74. [Google Scholar]
- Wu, Z.; Liu, J.L. Cytoophidia respond to nutrient stress in Drosophila. Exp. Cell Res. 2019, 376, 159–167. [Google Scholar] [CrossRef]
- Li, Y.L.; Liu, J.L. Hypoosmolality impedes cytoophidium integrity during nitrogen starvation. Yeast 2021, 38, 276–289. [Google Scholar] [PubMed]
- Chang, C.C.; Peng, M.; Zhong, J.; Zhang, Z.; Keppeke, G.D.; Sung, L.-Y.; Liu, J.-L. Molecular crowding facilitates bundling of IMPDH polymers and cytoophidium formation. Cell. Mol. Life Sci. 2022, 79, 14–19. [Google Scholar] [CrossRef]
- Aughey, G.N.; Grice, S.J.; Shen, Q.J.; Xu, Y.; Chang, C.C.; Azzam, G.; Wang, P.Y.; Freeman-Mills, L.; Pai, L.M.; Sung, L.Y.; et al. Nucleotide synthesis is regulated by cytoophidium formation during neurodevelopment and adaptive metabolism. Biol. Open 2014, 3, 1045–1056. [Google Scholar]
- Shen, Q.J.; Kassim, H.; Huang, Y.; Li, H.; Zhang, J.; Li, G.; Wang, P.Y.; Yan, J.; Ye, F.; Liu, J.L. Filamentation of Metabolic Enzymes in Saccharomyces cerevisiae. J. Genet. Genom. 2016, 43, 393–404. [Google Scholar]
- Deng, R.; Li, Y.L.; Liu, J.L. Cytoophidia Influence Cell Cycle and Size in Schizosaccharomyces pombe. Int. J. Mol. Sci. 2024, 25, 608. [Google Scholar] [CrossRef]
- Azzam, G.; Liu, J.L. Only One Isoform of Drosophila melanogaster CTP Synthase Forms the Cytoophidium. PLoS Genet. 2013, 9, e1003256. [Google Scholar]
- Weng, R.Y.; Zhang, L.; Liu, J.L. Connecting Hippo Pathway and Cytoophidia in Drosophila Posterior Follicle Cells. Int. J. Mol. Sci. 2024, 25, 1453. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, Y.; Zhou, Y.; Wang, Q.Q.; Ding, K.; Zhao, S.; Lu, P.; Liu, J.L. Cytoophidia coupling adipose architecture and metabolism. Cell. Mol. Life Sci. 2022, 79, 534. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, Y.; Mori, H.; Kubota, T.; Takegawa, K. Analysis of ambient pH stress response mediated by iron and copper intake in Schizosaccharomyces pombe. J. Biosci. Bioeng. 2018, 125, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Chen, W.; Pan, Y.; Liu, C.; Zhang, Z.; Shao, S.; Zhang, W. CTPS cytoophidia formation affects cell cycle progression and promotes TSN-induced apoptosis of MKN45 cells. Mol. Med. Rep. 2022, 26, 319. [Google Scholar] [CrossRef] [PubMed]
- Bähler, J.; Wu, J.Q.; Longtine, M.S.; Shah, N.G.; Mckenzie III, A.; Steever, A.B.; Wach, A.; Philippsen, P.; Pringle, J.R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 1998, 14, 943–951. [Google Scholar] [CrossRef]
- Zhang, S.; Li, H.; Liu, J.-L. Long-Term Imaging and Dynamic Analysis of Cytoophidia in Yeast. Methods Mol. Biol. 2021, 2196, 235–244. [Google Scholar]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, R.; Li, Y.-L.; Liu, J.-L. Differential Cytoophidium Assembly between Saccharomyces cerevisiae and Schizosaccharomyces pombe. Int. J. Mol. Sci. 2024, 25, 10092. https://doi.org/10.3390/ijms251810092
Deng R, Li Y-L, Liu J-L. Differential Cytoophidium Assembly between Saccharomyces cerevisiae and Schizosaccharomyces pombe. International Journal of Molecular Sciences. 2024; 25(18):10092. https://doi.org/10.3390/ijms251810092
Chicago/Turabian StyleDeng, Ruolan, Yi-Lan Li, and Ji-Long Liu. 2024. "Differential Cytoophidium Assembly between Saccharomyces cerevisiae and Schizosaccharomyces pombe" International Journal of Molecular Sciences 25, no. 18: 10092. https://doi.org/10.3390/ijms251810092
APA StyleDeng, R., Li, Y.-L., & Liu, J.-L. (2024). Differential Cytoophidium Assembly between Saccharomyces cerevisiae and Schizosaccharomyces pombe. International Journal of Molecular Sciences, 25(18), 10092. https://doi.org/10.3390/ijms251810092






