Baicalin Inhibits FIPV Infection In Vitro by Modulating the PI3K-AKT Pathway and Apoptosis Pathway
Abstract
1. Introduction
2. Results
2.1. In Vitro Screening of Nine Natural Compounds Revealed That Baicalin Can Inhibit FIPV Infection
2.2. Impact of Baicalin on FIPV Replication, Adsorption, and Its Direct Inactivation of FIPV
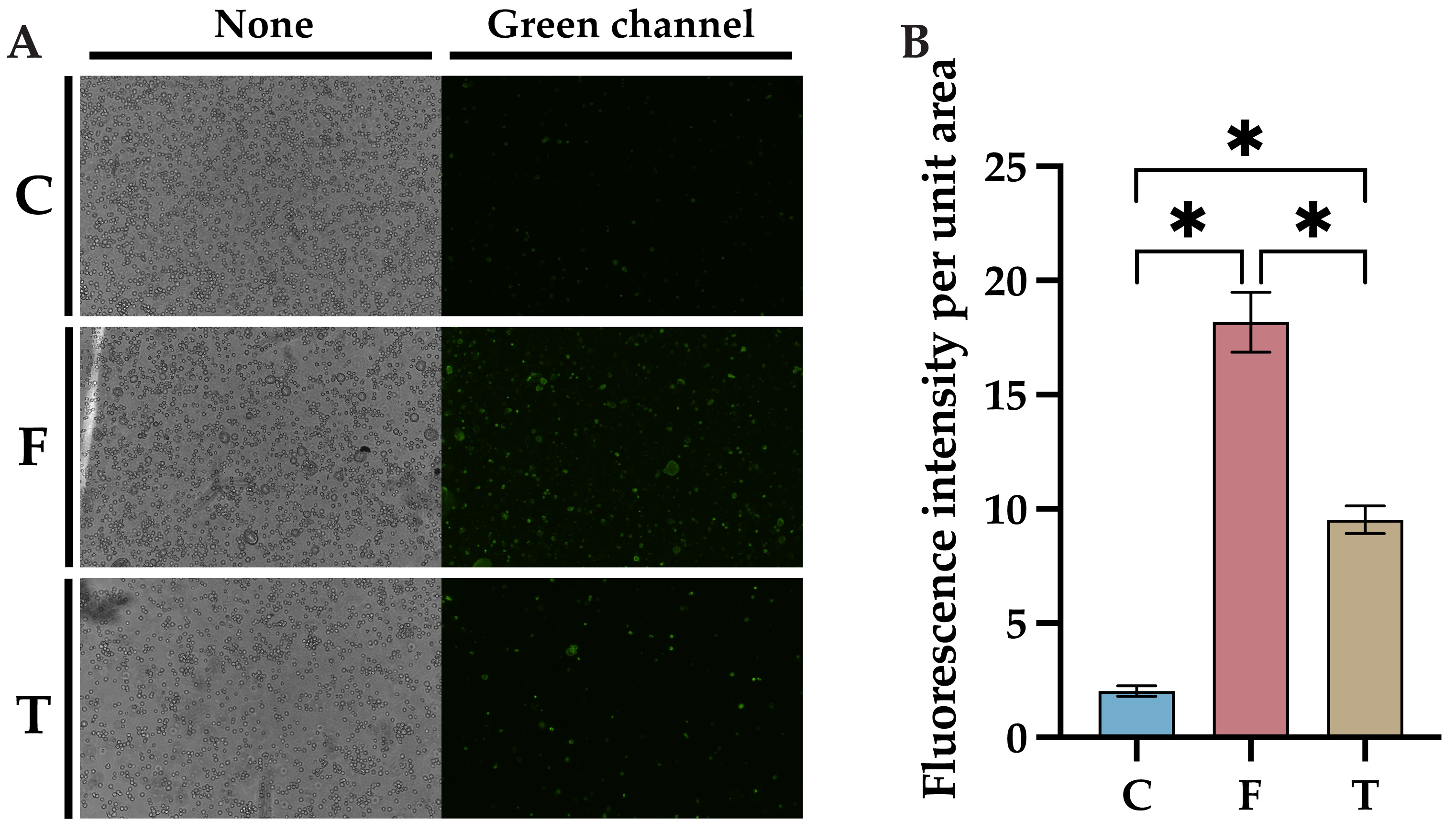
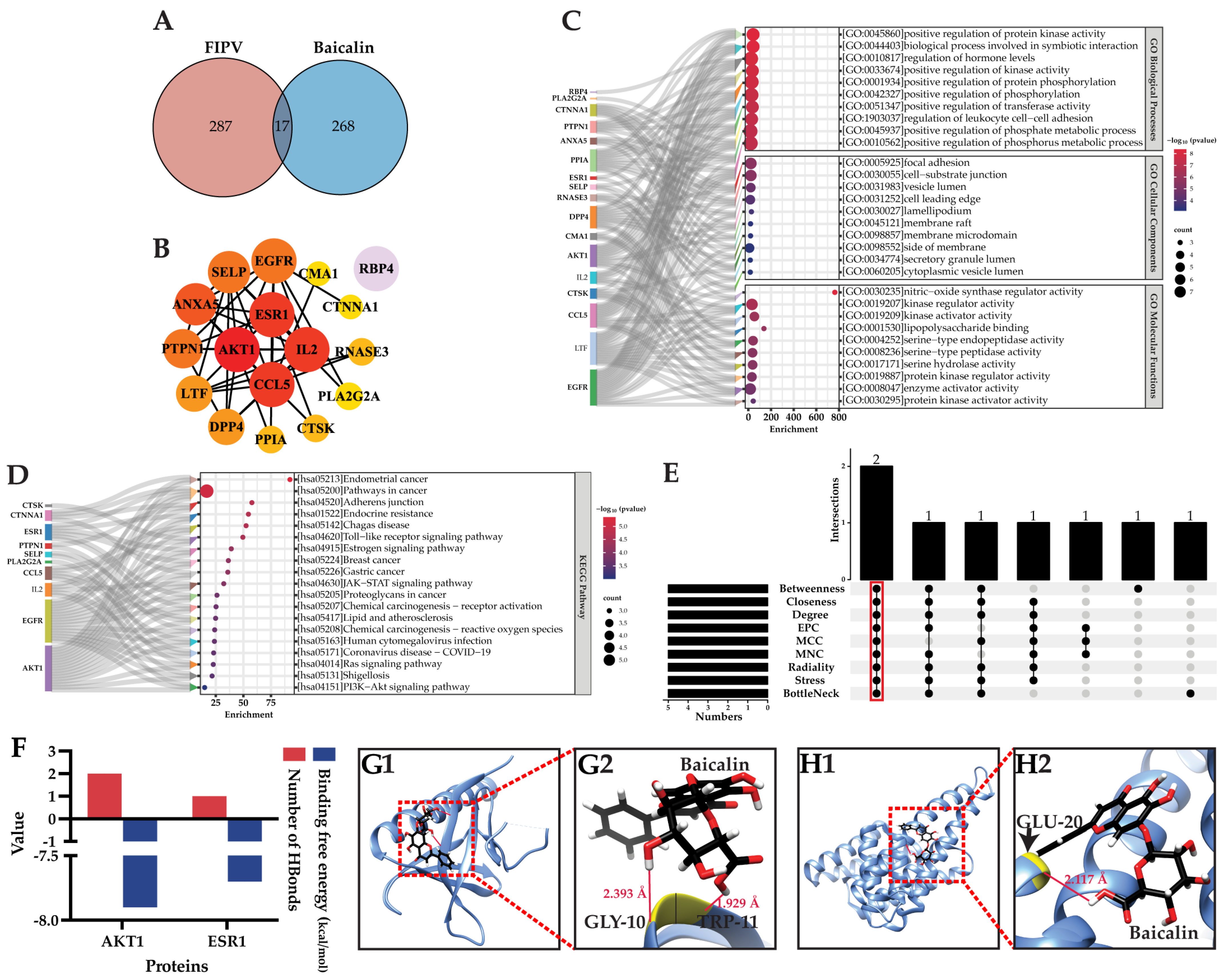
2.3. Baicalin Inhibits FIPV-Induced Apoptosis Network Pharmacology and Molecular Docking Analyses Identify AKT1 as the Best Target of Action for Baicalin against FIPV Infection
2.4. NP and MD Analyses Showed That AKT1 Is the Best Target of Baicalin for Inhibiting FIPV Infection
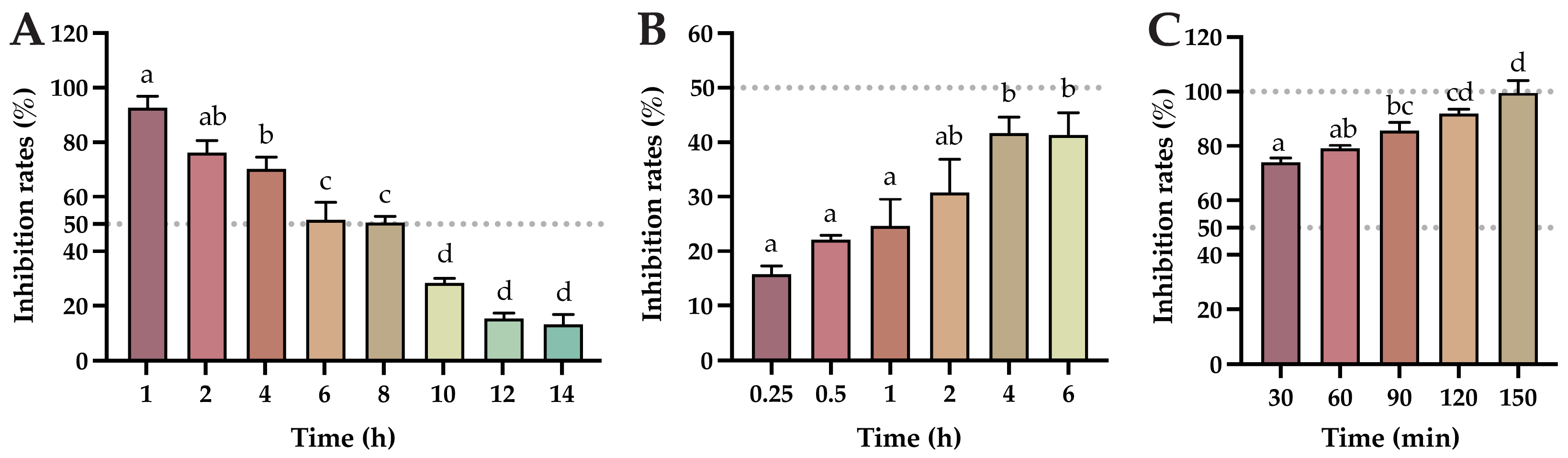
2.5. Determination of the Cellular Viral Load at Different Times of Baicalin Action
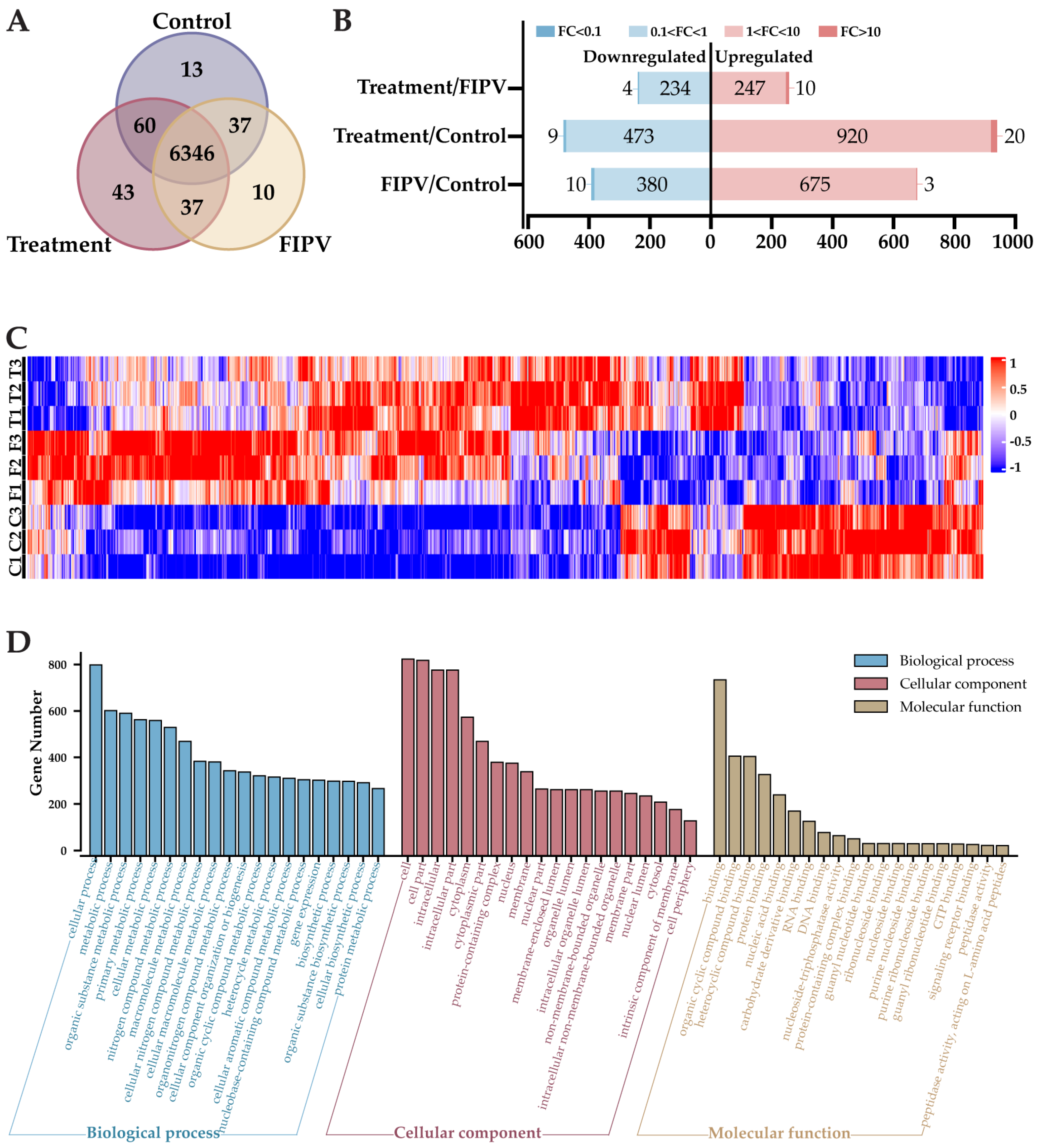
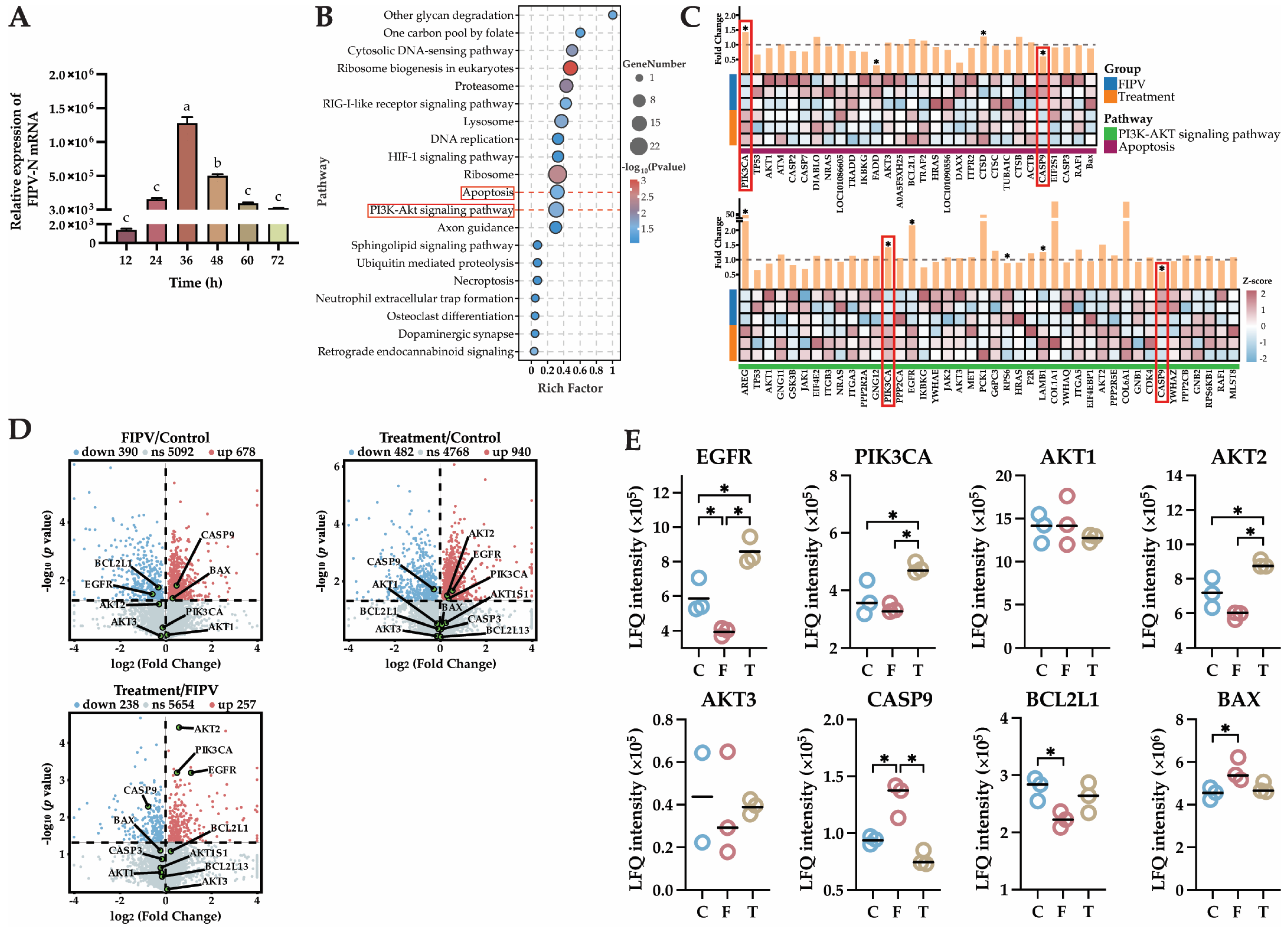
2.6. The 4D-LFQ Proteomics Analyses Suggested That Baicalin May Inhibit FIPV Infection by Regulating the PI3K-AKT Signaling Pathway and the Apoptosis Pathway
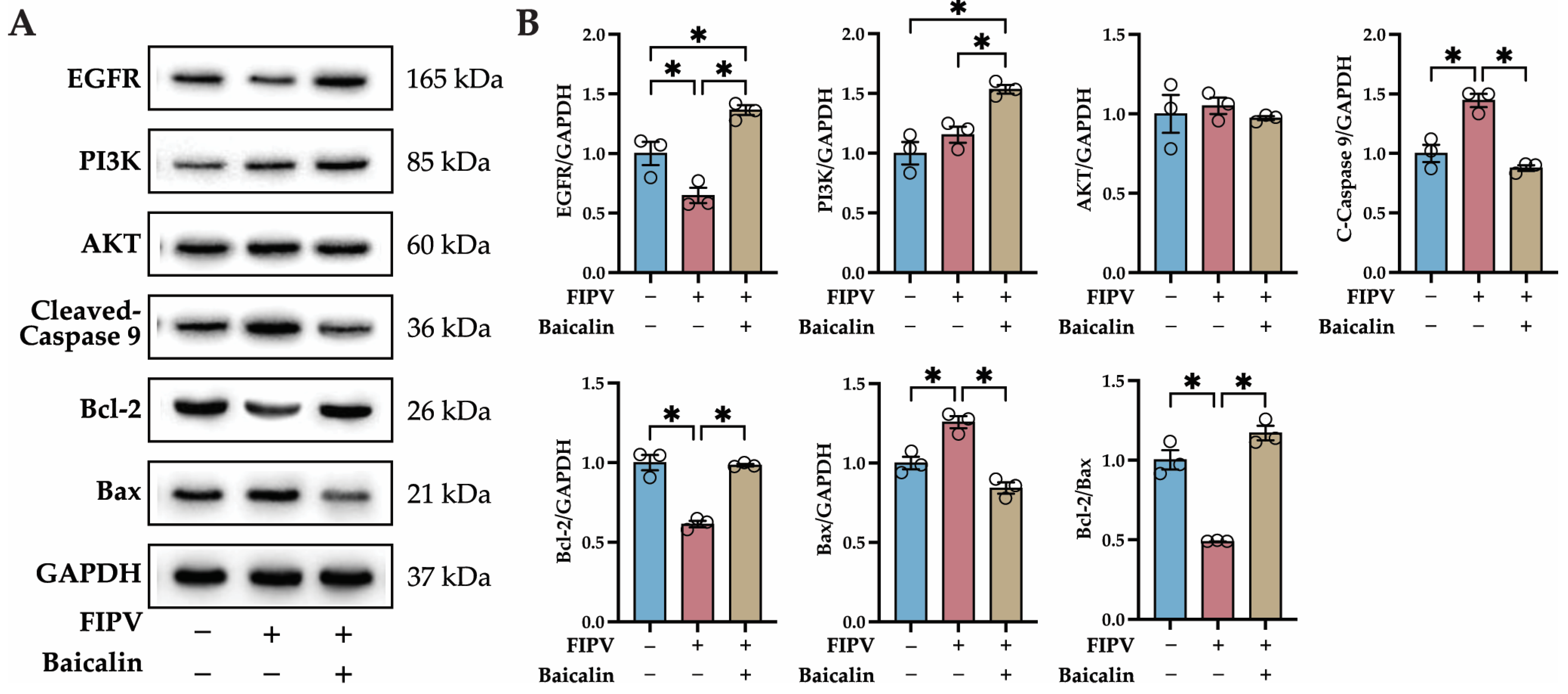
2.7. Identification of Key Proteins in the PI3K-AKT Signaling Pathway and Apoptosis Pathway by Western Blot
3. Discussion
4. Materials and Methods
4.1. Virus, Cells, Compounds, and Antibodies
4.1.1. Virus
4.1.2. Cells
4.1.3. Compounds
4.1.4. Antibodies
4.2. Screening of Natural Compounds That Inhibit FIPV Infection In Vitro
4.2.1. Cytotoxicity Assay of the Compounds
4.2.2. Inhibitory Effects of Compounds on FIPV-Infected Cells
4.3. Impact of the Compound on FIPV Replication, Adsorption, and Direct Inactivation
4.3.1. Impact of the Compound on FIPV Replication
4.3.2. Impact of the Compound on FIPV Adsorption
4.3.3. Impact of the Compound on FIPV Direct Inactivation
4.4. Effect of the Compounds on FIPV-Induced Apoptosis
4.5. NP and MD Were Used to Determine the Best Targets of the Compounds for Inhibiting FIPV Infection
4.6. Determination of the Cellular Viral Load at Different Times of Compound Action
4.7. 4D-LFQ Proteomics Analysis of the Mechanisms by Which Compounds Inhibit FIPV Infection
4.8. Verification of Key Proteins of the Pathway Using Western Blot
4.9. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| FIP | Feline infectious peritonitis |
| FIPV | Feline infectious peritonitis virus |
| CRFK | Crandell Reese feline kidney (cell) |
| MNTC | Maximum noncytotoxic concentration |
| CC50 | 50% cytotoxic concentration |
| EC50 | 50% effective concentration |
| SI | Selection index |
| MIR | Maximum inhibition ratio |
| NP | Network pharmacology |
| MD | Molecular docking |
| 4D-LFQ | 4D label-free quantitative |
| TCM | Traditional Chinese medicine |
| PPI | Protein-protein interaction |
| DEPs | Differentially expressed proteins |
| BP | Biological process |
| CC | Cellular component |
| MF | Molecular function |
| WB | Western blot |
| FBS | Fetal bovine serum |
| MM | Maintenance medium |
Appendix A
References
- Drechsler, Y.; Alcaraz, A.; Bossong, F.J.; Collisson, E.W.; Diniz, P.P.V.P.; Drechsler, Y.; Alcaraz, A.; Bossong, F.J.; Collisson, E.W.; Diniz, P.P.V.P. Feline Coronavirus in Multicat Environments. Vet. Clin. N. Am. Small Anim. Pract. 2011, 41, 1133–1169. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, M.A.; Kennedy, M.A. Feline Infectious Peritonitis. Vet. Clin. N. Am. Small Anim. Pract. 2020, 50, 1001–1011. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.-Y.; Wang, Q.; Liang, X.-Y.; Zhang, S.; Bao, D.; Zhao, H.; Li, S.-B.; Wang, K.; Hu, G.-X.; Gao, F.-S. An Updated Review of Feline Coronavirus: Mind the Two Biotypes. Virus Res. 2023, 326, 199059. [Google Scholar] [CrossRef] [PubMed]
- Felten, S.; Hartmann, K. Diagnosis of Feline Infectious Peritonitis: A Review of the Current Literature. Viruses 2019, 11, 1068. [Google Scholar] [CrossRef] [PubMed]
- Murphy, B.G.; Perron, M.; Murakami, E.; Bauer, K.; Park, Y.; Eckstrand, C.; Liepnieks, M.; Pedersen, N.C. The Nucleoside Analog GS-441524 Strongly Inhibits Feline Infectious Peritonitis (FIP) Virus in Tissue Culture and Experimental Cat Infection Studies. Vet. Microbiol. 2018, 219, 226–233. [Google Scholar] [CrossRef]
- Abaidullah, M.; Peng, S.; Song, X.; Zou, Y.; Li, L.; Jia, R.; Yin, Z. Chlorogenic Acid Is a Positive Regulator of MDA5, TLR7 and NF-κB Signaling Pathways Mediated Antiviral Responses against Gammacoronavirus Infection. Int. Immunopharmacol. 2021, 96, 107671. [Google Scholar] [CrossRef]
- Feng, H.; Zhang, K.; Zhang, K.; Guo, Z.; Liu, Q.; Wang, L.; Wang, X.; Qiu, Z.; Wang, G.; Zhang, J.; et al. Antiviral Activity and Underlying Mechanisms of Baicalin against Avian Infectious Bronchitis Virus in Vitro. Avian Pathol. 2022, 51, 574–589. [Google Scholar] [CrossRef]
- Zhao, Y.; Ling, X.; Zhang, H.; Sun, P.; Sun, Y.; Yin, W.; Fan, K.; Yang, H.; Zhong, J.; Zhang, Z.; et al. Network Pharmacology and Experimental Validation to Reveal the Target of Matrine against PRRSV. iScience 2023, 26, 106371. [Google Scholar] [CrossRef]
- Shirasago, Y.; Inamori, Y.; Suzuki, T.; Tanida, I.; Suzuki, T.; Sugiyama, K.; Wakita, T.; Hanada, K.; Fukasawa, M. Inhibition Mechanisms of Hepatitis C Virus Infection by Caffeic Acid and Tannic Acid. Biol. Pharm. Bull. 2019, 42, 770–777. [Google Scholar] [CrossRef]
- Lin, J.-C. Mechanism of Action of Glycyrrhizic Acid in Inhibition of Epstein-Barr Virus Replication in Vitro. Antivir. Res. 2003, 59, 41–47. [Google Scholar] [CrossRef]
- Lim, C.T.; Tan, K.W.; Wu, M.; Ulferts, R.; Armstrong, L.A.; Ozono, E.; Drury, L.S.; Milligan, J.C.; Zeisner, T.U.; Zeng, J.; et al. Identifying SARS-CoV-2 Antiviral Compounds by Screening for Small Molecule Inhibitors of Nsp3 Papain-like Protease. Biochem. J. 2021, 478, 2517–2531. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Li, X.; Wang, H.; Nie, K.; Meng, Q.; He, J.; Zheng, C. Puerarin: A Potential Therapeutic for SARS-CoV-2 and Hantavirus Co-Infection. Front. Immunol. 2022, 13, 892350. [Google Scholar] [CrossRef] [PubMed]
- Elebeedy, D.; Elkhatib, W.F.; Kandeil, A.; Ghanem, A.; Kutkat, O.; Alnajjar, R.; Saleh, M.A.; Abd El Maksoud, A.I.; Badawy, I.; Al-Karmalawy, A.A. Anti-SARS-CoV-2 Activities of Tanshinone IIA, Carnosic Acid, Rosmarinic Acid, Salvianolic Acid, Baicalein, and Glycyrrhetinic Acid between Computational and in Vitro Insights. RSC Adv. 2021, 11, 29267–29286. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhang, F.; Li, Z.; Jin, W.; Shi, Y. Integration Strategy of Network Pharmacology in Traditional Chinese Medicine: A Narrative Review. J. Tradit. Chin. Med. 2022, 42, 479–486. [Google Scholar] [CrossRef]
- Pinzi, L.; Rastelli, G. Molecular Docking: Shifting Paradigms in Drug Discovery. Int. J. Mol. Sci. 2019, 20, 4331. [Google Scholar] [CrossRef]
- Ding, Y.; Li, J.; Shan, H.; Yang, S.; Wang, X.; Zhao, D. Biomarker Study of Symptomatic Intracranial Atherosclerotic Stenosis in Patients with Acute Ischemic Stroke. Front. Neurol. 2023, 14, 1291929. [Google Scholar] [CrossRef]
- Wei, Z.; Gao, R.; Sun, Z.; Yang, W.; He, Q.; Wang, C.; Zhang, J.; Zhang, X.; Guo, L.; Wang, S. Baicalin Inhibits Influenza A (H1N1)-induced Pyroptosis of Lung Alveolar Epithelial Cells via Caspase-3/GSDME Pathway. J. Med. Virol. 2023, 95, e28790. [Google Scholar] [CrossRef]
- Guo, L.-T.; Wang, S.-Q.; Su, J.; Xu, L.-X.; Ji, Z.-Y.; Zhang, R.-Y.; Zhao, Q.-W.; Ma, Z.-Q.; Deng, X.-Y.; Ma, S.-P. Baicalin Ameliorates Neuroinflammation-Induced Depressive-like Behavior through Inhibition of Toll-like Receptor 4 Expression via the PI3K/AKT/FoxO1 Pathway. J. Neuroinflamm. 2019, 16, 95. [Google Scholar] [CrossRef]
- Boozari, M.; Hosseinzadeh, H. Natural Products for COVID-19 Prevention and Treatment Regarding to Previous Coronavirus Infections and Novel Studies. Phytother. Res. 2021, 35, 864–876. [Google Scholar] [CrossRef]
- Dinda, B.; Dinda, M.; Dinda, S.; De, U.C. An Overview of Anti-SARS-CoV-2 and Anti-Inflammatory Potential of Baicalein and Its Metabolite Baicalin: Insights into Molecular Mechanisms. Eur. J. Med. Chem. 2023, 258, 115629. [Google Scholar] [CrossRef]
- Shuid, A.N.; Safi, N.; Haghani, A.; Mehrbod, P.; Haron, M.S.R.; Tan, S.W.; Omar, A.R. Apoptosis Transcriptional Mechanism of Feline Infectious Peritonitis Virus Infected Cells. Apoptosis 2015, 20, 1457–1470. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Chen, B.; Ren, Q. Baicalin Relieves Hypoxia-Aroused H9c2 Cell Apoptosis by Activating Nrf2/HO-1-Mediated HIF1α/BNIP3 Pathway. Artif. Cells Nanomed. Biotechnol. 2019, 47, 3657–3663. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, M.; Li, W.; Ouyang, J.; Gou, X.; Huang, Y. Mechanism of Action of Daqinjiao Decoction in Treating Cerebral Small Vessel Disease Explored Using Network Pharmacology and Molecular Docking Technology. Phytomedicine 2023, 108, 154538. [Google Scholar] [CrossRef]
- Duan, Z.; Wang, Y.; Lu, Z.; Tian, L.; Xia, Z.-Q.; Wang, K.; Chen, T.; Wang, R.; Feng, Z.; Shi, G.; et al. Wumei Wan Attenuates Angiogenesis and Inflammation by Modulating RAGE Signaling Pathway in IBD: Network Pharmacology Analysis and Experimental Evidence. Phytomedicine 2023, 111, 154658. [Google Scholar] [CrossRef] [PubMed]
- Suo, T.; Wang, H.; Li, Z.; Suo, T.; Wang, H.; Li, Z. Application of Proteomics in Research on Traditional Chinese Medicine. Expert Rev. Proteom. 2016, 13, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Zhong, M.; Zhu, E.; Li, N.; Gong, L.; Xu, H.; Zhong, Y.; Gong, K.; Jiang, S.; Wang, X.; Fei, L.; et al. Identification of Diagnostic Markers Related to Oxidative Stress and Inflammatory Response in Diabetic Kidney Disease by Machine Learning Algorithms: Evidence from Human Transcriptomic Data and Mouse Experiments. Front. Endocrinol. 2023, 14, 1134325. [Google Scholar] [CrossRef]
- Duan, S.; Zhang, M.; Zeng, H.; Song, J.; Zhang, M.; Gao, S.; Yang, H.; Ding, M.; Li, P. Integrated Proteomics and Phosphoproteomics Profiling Reveals the Cardioprotective Mechanism of Bioactive Compounds Derived from Salvia Miltiorrhiza Burge. Phytomedicine 2023, 117, 154897. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, X. The Role of PI3K/AKT/FOXO Signaling in Psoriasis. Arch. Dermatol. Res. 2019, 311, 83–91. [Google Scholar] [CrossRef]
- Tang, Q.; Luan, F.; Yuan, A.; Sun, J.; Rao, Z.; Wang, B.; Liu, Y.; Zeng, N.; Tang, Q.; Luan, F.; et al. Sophoridine Suppresses Herpes Simplex Virus Type 1 Infection by Blocking the Activation of Cellular PI3K/Akt and P38 MAPK Pathways. Front. Microbiol. 2022, 13, 872505. [Google Scholar] [CrossRef]
- Lu, X.; Masic, A.; Li, Y.; Shin, Y.; Liu, Q.; Zhou, Y.; Lu, X.; Masic, A.; Li, Y.; Shin, Y.; et al. The PI3K/Akt Pathway Inhibits Influenza A Virus-Induced Bax-Mediated Apoptosis by Negatively Regulating the JNK Pathway via ASK1. J. Gen. Virol. 2010, 91, 1439–1449. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhen, H.; Mei, Y.; Wang, Y.; Feng, J.; Xu, S.; Fu, X. PI3K/AKT Mediated P53 Down-Regulation Participates in CpG DNA Inhibition of Spontaneous B Cell Apoptosis. Cell. Mol. Immunol. 2009, 6, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xia, J.; Jiang, N.; Xian, Y.; Ju, H.; Wei, Y.; Zhang, X. Corin Protects H2O2-Induced Apoptosis through PI3K/AKT and NF-κB Pathway in Cardiomyocytes. Biomed. Pharmacother. 2018, 97, 594–599. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Bie, M.; Wang, X.; Fan, M.; Chen, B.; Shi, Q.; Jiang, Y. PGRN Exacerbates the Progression of Non-Small Cell Lung Cancer via PI3K/AKT/Bcl-2 Antiapoptotic Signaling. Genes Dis. 2022, 9, 1650–1661. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Zhang, G.; Yang, X.; Wang, M.; Wang, R.; Wan, M.; Wang, J.; Wu, B.; Yan, T.; Jia, Y. A Network Pharmacology Approach and Experimental Validation to Investigate the Anticancer Mechanism and Potential Active Targets of Ethanol Extract of Wei-Tong-Xin against Colorectal Cancer through Induction of Apoptosis via PI3K/AKT Signaling Pathway. J. Ethnopharmacol. 2023, 303, 115933. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Pi, C.; Feng, X.; Wang, Y.; Fu, S.; Zhang, X.; Zhao, L.; Wei, Y.; Hou, Y.; Pi, C.; et al. Antitumor Activity In Vivo and Vitro of New Chiral Derivatives of Baicalin and Induced Apoptosis via the PI3K/Akt Signaling Pathway. Mol. Ther.-Oncolytics 2020, 19, 67–78. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhao, R.; Wu, X.; Bi, X.-Y.; Yang, H.; Zhang, Q.; Zhao, R.; Wu, X.; Bi, X.-Y.; Yang, H. Baicalin Attenuates Blood-Spinal Cord Barrier Disruption and Apoptosis through PI3K/Akt Signaling Pathway after Spinal Cord Injury. Neural Regen. Res. 2022, 17, 1080. [Google Scholar] [CrossRef]
- Zhao, N.; Shi, J.; Xu, H.; Luo, Q.; Li, Q.; Liu, M.; Zhao, N.; Shi, J.; Xu, H.; Luo, Q.; et al. Baicalin Suppresses Glaucoma Pathogenesis by Regulating the PI3K/AKT Signaling in Vitro and in Vivo. Bioengineered 2021, 12, 10187–10198. [Google Scholar] [CrossRef]
- Li, S.; Chen, C.; Zhang, H.; Guo, H.; Wang, H.; Wang, L.; Zhang, X.; Hua, S.; Yu, J.; Xiao, P. Identification of Natural Compounds with Antiviral Activities against SARS-Associated Coronavirus. Antivir. Res. 2005, 67, 18–23. [Google Scholar] [CrossRef]
- Álvarez, Á.L.; Habtemariam, S.; Juan-Badaturuge, M.; Jackson, C.; Parra, F. In Vitro Anti HSV-1 and HSV-2 Activity of Tanacetum Vulgare Extracts and Isolated Compounds: An Approach to Their Mechanisms of Action. Phytother. Res. 2011, 25, 296–301. [Google Scholar] [CrossRef]
- Xiang, Y.; Pei, Y.; Qu, C.; Lai, Z.; Ren, Z.; Yang, K.; Xiong, S.; Zhang, Y.; Yang, C.; Wang, D.; et al. In Vitro Anti-Herpes Simplex Virus Activity of 1,2,4,6-Tetra-O-galloyl-β-d-glucose from Phyllanthus emblica L. (Euphorbiaceae). Phytother. Res. 2011, 25, 975–982. [Google Scholar] [CrossRef]
- Dong, C.-X.; Hayashi, K.; Mizukoshi, Y.; Lee, J.-B.; Hayashi, T. Structures of Acidic Polysaccharides from Basella rubra L. and Their Antiviral Effects. Carbohydr. Polym. 2011, 84, 1084–1092. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New Data Content and Improved Web Interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Shen, Y.; Wang, S.; Li, S.; Zhang, W.; Liu, X.; Lai, L.; Pei, J.; Li, H. PharmMapper 2017 Update: A Web Server for Potential Drug Target Identification with a Comprehensive Target Pharmacophore Database. Nucleic Acids Res. 2017, 45, W356–W360. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ouyang, S.; Yu, B.; Liu, Y.; Huang, K.; Gong, J.; Zheng, S.; Li, Z.; Li, H.; Jiang, H.; et al. PharmMapper Server: A Web Server for Potential Drug Target Identification Using Pharmacophore Mapping Approach. Nucleic Acids Res. 2010, 38, W609–W614. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Pan, C.; Gong, J.; Liu, X.; Li, H. Enhancing the Enrichment of Pharmacophore-Based Target Prediction for the Polypharmacological Profiles of Drugs. J. Chem. Inf. Model. 2016, 56, 1175–1183. [Google Scholar] [CrossRef]
- Safran, M.; Rosen, N.; Twik, M.; BarShir, R.; Stein, T.I.; Dahary, D.; Fishilevich, S.; Lancet, D. The GeneCards Suite. In Practical Guide to Life Science Databases; Abugessaisa, I., Kasukawa, T., Eds.; Springer Nature: Singapore, 2021; pp. 27–56. ISBN 9789811658129. [Google Scholar]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef]
- Paltrinieri, S. Human Severe Acute Respiratory Syndrome (SARS) and Feline Coronaviruses. J. Feline Med. Surg. 2004, 6, 131–132. [Google Scholar] [CrossRef]
- Yang, C.-W.; Peng, T.-T.; Hsu, H.-Y.; Lee, Y.-Z.; Wu, S.-H.; Lin, W.-H.; Ke, Y.-Y.; Hsu, T.-A.; Yeh, T.-K.; Huang, W.-Z.; et al. Repurposing Old Drugs as Antiviral Agents for Coronaviruses. Biomed. J. 2020, 43, 368–374. [Google Scholar] [CrossRef]
- UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING Database in 2023: Protein–Protein Association Networks and Functional Enrichment Analyses for Any Sequenced Genome of Interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Li, M.; Wang, J.; Pan, Y.; Wu, F.-X.; Tang, Y.; Li, M.; Wang, J.; Pan, Y.; Wu, F.-X. CytoNCA: A Cytoscape Plugin for Centrality Analysis and Evaluation of Protein Interaction Networks. Biosystems 2015, 127, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.-H.; Chen, S.-H.; Wu, H.-H.; Ho, C.-W.; Ko, M.-T.; Lin, C.-Y.; Chin, C.-H.; Chen, S.-H.; Wu, H.-H.; Ho, C.-W.; et al. cytoHubba: Identifying Hub Objects and Sub-Networks from Complex Interactome. BMC Syst. Biol. 2014, 8, S11. [Google Scholar] [CrossRef]

| S/N | Compounds | MNTC (μg/mL) | CC50 (μg/mL) |
|---|---|---|---|
| 1 | Baicalin | 31.36 | 114.5 |
| 2 | Matrine | 699.8 | 1110 |
| 3 | Ligustrazine hydrochloride | 969.6 | 2709 |
| 4 | Caffeic acid | 53.43 | 58.78 |
| 5 | Glycyrrhizic acid | 1412 | 3324 |
| 6 | Puerarin | 83.77 | 208.8 |
| 7 | Chlorogenic acid | 1605 | 2170 |
| 8 | Tanshinone IIA | 290.9 | 806.6 |
| 9 | Dihydrotanshinone I | 246.4 | 426.4 |
| 10 | GS-441524 | 36.20 | 70.31 |
| S/N | UniProt ID | Gene Name | Protein Name |
|---|---|---|---|
| 1 | P31749 | AKT1 | RAC-alpha serine/threonine-protein kinase |
| 2 | P08758 | ANXA5 | Annexin A5 |
| 3 | P13501 | CCL5 | C-C motif chemokine 5 |
| 4 | P23946 | CMA1 | Chymase |
| 5 | P35221 | CTNNA1 | Catenin alpha-1 |
| 6 | P43235 | CTSK | Cathepsin K |
| 7 | P27487 | DPP4 | Dipeptidyl peptidase 4 |
| 8 | P00533 | EGFR | Epidermal growth factor receptor |
| 9 | P03372 | ESR1 | Estrogen receptor |
| 10 | P60568 | IL2 | Interleukin-2 |
| 11 | P02788 | LTF | Lactotransferrin |
| 12 | P14555 | PLA2G2A | Phospholipase A2, membrane associated |
| 13 | P62937 | PPIA | Peptidyl-prolyl cis-trans isomerase A |
| 14 | P18031 | PTPN1 | Tyrosine-protein phosphatase non-receptor type 1 |
| 15 | P02753 | RBP4 | Retinol-binding protein 4 |
| 16 | P12724 | RNASE3 | Eosinophil cationic protein |
| 17 | P16109 | SELP | P-selectin |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, Z.; Ma, N.; Shan, M.; Wang, S.; Du, J.; Cheng, J.; Sun, P.; Sun, N.; Jin, L.; Fan, K.; et al. Baicalin Inhibits FIPV Infection In Vitro by Modulating the PI3K-AKT Pathway and Apoptosis Pathway. Int. J. Mol. Sci. 2024, 25, 9930. https://doi.org/10.3390/ijms25189930
Cao Z, Ma N, Shan M, Wang S, Du J, Cheng J, Sun P, Sun N, Jin L, Fan K, et al. Baicalin Inhibits FIPV Infection In Vitro by Modulating the PI3K-AKT Pathway and Apoptosis Pathway. International Journal of Molecular Sciences. 2024; 25(18):9930. https://doi.org/10.3390/ijms25189930
Chicago/Turabian StyleCao, Zhongda, Nannan Ma, Maoyang Shan, Shiyan Wang, Jige Du, Jia Cheng, Panpan Sun, Na Sun, Lin Jin, Kuohai Fan, and et al. 2024. "Baicalin Inhibits FIPV Infection In Vitro by Modulating the PI3K-AKT Pathway and Apoptosis Pathway" International Journal of Molecular Sciences 25, no. 18: 9930. https://doi.org/10.3390/ijms25189930
APA StyleCao, Z., Ma, N., Shan, M., Wang, S., Du, J., Cheng, J., Sun, P., Sun, N., Jin, L., Fan, K., Yin, W., Li, H., Yin, C., & Sun, Y. (2024). Baicalin Inhibits FIPV Infection In Vitro by Modulating the PI3K-AKT Pathway and Apoptosis Pathway. International Journal of Molecular Sciences, 25(18), 9930. https://doi.org/10.3390/ijms25189930





