Discovery and Prediction Study of the Dominant Pharmacological Action Organ of Aconitum carmichaeli Debeaux Using Multiple Bioinformatic Analyses
Abstract
1. Introduction
2. Results
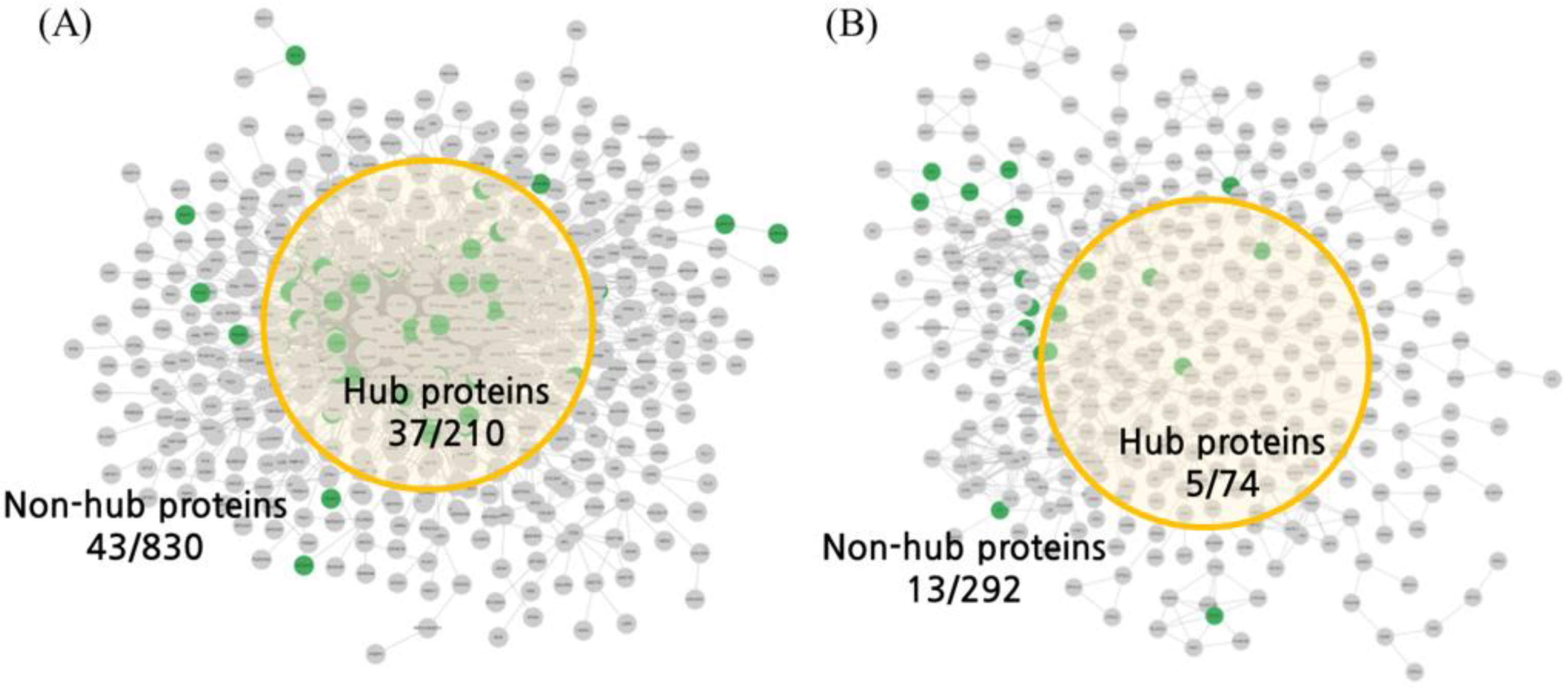
2.1. Organ Network Construction Results
2.2. NP Analysis Results
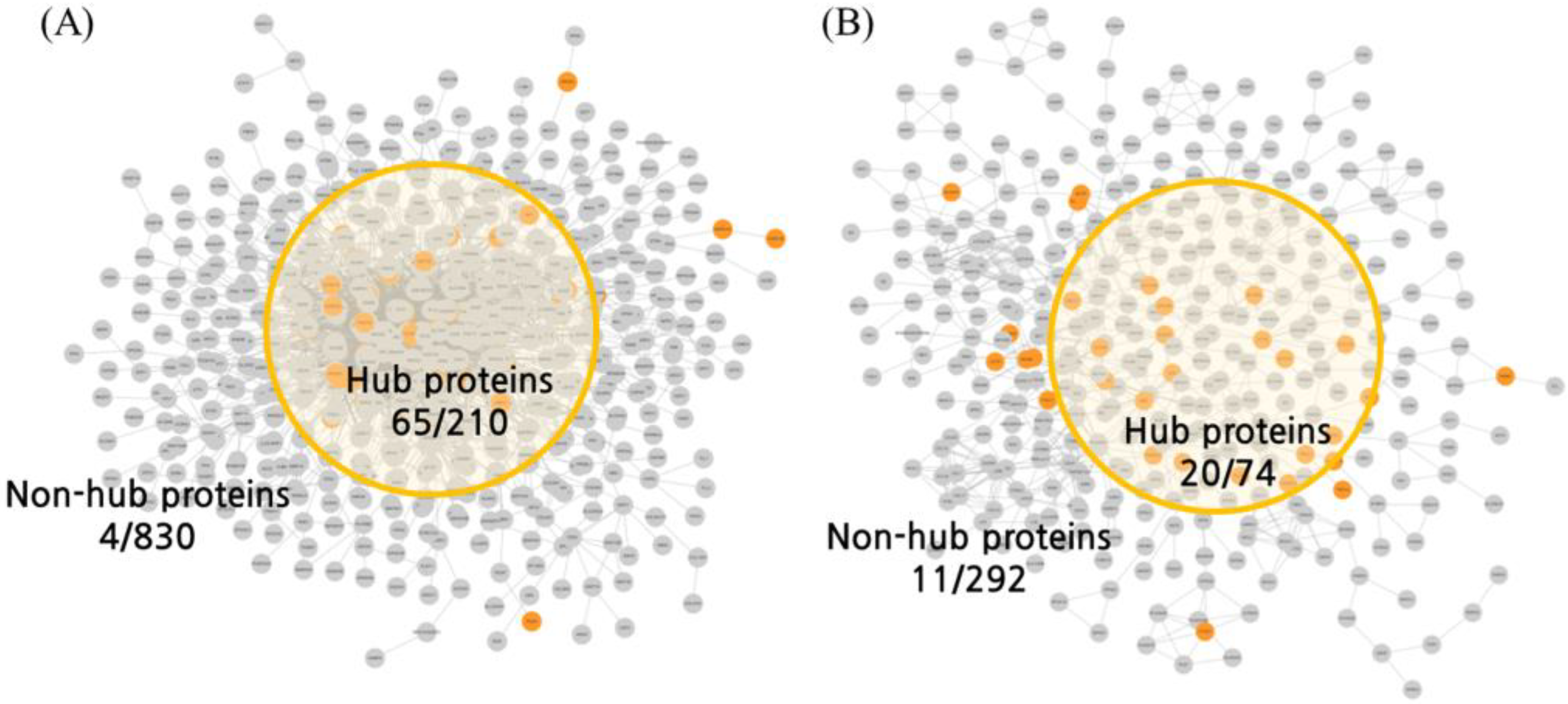
2.3. Molecular Docking Analysis
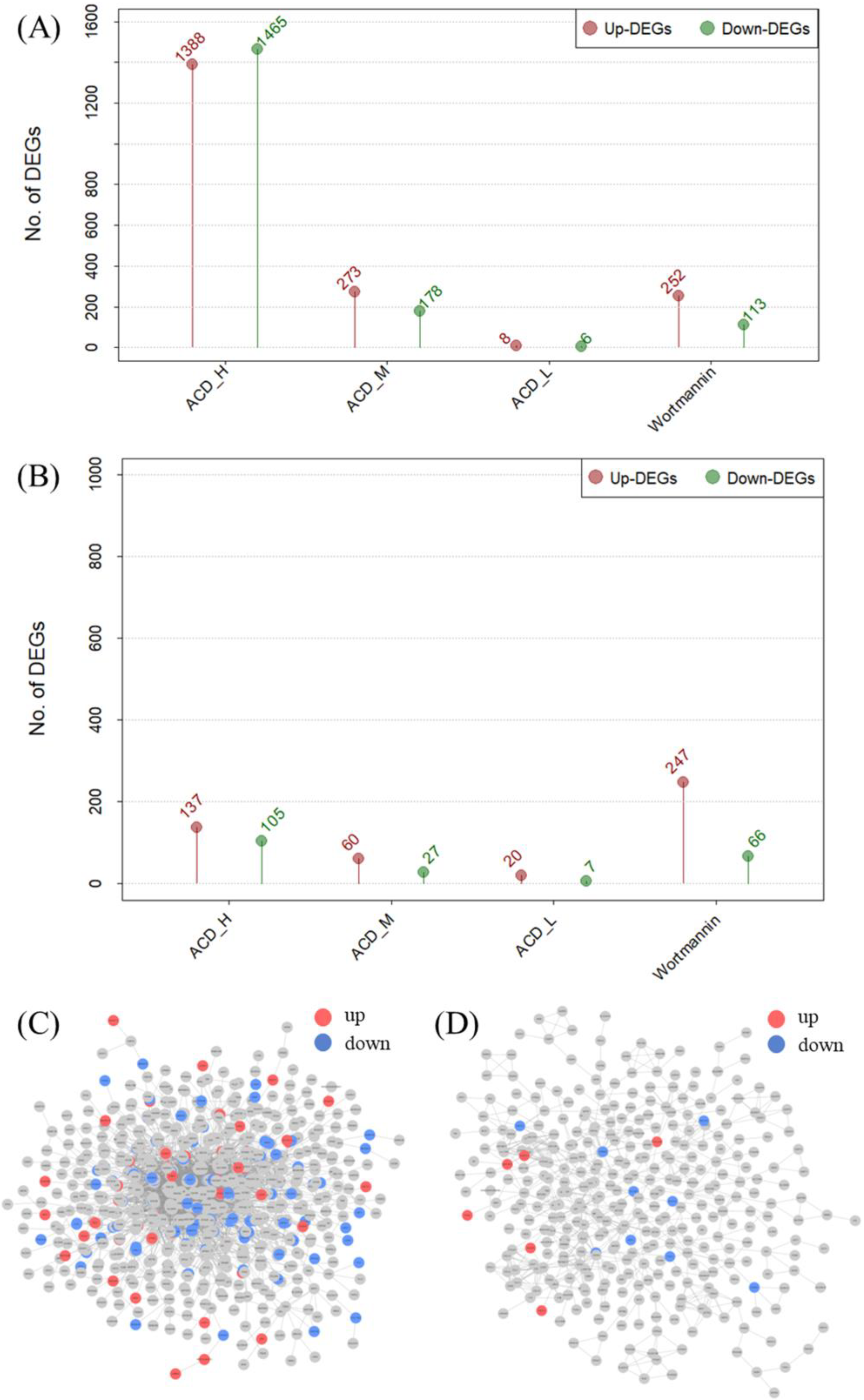
2.4. Transcriptome Verification Experiments
2.4.1. Comparison of the Number of Differentially Expressed Genes (DEGs) by Cell Line
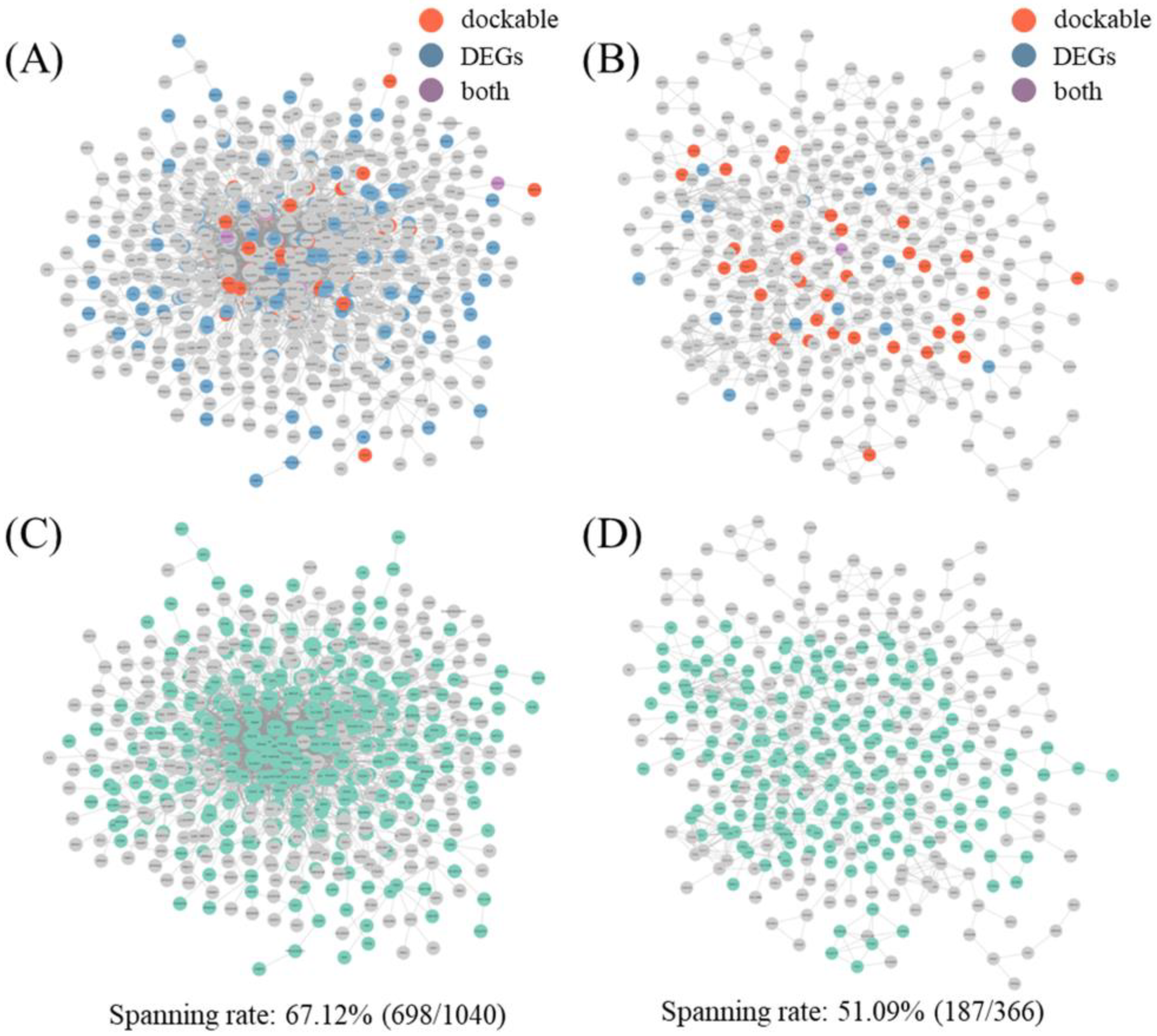
2.4.2. Docking and Transcript-Based Hub Analysis and Network Spanning Results
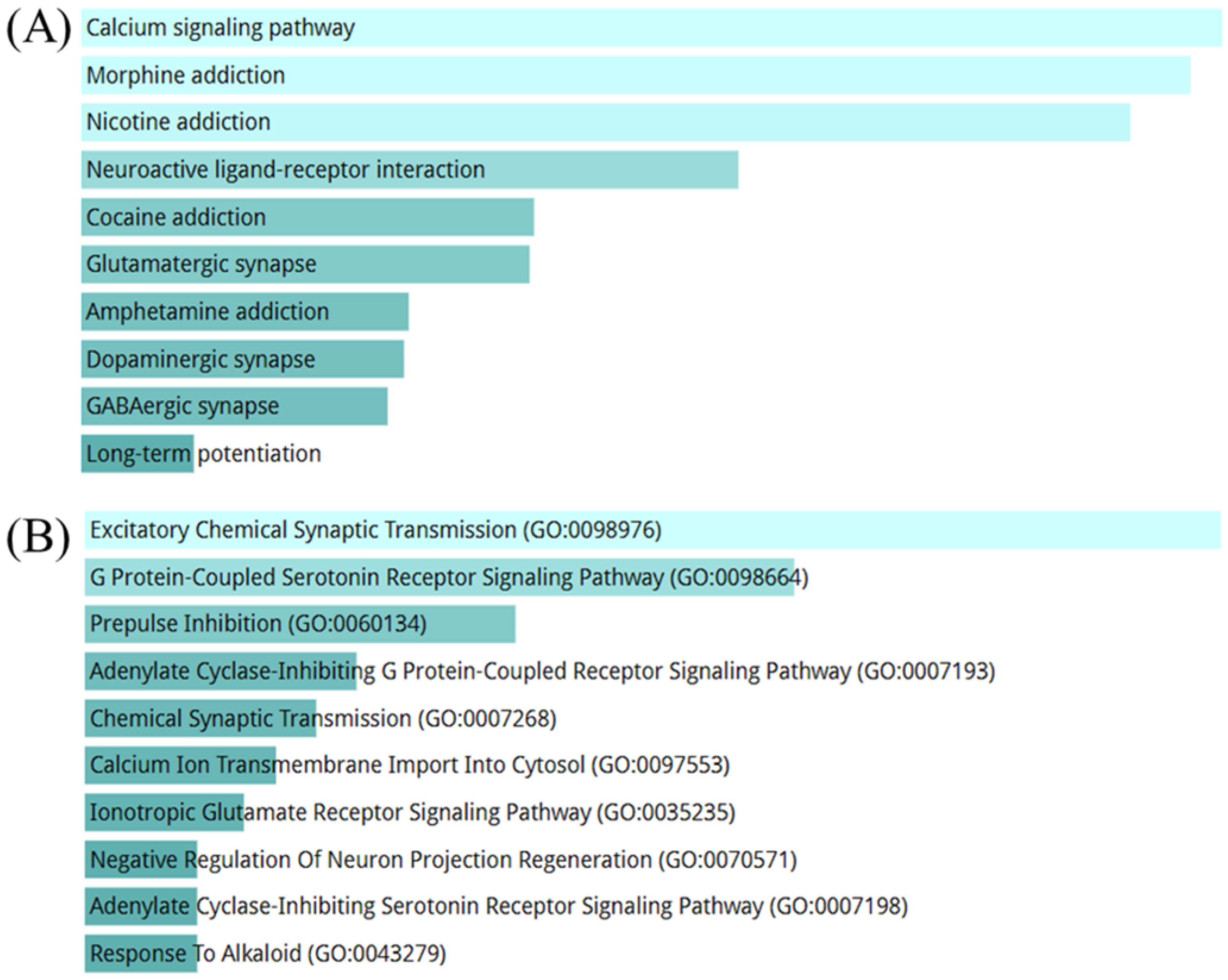
2.4.3. Over-Representation Analysis (ORA) and Gene Set Enrichment Analysis (GSEA) Results Using Transcriptome Analysis Results
3. Discussion
4. Materials and Methods
4.1. Organ Network Construction
4.2. NP Analysis
4.3. Large-Scale Molecular Docking Analysis
4.4. Producing ACD-Induced Transcriptomes in SW1783 and HT29 Cell Lines
4.4.1. Chemicals and Reagents
4.4.2. Preparation of Hot Water Extracts of ACD
4.4.3. Cell Culture
4.4.4. Drug Treatment
4.4.5. RNA Preparation for RNA-Seq
4.4.6. RNA-Seq Preprocessing Method
4.5. Transcriptome Verification Experiments and Analysis
4.5.1. DEG Selection and Comparison of the Number of DEGs
4.5.2. Docking and Transcript-Based Hub Analysis and Network Spanning Analysis
4.5.3. ORA and GSEA Results Using Transcriptome Analysis Results
4.6. Phytochemical Analysis of ACD Extract by LC/MSMS
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gao, Y.B.; Fan, H.; Nie, A.Z.; Yang, K.; Xing, H.Y.; Gao, Z.Q.; Yang, L.J.; Wang, Z.; Zhang, L.Q. Aconitine: A review of its pharmacokinetics, pharmacology, toxicology and detoxification. J. Ethnopharmacol. 2022, 293, 115270. [Google Scholar] [CrossRef]
- Xing, Z.W.; Yang, C.; He, J.Y.; Feng, Y.Q.; Li, X.; Peng, C.; Li, D. Cardioprotective Effects of Aconite in Isoproterenol-Induced Myocardial Infarction in Rats. Oxid. Med. Cell. Longev. 2022, 2022, 1090893. [Google Scholar] [CrossRef]
- Zhao, P.Y.; Tian, Y.; Geng, Y.F.; Zeng, C.J.; Ma, X.Y.; Kang, J.; Lu, L.; Zhang, X.; Tang, B.; Geng, F.N. Aconitine and its derivatives: Bioactivities, structure-activity relationships and preliminary molecular mechanisms. Front. Chem. 2024, 12, 1339364. [Google Scholar] [CrossRef]
- Park, T.E.; Hwang, M.S.; Kim, K.S. Transdermal Delivery of Polymeric Nanoparticles Containing Aconite Root for the Treatment of Chemotherapy-Induced Peripheral Neuropathy. Adv. NanoBiomed Res. 2024, 4, 2400006. [Google Scholar] [CrossRef]
- Tanimura, Y.; Yoshida, M.; Ishiuchi, K.; Ohsawa, M.; Makino, T. Neoline is the active ingredient of processed aconite root against murine peripheral neuropathic pain model, and its pharmacokinetics in rats. J. Ethnopharmacol. 2019, 241, 111859. [Google Scholar] [CrossRef]
- Kondo, T.; Ishida, T.; Ye, K.; Muraguchi, M.; Tanimura, Y.; Yoshida, M.; Ishiuchi, K.; Abe, T.; Nikawa, T.; Hagihara, K.; et al. Suppressive effects of processed aconite root on dexamethasone-induced muscle ring finger protein-1 expression and its active ingredients. J. Nat. Med. 2022, 76, 594–604. [Google Scholar] [CrossRef]
- Xu, W.M.; Zhang, M.; Liu, H.W.; Wei, K.; He, M.; Li, X.Y.; Hu, D.Y.; Yang, S.; Zheng, Y.G. Antiviral activity of aconite alkaloids from Aconitum carmichaelii Debx. Nat. Prod. Res. 2019, 33, 1486–1490. [Google Scholar] [CrossRef]
- World Health Organization. WHO International Standard Terminologies on Traditional Chinese Medicine; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- Yang, J.M.; Li, G.; Wang, M.; Jin, Y.X.; Zheng, F.J.; Sun, Y.; Gao, Y.S.; Zhang, S.J.; Kang, P.F.; Chen, L.; et al. The Effects of Natural Chinese Medicine Aconite Root, Dried Ginger Rhizome, and on Rectal and Skin Temperatures at Acupuncture Points. Evid-Based Compl. Alt. 2017, 2017, 7250340. [Google Scholar] [CrossRef]
- Chan, T.Y.K. Aconite poisoning. Clin. Toxicol. 2009, 47, 279–285. [Google Scholar] [CrossRef]
- Gros, M.; Gros, B.; Mesonero, J.E.; Latorre, E. Neurotransmitter Dysfunction in Irritable Bowel Syndrome: Emerging Approaches for Management. J. Clin. Med. 2021, 10, 3429. [Google Scholar] [CrossRef]
- Barabási, A.L.; Oltvai, Z.N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef]
- Baek, S.J.; Lee, H.; Park, S.M.; Park, M.; Yi, J.M.; Kim, N.S.; Kim, A.; Cha, S. Identification of a novel anticancer mechanism of extracts based on systematic transcriptome analysis. Biomed. Pharmacother. 2022, 148, 112748. [Google Scholar] [CrossRef]
- Park, S.M.; Kim, A.; Lee, H.; Baek, S.J.; Kim, N.S.; Park, M.; Yi, J.M.; Cha, S.W. Systematic transcriptome analysis reveals molecular mechanisms and indications of bupleuri radix. Front. Pharmacol. 2022, 13, 1010520. [Google Scholar] [CrossRef]
- Kim, A.; Kim, Y.R.; Park, S.M.; Lee, H.; Park, M.; Yi, J.M.; Cha, S.; Kim, N.S. Jakyak-gamcho-tang, a decoction of Paeoniae Radix and Glycyrrhizae Radix et Rhizoma, ameliorates dexamethasone-induced muscle atrophy and muscle dysfunction. Phytomedicine 2024, 123, 155057. [Google Scholar] [CrossRef]
- Hopkins, A.L. Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 2008, 4, 682–690. [Google Scholar] [CrossRef]
- Dogrusoz, U.; Giral, E.; Cetintas, A.; Civril, A.; Demir, E. A layout algorithm for undirected compound graphs. Inform. Sci. 2009, 179, 980–994. [Google Scholar] [CrossRef]
- Park, M.; Baek, S.J.; Park, S.M.; Yi, J.M.; Cha, S. Comparative study of the mechanism of natural compounds with similar structures using docking and transcriptome data for improving in silico herbal medicine experimentations. Brief Bioinform. 2023, 24, bbad344. [Google Scholar] [CrossRef]
- Park, M.; Yi, J.M.; Kim, N.S.; Lee, S.Y.; Lee, H.S. Effect of Terpenes: Verifying Modes of Action Using Molecular Docking, Drug-Induced Transcriptomes, and Diffusion Network Analyses. Int. J. Mol. Sci. 2024, 25, 4636. [Google Scholar] [CrossRef]
- Crosara, K.T.B.; Moffa, E.B.; Xiao, Y.Z.; Siqueira, W.L. Merging and salivary protein complex partners using the STRING database: A tutorial. J. Proteom. 2018, 171, 87–94. [Google Scholar] [CrossRef]
- Moosavi, S.; Rahgozar, M.; Rahimi, A. Protein function prediction using neighbor relativity in protein-protein interaction network. Comput. Biol. Chem. 2013, 43, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Sandler, T.; Blitzer, J.; Talukdar, P.; Ungar, L. Regularized learning with networks of features. In Advances in Neural Information Processing Systems; Neural Information Processing Systems Foundation, Inc.: La Jolla, CA, USA, 2008; Volume 21. [Google Scholar]
- Strang, G. Introduction to Linear Algebra; SIAM: Philadelphia, PA, USA, 2022. [Google Scholar]
- Masuda, N.; Porter, M.A.; Lambiotte, R. Random walks and diffusion on networks. Phys. Rep. 2017, 716, 1–58. [Google Scholar] [CrossRef]
- Bliss, T.V.; Collingridge, G.L. A synaptic model of memory: Long-term potentiation in the hippocampus. Nature 1993, 361, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Malenka, R.C.; Bear, M.F. LTP and LTD: An embarrassment of riches. Neuron 2004, 44, 5–21. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.-S.; Liang, W.-Z. Cytotoxic Effects of Mesaconitine, the Aconitum carmichaelii Debx Bioactive Compound, on HBEC-5i Human Brain Microvascular Endothelial Cells: Role of Ca2+ Signaling-Mediated Pathway. Neurotox. Res. 2021, 39, 256–265. [Google Scholar] [CrossRef]
- Li, M.T.; Xie, X.F.; Chen, H.M.; Xiong, Q.Y.; Tong, R.S.; Peng, C.; Peng, F. Aconitine induces cardiotoxicity through regulation of calcium signaling pathway in zebrafish embryos and in H9c2 cells. J. Appl. Toxicol. 2020, 40, 780–793. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Xie, X.F.; Zhang, H.Q.; Wang, P.; Li, G.M.; Chen, J.R.; Chen, G.R.; Cao, X.Y.; Xiong, L.; Peng, F.; et al. Water-soluble alkaloids extracted from Aconiti Radix lateralis praeparata protect against chronic heart failure in rats via a calcium signaling pathway. Biomed. Pharmacother. 2021, 135, 111184. [Google Scholar] [CrossRef]
- Wang, Y.T.; Zhang, H.X.; Wang, J.; Yu, M.; Zhang, Q.Q.; Yan, S.; You, D.Y.; Shi, L.L.; Zhang, L.H.; Wang, L.M.; et al. Aconiti lateralis Radix Praeparata inhibits Alzheimer’s disease by regulating the complex regulation network with the core of GRIN1 and MAPK1. Pharm. Biol. 2021, 59, 309–318. [Google Scholar] [CrossRef]
- Uhlen, M.; Oksvold, P.; Fagerberg, L.; Lundberg, E.; Jonasson, K.; Forsberg, M.; Zwahlen, M.; Kampf, C.; Wester, K.; Hober, S.; et al. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 2010, 28, 1248–1250. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Liu, Z.Y.; Guo, F.F.; Wang, Y.; Li, C.; Zhang, X.L.; Li, H.L.; Diao, L.H.; Gu, J.Y.; Wang, W.; Li, D.; et al. BATMAN-TCM: A Bioinformatics Analysis Tool for Molecular mechANism of Traditional Chinese Medicine. Sci. Rep. 2016, 6, 21146. [Google Scholar] [CrossRef] [PubMed]
- Pesarin, F.; Salmaso, L. The permutation testing approach: A review. Statistica 2010, 70, 481–509. [Google Scholar]
- Ru, J.L.; Li, P.; Wang, J.N.; Zhou, W.; Li, B.H.; Huang, C.; Li, P.D.; Guo, Z.H.; Tao, W.Y.; Yang, Y.F.; et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J. Cheminformatics 2014, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Lee, M.K.; Jang, H.; Lee, J.J.; Lee, S.H.; Jang, Y.; Jang, H.; Kim, A. TM-MC 2.0: An enhanced chemical database of medicinal materials in Northeast Asian traditional medicine. BMC Complement. Med. 2024, 24, 40. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.J.; Gindulyte, A.; He, J.; He, S.Q.; Li, Q.L.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminformatics 2011, 3, 33. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. Software News and Update AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 22 May 2024).
- Krueger, F. Trim Galore: A Wrapper Tool around Cutadapt and FastQC to Consistently Apply Quality and Adapter Trimming to FastQ Files. Available online: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 22 May 2024).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Hagberg, A.; Swart, P.J.; Schult, D.A. Exploring Network Structure, Dynamics, and Function Using NetworkX; Los Alamos National Laboratory (LANL): Los Alamos, NM, USA, 2008. [Google Scholar]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.N.; Wang, Z.C.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed]
- Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.L.; Hill, D.P.; et al. The Gene Ontology knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Korotkevich, G.; Sukhov, V.; Budin, N.; Shpak, B.; Artyomov, M.N.; Sergushichev, A. Fast gene set enrichment analysis. BioRxiv 2016, 060012. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
| Analysis | Organ | #Whole Proteins | #Hit Proteins | Hit Ratio |
|---|---|---|---|---|
| Network pharmacology | BSN | 1040 | 80 | 7.69% |
| ISN | 366 | 18 | 4.92% | |
| Molecular docking | BSN | 1040 | 69 | 6.39% |
| ISN | 366 | 31 | 8.47% | |
| DEG count | BSN | 1040 | 172 | 16.54% |
| ISN | 366 | 15 | 4.10% |
| Analysis | Organ | CCM | #Hub | HDT | #Hit | Hit Ratio | p-Value |
|---|---|---|---|---|---|---|---|
| NP | BSN | Degree | 210 | >11 | 37 | 17.62% | <0.00001 |
| ISN | Degree | 74 | >6 | 5 | 4.92% | 0.13133 | |
| MD | BSN | Degree | 210 | >11 | 28 | 13.33% | 0.00004 |
| CoSE | - | 65 | 30.95% | - | |||
| ISN | Degree | 74 | >6 | 5 | 13.51% | 0.08452 | |
| CoSE | - | 20 | 27.03% | - | |||
| IP | BSN | Degree | 210 | >11 | 55 | 26.19% | 0.04705 |
| ISN | Degree | 74 | >6 | 15 | 20.27% | 0.02332 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, M.; Seo, E.-H.; Yi, J.-M.; Cha, S. Discovery and Prediction Study of the Dominant Pharmacological Action Organ of Aconitum carmichaeli Debeaux Using Multiple Bioinformatic Analyses. Int. J. Mol. Sci. 2024, 25, 10219. https://doi.org/10.3390/ijms251810219
Park M, Seo E-H, Yi J-M, Cha S. Discovery and Prediction Study of the Dominant Pharmacological Action Organ of Aconitum carmichaeli Debeaux Using Multiple Bioinformatic Analyses. International Journal of Molecular Sciences. 2024; 25(18):10219. https://doi.org/10.3390/ijms251810219
Chicago/Turabian StylePark, Musun, Eun-Hye Seo, Jin-Mu Yi, and Seongwon Cha. 2024. "Discovery and Prediction Study of the Dominant Pharmacological Action Organ of Aconitum carmichaeli Debeaux Using Multiple Bioinformatic Analyses" International Journal of Molecular Sciences 25, no. 18: 10219. https://doi.org/10.3390/ijms251810219
APA StylePark, M., Seo, E.-H., Yi, J.-M., & Cha, S. (2024). Discovery and Prediction Study of the Dominant Pharmacological Action Organ of Aconitum carmichaeli Debeaux Using Multiple Bioinformatic Analyses. International Journal of Molecular Sciences, 25(18), 10219. https://doi.org/10.3390/ijms251810219






