Transglutaminase 1: Emerging Functions beyond Skin
Abstract
:1. Overview of Transglutaminases (TGs)
2. Structure of TG1
3. Activation of TG1
4. Identification of Substrates and Activity of TG1
5. TG1 in Skin
6. TG1 in Vasculature
7. TG1 in Lung and Nasal Systems
8. TG1 in Hemostasis
9. TG1 in Neurodegenerative Diseases
10. TG1 in Cancer
11. TG1 in Fibrosis
12. TG1 in Innate Immunity: Neutrophils
13. TG1 and Regulation of Bone Mass
14. TG1 in the Visual System
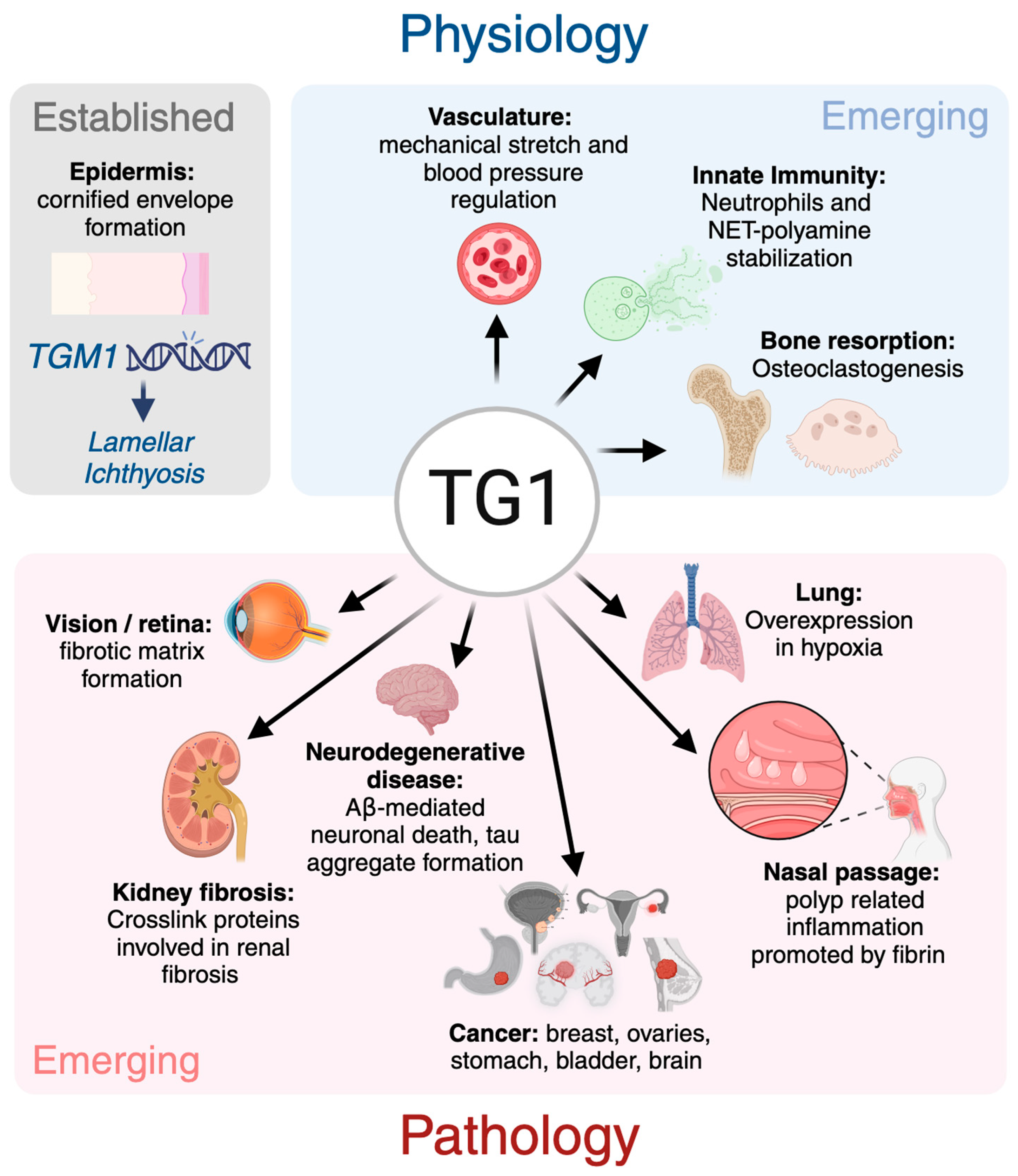
15. Future Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eckert, R.L.; Kaartinen, M.T.; Nurminskaya, M.; Belkin, A.M.; Colak, G.; Johnson, G.V.; Mehta, K. Transglutaminase regulation of cell function. Physiol. Rev. 2014, 94, 383–417. [Google Scholar] [CrossRef] [PubMed]
- Hiiragi, T.; Sasaki, H.; Nagafuchi, A.; Sabe, H.; Shen, S.C.; Matsuki, M.; Yamanishi, K.; Tsukita, S. Transglutaminase type 1 and its cross-linking activity are concentrated at adherens junctions in simple epithelial cells. J. Biol. Chem. 1999, 274, 34148–34154. [Google Scholar] [CrossRef] [PubMed]
- Lorand, L.; Graham, R.M. Transglutaminases: Crosslinking enzymes with pleiotropic functions. Nat. Rev. Mol. Cell Biol. 2003, 4, 140–156. [Google Scholar] [CrossRef]
- Elli, L.; Bergamini, C.; Bardella, M.; Schuppan, D. Transglutaminases in inflammation and fibrosis of the gastrointestinal tract and the liver. Dig. Liver Dis. 2009, 41, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Tabolacci, C.; Lentini, A.; Provenzano, B.; Beninati, S. Evidences for a role of protein cross-links in transglutaminase-related disease. Amino Acids 2012, 42, 975–986. [Google Scholar] [CrossRef]
- Folk, J.; Park, M.; Chung, S.; Schrode, J.; Lester, E.; Cooper, H. Polyamines as physiological substrates for transglutaminases. J. Biol. Chem. 1980, 255, 3695–3700. [Google Scholar] [CrossRef] [PubMed]
- Hummerich, R.; Thumfart, J.-O.; Findeisen, P.; Bartsch, D.; Schloss, P. Transglutaminase-mediated transamidation of serotonin, dopamine and noradrenaline to fibronectin: Evidence for a general mechanism of monoaminylation. FEBS Lett. 2012, 586, 3421–3428. [Google Scholar] [CrossRef]
- Schuppan, D.; Mäki, M.; Lundin, K.E.; Isola, J.; Friesing-Sosnik, T.; Taavela, J.; Popp, A.; Koskenpato, J.; Langhorst, J.; Hovde, Ø. A randomized trial of a transglutaminase 2 inhibitor for celiac disease. N. Engl. J. Med. 2021, 385, 35–45. [Google Scholar] [CrossRef]
- Grenard, P.; Bates, M.K.; Aeschlimann, D. Evolution of transglutaminase genes: Identification of a transglutaminase gene cluster on human chromosome 15q15: Structure of the gene encoding transglutaminase X and a novel gene family member, transglutaminase Z. J. Biol. Chem. 2001, 276, 33066–33078. [Google Scholar] [CrossRef]
- Yokosaki, Y. A New Integrin-Binding Site on a Transglutaminase-Catalyzed Polymer. In Transglutaminases: Multiple Functional Modifiers and Targets for New Drug Discovery; Springer: Tokyo, Japan, 2015; pp. 129–151. [Google Scholar]
- Iismaa, S.E.; Mearns, B.M.; Lorand, L.; Graham, R.M. Transglutaminases and disease: Lessons from genetically engineered mouse models and inherited disorders. Physiol. Rev. 2009, 89, 991–1023. [Google Scholar] [CrossRef]
- Chung, S.; Folk, J. Transglutaminase from hair follicle of guinea pig. Proc. Natl. Acad. Sci. USA 1972, 69, 303–307. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, L.A.; Martin, C.M. Human epidermal transamidase. J. Investig. Dermatol. 1975, 64, 316–321. [Google Scholar] [CrossRef] [PubMed]
- Buxman, M.M.; Wuepper, K.D. Keratin cross-linking and epidermal transglutaminase. A review with observations on the histochemical and immunochemical localization of the enzyme. J. Investig. Dermatol. 1975, 65, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Rice, R.H.; Green, H. The cornified envelope of terminally differentiated human epidermal keratinocytes consists of cross-linked protein. Cell 1977, 11, 417–422. [Google Scholar] [CrossRef]
- Thacher, S.M.; Rice, R.H. Keratinocyte-specific transglutaminase of cultured human epidermal cells: Relation to cross-linked envelope formation and terminal differentiation. Cell 1985, 40, 685–695. [Google Scholar] [CrossRef]
- Rice, R.; Chakravarty, R.; Chen, J.; O’Callahan, W.; Rubin, A. Keratinocyte transglutaminase: Regulation and release. In Advances in Post-Translational Modifications of Proteins and Aging; Springer: Boston, MA, USA, 1988; pp. 51–61. [Google Scholar]
- Thacher, S. Purification of keratinocyte transglutaminase and its expression during squamous differentiation. J. Investig. Dermatol. 1989, 92, 578–584. [Google Scholar]
- Egberts, F.; Heinrich, M.; Jensen, J.-M.; Winoto-Morbach, S.; Pfeiffer, S.; Wickel, M.; Schunck, M.; Steude, J.; Saftig, P.; Proksch, E. Cathepsin D is involved in the regulation of transglutaminase 1 and epidermal differentiation. J. Cell Sci. 2004, 117, 2295–2307. [Google Scholar] [CrossRef]
- Elias, P.M.; Schmuth, M.; Uchida, Y.; Rice, R.H.; Behne, M.; Crumrine, D.; Feingold, K.R.; Holleran, W.M.; Pharm, D. Basis for the permeability barrier abnormality in lamellar ichthyosis. Exp. Dermatol. 2002, 11, 248–256. [Google Scholar] [CrossRef]
- Terrinoni, A.; Serra, V.; Codispoti, A.; Talamonti, E.; Bui, L.; Palombo, R.; Sette, M.; Campione, E.; Didona, B.; Annicchiarico-Petruzzelli, M. Novel transglutaminase 1 mutations in patients affected by lamellar ichthyosis. Cell Death Dis. 2012, 3, e416. [Google Scholar] [CrossRef]
- Matsuki, M.; Yamashita, F.; Ishida-Yamamoto, A.; Yamada, K.; Kinoshita, C.; Fushiki, S.; Ueda, E.; Morishima, Y.; Tabata, K.; Yasuno, H. Defective stratum corneum and early neonatal death in mice lacking the gene for transglutaminase 1 (keratinocyte transglutaminase). Proc. Natl. Acad. Sci. USA 1998, 95, 1044–1049. [Google Scholar] [CrossRef]
- Russell, L.; DiGiovanna, J.; Hashem, N.; Compton, J.; Bale, S. Linkage of autosomal recessive lamellar ichthyosis to chromosome 14q. Am. J. Hum. Genet. 1994, 55, 1146. [Google Scholar] [PubMed]
- Polakowska, R.; Eddy, R.; Shows, T.; Goldsmith, L. Epidermal type I transglutaminase (TGM1) is assigned to human chromosome 14. Cytogenet. Genome Res. 1991, 56, 105–107. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.-G.; McBride, O.; Wang, M.; Kim, S.; Idler, W.; Steinert, P. Structure and organization of the human transglutaminase 1 gene. J. Biol. Chem. 1992, 267, 7710–7717. [Google Scholar] [CrossRef] [PubMed]
- Yamanishi, K.; Inazawa, J.; Liew, F.; Nonomura, K.; Ariyama, T.; Yasuno, H.; Abe, T.; Doi, H.; Hirano, J.; Fukushima, S. Structure of the gene for human transglutaminase 1. J. Biol. Chem. 1992, 267, 17858–17863. [Google Scholar] [CrossRef] [PubMed]
- Demény, M.Á.; Korponay-Szabó, I.; Fésüs, L. Structure of transglutaminases: Unique features serve diverse functions. In Transglutaminases: Multiple Functional Modifiers and Targets for New Drug Discovery; Springer: Tokyo, Japan, 2015; pp. 1–41. [Google Scholar]
- Herman, M.L.; Farasat, S.; Steinbach, P.J.; Wei, M.H.; Toure, O.; Fleckman, P.; Blake, P.; Bale, S.J.; Toro, J.R. Transglutaminase-1 gene mutations in autosomal recessive congenital ichthyosis: Summary of mutations (including 23 novel) and modeling of TGase-1. Hum. Mutat. 2009, 30, 537–547. [Google Scholar] [CrossRef]
- Eckert, R.L.; Sturniolo, M.T.; Jans, R.; Kraft, C.A.; Jiang, H.; Rorke, E.A. TIG3: A regulator of type I transglutaminase activity in epidermis. Amino Acids 2009, 36, 739–746. [Google Scholar] [CrossRef] [PubMed]
- Sugimura, Y.; Hosono, M.; Kitamura, M.; Tsuda, T.; Yamanishi, K.; Maki, M.; Hitomi, K. Identification of preferred substrate sequences for transglutaminase 1–development of a novel peptide that can efficiently detect cross-linking enzyme activity in the skin. FEBS J. 2008, 275, 5667–5677. [Google Scholar] [CrossRef]
- Muszbek, L.; Bereczky, Z.; Bagoly, Z.; Komáromi, I.; Katona, É. Factor XIII: A coagulation factor with multiple plasmatic and cellular functions. Physiol. Rev. 2011, 91, 931–972. [Google Scholar] [CrossRef]
- Bagoly, Z.; Koncz, Z.; Hársfalvi, J.; Muszbek, L. Factor XIII, clot structure, thrombosis. Thromb. Res. 2012, 129, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-Y.; Chung, S.-I.; Steinert, P.M. Highly active soluble processed forms of the transglutaminase 1 enzyme in epidermal keratinocytes. J. Biol. Chem. 1995, 270, 18026–18035. [Google Scholar] [CrossRef]
- Steinert, P.M.; Kim, S.-Y.; Chung, S.-I.; Marekov, L.N. The transglutaminase 1 enzyme is variably acylated by myristate and palmitate during differentiation in epidermal keratinocytes. J. Biol. Chem. 1996, 271, 26242–26250. [Google Scholar] [CrossRef] [PubMed]
- Klöck, C.; Khosla, C. Regulation of the activities of the mammalian transglutaminase family of enzymes. Protein Sci. 2012, 21, 1781–1791. [Google Scholar] [CrossRef]
- Boeshans, K.M.; Mueser, T.C.; Ahvazi, B. A three-dimensional model of the human transglutaminase 1: Insights into the understanding of lamellar ichthyosis. J. Mol. Model. 2007, 13, 233–246. [Google Scholar] [CrossRef]
- Steinert, P.M.; Chung, S.-I.; Kim, S.-Y. Inactive zymogen and highly active proteolytically processed membrane-bound forms of the transglutaminase 1 enzyme in human epidermal keratinocytes. Biochem. Biophys. Res. Commun. 1996, 221, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Negi, M.; Matsui, T.; Ogawa, H. Mechanism of regulation of human epidermal transglutaminase. J. Investig. Dermatol. 1981, 77, 389–392. [Google Scholar] [CrossRef]
- Sugimura, Y.; Hosono, M.; Wada, F.; Yoshimura, T.; Maki, M.; Hitomi, K. Screening for the preferred substrate sequence of transglutaminase using a phage-displayed peptide library: Identification of peptide substrates for TGASE 2 and Factor XIIIA. J. Biol. Chem. 2006, 281, 17699–17706. [Google Scholar] [CrossRef]
- Hitomi, K.; Kitamura, M.; Sugimura, Y. Preferred substrate sequences for transglutaminase 2: Screening using a phage-displayed peptide library. Amino Acids 2009, 36, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Yamane, A.; Fukui, M.; Sugimura, Y.; Itoh, M.; Alea, M.P.; Thomas, V.; El Alaoui, S.; Akiyama, M.; Hitomi, K. Identification of a preferred substrate peptide for transglutaminase 3 and detection of in situ activity in skin and hair follicles. FEBS J. 2010, 277, 3564–3574. [Google Scholar] [CrossRef] [PubMed]
- Yamane, M.; Sugimura, K.; Kawasaki, H.; Tatsukawa, H.; Hitomi, K. Analysis on transglutaminase 1 and its substrates using specific substrate peptide in cultured keratinocytes. Biochem. Biophys. Res. Commun. 2016, 478, 343–348. [Google Scholar] [CrossRef]
- Tatsukawa, H.; Otsu, R.; Tani, Y.; Wakita, R.; Hitomi, K. Isozyme-specific comprehensive characterization of transglutaminase-crosslinked substrates in kidney fibrosis. Sci. Rep. 2018, 8, 7306. [Google Scholar] [CrossRef]
- Tatsukawa, H.; Tani, Y.; Otsu, R.; Nakagawa, H.; Hitomi, K. Global identification and analysis of isozyme-specific possible substrates crosslinked by transglutaminases using substrate peptides in mouse liver fibrosis. Sci. Rep. 2017, 7, 45049. [Google Scholar] [CrossRef] [PubMed]
- Martinet, N.; Bonnard, L.; Regnault, V.; Picard, E.; Burke, L.; Siat, J.; Grosdidier, G.; Martinet, Y.; Vignaud, J.-M. In vivo transglutaminase type 1 expression in normal lung, preinvasive bronchial lesions, and lung cancer. Am. J. Respir. Cell Mol. Biol. 2003, 28, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Baumgartner, W.; Golenhofen, N.; Weth, A.; Hiiragi, T.; Saint, R.; Griffin, M.; Drenckhahn, D. Role of transglutaminase 1 in stabilisation of intercellular junctions of the vascular endothelium. Histochem. Cell Biol. 2004, 122, 17–25. [Google Scholar] [CrossRef]
- Ponnusamy, M.; Pang, M.; Annamaraju, P.K.; Zhang, Z.; Gong, R.; Chin, Y.E.; Zhuang, S. Transglutaminase-1 protects renal epithelial cells from hydrogen peroxide-induced apoptosis through activation of STAT3 and AKT signaling pathways. Am. J. Physiol.-Ren. Physiol. 2009, 297, F1361–F1370. [Google Scholar] [CrossRef]
- Itoh, M.; Tatsukawa, H.; Eun-Seo, L.; Yamanishi, K.; Kojima, S.; Hitomi, K. Variations in both TG1 and TG2 isozyme-specific in situ activities and protein expressions during mouse embryonic development. J. Histochem. Cytochem. 2013, 61, 793–801. [Google Scholar] [CrossRef]
- Kalinin, A.E.; Kajava, A.V.; Steinert, P.M. Epithelial barrier function: Assembly and structural features of the cornified cell envelope. Bioessays 2002, 24, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Huber, M.; Rettler, I.; Bernasconi, K.; Frenk, E.; Lavrijsen, S.P.; Ponec, M.; Bon, A.; Lautenschlager, S.; Schorderet, D.F.; Hohl, D. Mutations of keratinocyte transglutaminase in lamellar ichthyosis. Science 1995, 267, 525–528. [Google Scholar] [CrossRef]
- Russell, L.J.; DiGiovanna, J.J.; Rogers, G.R.; Steinert, P.M.; Hashem, N.; Compton, J.G.; Bale, S.J. Mutations in the gene for transglutaminase 1 in autosomal recessive lamellar ichthyosis. Nat. Genet. 1995, 9, 279–283. [Google Scholar] [CrossRef]
- Tripathy, D.; Migazzi, A.; Costa, F.; Roncador, A.; Gatto, P.; Fusco, F.; Boeri, L.; Albani, D.; Juárez-Hernández, J.L.; Musio, C. Increased transcription of transglutaminase 1 mediates neuronal death in in vitro models of neuronal stress and Aβ1–42-mediated toxicity. Neurobiol. Dis. 2020, 140, 104849. [Google Scholar] [CrossRef]
- Huang, H.; Chen, Z.; Ni, X. Tissue transglutaminase-1 promotes stemness and chemoresistance in gastric cancer cells by regulating Wnt/β-catenin signaling. Exp. Biol. Med. 2017, 242, 194–202. [Google Scholar] [CrossRef]
- Lichti, U.; Ben, T.; Yuspa, S. Retinoic acid-induced transglutaminase in mouse epidermal cells is distinct from epidermal transglutaminase. J. Biol. Chem. 1985, 260, 1422–1426. [Google Scholar] [CrossRef]
- Reichert, U.; Michel, S.; Schmidt, R. The cornified envelope: A key structure of terminally differentiating keratinocytes. In Molecular Biology of the Skin: The Keratinocyte; Academic Press: Cambridge, MA, USA, 1993; pp. 107–150. [Google Scholar]
- Candi, E.; Schmidt, R.; Melino, G. The cornified envelope: A model of cell death in the skin. Nat. Rev. Mol. Cell Biol. 2005, 6, 328–340. [Google Scholar] [CrossRef]
- Steinert, P.M. The complexity and redundancy of epithelial barrier function. J. Cell Biol. 2000, 151, F5–F8. [Google Scholar] [CrossRef] [PubMed]
- Buxman, M.M.; Wuepper, K.D. Isolation, purification and characterization of bovine epidermal transglutaminase. Biochim. Biophys. Acta (BBA)-Enzymol. 1976, 452, 356–369. [Google Scholar] [CrossRef]
- Sturniolo, M.T.; Chandraratna, R.A.; Eckert, R.L. A novel transglutaminase activator forms a complex with type 1 transglutaminase. Oncogene 2005, 24, 2963–2972. [Google Scholar] [CrossRef]
- Hitomi, K.; Tatsukawa, H. Preferred substrate structure of transglutaminases. In Transglutaminases: Multiple Functional Modifiers and Targets for New Drug Discovery; Springer: Tokyo, Japan, 2015; pp. 63–82. [Google Scholar]
- Phillips, M.A.; Jessen, B.A.; Lu, Y.; Qin, Q.; Stevens, M.E.; Rice, R.H. A distal region of the human TGM1 promoter is required for expression in transgenic mice and cultured keratinocytes. BMC Dermatol. 2004, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Surbek, M.; Van de Steene, T.; Sachslehner, A.P.; Golabi, B.; Griss, J.; Eyckerman, S.; Gevaert, K.; Eckhart, L. Cornification of keratinocytes is associated with differential changes in the catalytic activity and the immunoreactivity of transglutaminase-1. Sci. Rep. 2023, 13, 21550. [Google Scholar] [CrossRef]
- Pitolli, C.; Pietroni, V.; Marekov, L.; Terrinoni, A.; Yamanishi, K.; Mazzanti, C.; Melino, G.; Candi, E. Characterization of TG2 and TG1–TG2 double knock-out mouse epidermis. Amino Acids 2017, 49, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Rice, R.H.; Green, H. Presence in human epidermal cells of a soluble protein precursor of the cross-linked envelope: Activation of the cross-linking by calcium ions. Cell 1979, 18, 681–694. [Google Scholar] [CrossRef] [PubMed]
- Nemes, Z.; Marekov, L.N.; Fésüs, L.; Steinert, P.M. A novel function for transglutaminase 1: Attachment of long-chain ω-hydroxyceramides to involucrin by ester bond formation. Proc. Natl. Acad. Sci. USA 1999, 96, 8402–8407. [Google Scholar] [CrossRef]
- Sun, Q.; Burgren, N.M.; Cheraghlou, S.; Paller, A.S.; Larralde, M.; Bercovitch, L.; Levinsohn, J.; Ren, I.; Hu, R.H.; Zhou, J. The genomic and phenotypic landscape of ichthyosis: An analysis of 1000 kindreds. JAMA Dermatol. 2022, 158, 16–25. [Google Scholar] [CrossRef]
- Zaouak, A.; Abdessalem, G.; Mkaouar, R.; Messaoud, O.; Abdelhak, S.; Hammami, H.; Fenniche, S. Congenital lamellar ichthyosis in Tunisia associated with vitamin D rickets caused by a founder nonsense mutation in the TGM1 gene. Int. J. Dermatol. 2019, 58, e135–e137. [Google Scholar] [CrossRef] [PubMed]
- Torkamani, N.; Phal, P.; Savarirayan, R.; Simm, P.; Varigos, G.; Wark, J. Concomitant extraspinal hyperostosis and osteoporosis in a patient with congenital ichthyosis. Clin. Cases Miner. Bone Metab. 2016, 13, 157. [Google Scholar] [CrossRef]
- Sethuraman, G.; Marwaha, R.K. Vitamin D, bone health and congenital ichthyosis. Indian J. Dermatol. Venereol. Leprol. 2016, 82, 249. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Ibe, M.; Kinouchi, M.; Ishida-Yamamoto, A.; Hashimoto, Y.; Iizuka, H. Similarly potent action of 1, 25-dihydroxyvitamin D3 and its analogues, tacalcitol, calcipotriol, and maxacalcitol on normal human keratinocyte proliferation and differentiation. J. Dermatol. Sci. 2003, 31, 21–28. [Google Scholar] [CrossRef]
- John, S.; Thiebach, L.; Frie, C.; Mokkapati, S.; Bechtel, M.; Nischt, R.; Rosser-Davies, S.; Paulsson, M.; Smyth, N. Epidermal transglutaminase (TGase 3) is required for proper hair development, but not the formation of the epidermal barrier. PLoS ONE 2012, 7, e34252. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, A.J.; van Steensel, M.A.; Steijlen, P.M.; van Geel, M.; van der Velden, J.; Morley, S.M.; Terrinoni, A.; Melino, G.; Candi, E.; McLean, W.I. A homozygous missense mutation in TGM5 abolishes epidermal transglutaminase 5 activity and causes acral peeling skin syndrome. Am. J. Hum. Genet. 2005, 77, 909–917. [Google Scholar] [CrossRef]
- Pigors, M.; Kiritsi, D.; Cobzaru, C.; Schwieger-Briel, A.; Suárez, J.; Faletra, F.; Aho, H.; Mäkelä, L.; Kern, J.S.; Bruckner-Tuderman, L. TGM5 mutations impact epidermal differentiation in acral peeling skin syndrome. J. Investig. Dermatol. 2012, 132, 2422–2429. [Google Scholar] [CrossRef]
- Wu, X.; Wang, R.; Jiao, J.; Li, S.; Yu, J.; Yin, Z.; Zhou, L.; Gong, Z. Transglutaminase 3 contributes to malignant transformation of oral leukoplakia to cancer. Int. J. Biochem. Cell Biol. 2018, 104, 34–42. [Google Scholar] [CrossRef]
- Chermnykh, E.S.; Alpeeva, E.V.; Vorotelyak, E.A. Transglutaminase 3: The involvement in epithelial differentiation and cancer. Cells 2020, 9, 1996. [Google Scholar] [CrossRef]
- Inada, R.; Matsuki, M.; Yamada, K.; Morishima, Y.; Shen, S.-C.; Kuramoto, N.; Yasuno, H.; Takahashi, K.; Miyachi, Y.; Yamanishi, K. Facilitated wound healing by activation of the Transglutaminase 1 gene. Am. J. Pathol. 2000, 157, 1875–1882. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Wang, C.; Zhao, Y.; Bo, T.; Han, L.; Yao, D.; Wang, Y.; Wang, X.; Shi, L.; Zhao, A. Superior COL7A1 and TGM1 gene expression in difficult-to-transfect skin cell mediated by highly branched poly (β-amino esters) through stepwise fractionation. J. Control. Release 2024, 370, 82–94. [Google Scholar] [CrossRef]
- Lin, Y.-C.; Hong, Y.-K.; Aala, W.J.F.; Hitomi, K.; Akiyama, M.; McGrath, J.A.; Hsu, C.-K. Tofacitinib ameliorates skin inflammation in a patient with severe autosomal recessive congenital ichthyosis. Clin. Exp. Dermatol. 2024, 49, 887–892. [Google Scholar] [CrossRef] [PubMed]
- Landegren, N.; Ishii, N.; Aranda-Guillén, M.; Gunnarsson, H.I.; Sardh, F.; Hallgren, Å.; Ståhle, M.; Hagforsen, E.; Bradley, M.; Edqvist, P.-H.D. A gene-centric approach to biomarker discovery identifies transglutaminase 1 as an epidermal autoantigen. Proc. Natl. Acad. Sci. USA 2021, 118, e2100687118. [Google Scholar] [CrossRef] [PubMed]
- Alharbi, N.; Almami, I. Activation and upregulation of keratinocyte and epidermal transglutaminases are associated with depletion of their substrates in psoriatic lesions. Eur. Rev. Med. Pharmacol. Sci. 2023, 27, 11281–11293. [Google Scholar]
- Hamdy, S.M.; Sayed, O.N.; Ibrahim, H.A.; Ayoub, S.E. Evaluation of serum long non-coding RNA (Gas5) level and keratinocyte transglutaminase 1 (TGM1) activity as novel biomarkers in psoriasis patients. Gene Rep. 2021, 25, 101421. [Google Scholar] [CrossRef]
- Ekman, A.-K.; Vegfors, J.; Bivik, C.; Enerbäck, C. Overexpression of psoriasin (S100A7) contributes to dysregulated differentiation in psoriasis. Acta Derm.-Venereol. 2017, 97, 441–448. [Google Scholar] [CrossRef]
- Cork, M.J.; Robinson, D.A.; Vasilopoulos, Y.; Ferguson, A.; Moustafa, M.; MacGowan, A.; Duff, G.W.; Ward, S.J.; Tazi-Ahnini, R. New perspectives on epidermal barrier dysfunction in atopic dermatitis: Gene–environment interactions. J. Allergy Clin. Immunol. 2006, 118, 3–21. [Google Scholar] [CrossRef]
- Sääf, A.M.; Tengvall-Linder, M.; Chang, H.Y.; Adler, A.S.; Wahlgren, C.-F.; Scheynius, A.; Nordenskjöld, M.; Bradley, M. Global expression profiling in atopic eczema reveals reciprocal expression of inflammatory and lipid genes. PLoS ONE 2008, 3, e4017. [Google Scholar] [CrossRef]
- Lieden, A.; Winge, M.C.; Sääf, A.; Kockum, I.; Ekelund, E.; Rodriguez, E.; Fölster-Holst, R.; Franke, A.; Illig, T.; Tengvall-Linder, M. Genetic variation in the epidermal transglutaminase genes is not associated with atopic dermatitis. PLoS ONE 2012, 7, e49694. [Google Scholar] [CrossRef]
- Murakami, M.; Simons, M. Regulation of vascular integrity. J. Mol. Med. 2009, 87, 571–582. [Google Scholar] [CrossRef] [PubMed]
- Petersen-Jones, H.G.; Johnson, K.B.; Hitomi, K.; Tykocki, N.R.; Thompson, J.M.; Watts, S.W. Transglutaminase activity is decreased in large arteries from hypertensive rats compared with normotensive controls. Am. J. Physiol.-Heart Circ. Physiol. 2015, 308, H592–H602. [Google Scholar] [CrossRef] [PubMed]
- Al-U’datt, D.a.G.; Tranchant, C.C.; Al-Husein, B.; Hiram, R.; Al-Dwairi, A.; AlQudah, M.; Al-Shboul, O.; Jaradat, S.; Alqbelat, J.; Almajwal, A. Involvement and possible role of transglutaminases 1 and 2 in mediating fibrotic signalling, collagen cross-linking and cell proliferation in neonatal rat ventricular fibroblasts. PLoS ONE 2023, 18, e0281320. [Google Scholar] [CrossRef]
- Baumgartner, W.; Weth, A. Transglutaminase 1 stabilizes β-actin in endothelial cells correlating with a stabilization of intercellular junctions. J. Vasc. Res. 2007, 44, 234–240. [Google Scholar] [CrossRef]
- Bakker, E.N.; Pistea, A.; VanBavel, E. Transglutaminases in vascular biology: Relevance for vascular remodeling and atherosclerosis. J. Vasc. Res. 2008, 45, 271–278. [Google Scholar] [CrossRef]
- Johnson, K.B.; Petersen-Jones, H.; Thompson, J.M.; Hitomi, K.; Itoh, M.; Bakker, E.N.; Johnson, G.V.; Colak, G.; Watts, S.W. Vena cava and aortic smooth muscle cells express transglutaminases 1 and 4 in addition to transglutaminase 2. Am. J. Physiol.-Heart Circ. Physiol. 2012, 302, H1355–H1366. [Google Scholar] [CrossRef]
- Frey, A.; Lunding, L.P.; Ehlers, J.C.; Weckmann, M.; Zissler, U.M.; Wegmann, M. More than just a barrier: The immune functions of the airway epithelium in asthma pathogenesis. Front. Immunol. 2020, 11, 761. [Google Scholar] [CrossRef]
- Bals, R.; Hiemstra, P. Innate immunity in the lung: How epithelial cells fight against respiratory pathogens. Eur. Respir. J. 2004, 23, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Watson, C.J.; Dubourd, M.; Bruton, A.; Xu, M.; Cooke, G.; Baugh, J.A. HIF-1-dependent TGM1 expression is associated with maintenance of airway epithelial junction proteins. Lung 2016, 194, 829–838. [Google Scholar] [CrossRef]
- Cocuzzi, E.; Chung, S. Cellular transglutaminase. Lung matrix-associated transglutaminase: Characterization and activation with sulfhydryls. J. Biol. Chem. 1986, 261, 8122–8127. [Google Scholar] [CrossRef]
- Lee, S.Y.; Kim, Y.J.; Kim, Y.J. Role of crosslinked protein in lung injury following total body irradiation and bone marrow transplantation. Exp. Mol. Med. 2003, 35, 565–571. [Google Scholar] [CrossRef]
- Sonoyama, T.; Ishino, T.; Takemoto, K.; Yamato, K.; Oda, T.; Nishida, M.; Horibe, Y.; Chikuie, N.; Kono, T.; Taruya, T. Deep association between transglutaminase 1 and tissue eosinophil infiltration leading to nasal polyp formation and/or maintenance with fibrin polymerization in chronic rhinosinusitis with nasal polyps. Int. J. Mol. Sci. 2022, 23, 12955. [Google Scholar] [CrossRef]
- Van Zele, T.; Claeys, S.; Gevaert, P.; Van Maele, G.; Holtappels, G.; Van Cauwenberge, P.; Bachert, C. Differentiation of chronic sinus diseases by measurement of inflammatory mediators. Allergy 2006, 61, 1280–1289. [Google Scholar] [CrossRef]
- D’Argenio, V.; Sarnataro, D. New insights into the molecular bases of familial Alzheimer’s disease. J. Personal. Med. 2020, 10, 26. [Google Scholar] [CrossRef]
- Johnson, G.; Bailey, C.; Tucholski, J.; Lesort, M. Tissue transglutaminase in neurodegenerative diseases. Minerva Biotecnol. 2002, 14, 171. [Google Scholar]
- André, W.; Nondier, I.; Valensi, M.; Guillonneau, F.; Federici, C.; Hoffner, G.; Djian, P. Identification of brain substrates of transglutaminase by functional proteomics supports its role in neurodegenerative diseases. Neurobiol. Dis. 2017, 101, 40–58. [Google Scholar] [CrossRef]
- Wilhelmus, M.M.; van Dam, A.-M.; Drukarch, B. Tissue transglutaminase: A novel pharmacological target in preventing toxic protein aggregation in neurodegenerative diseases. Eur. J. Pharmacol. 2008, 585, 464–472. [Google Scholar] [CrossRef]
- Johnson, G.V.; Cox, T.M.; Lockhart, J.P.; Zinnerman, M.D.; Miller, M.L.; Powers, R.E. Transglutaminase activity is increased in Alzheimer’s disease brain. Brain Res. 1997, 751, 323–329. [Google Scholar] [CrossRef]
- Wilhelmus, M.M.; Grunberg, S.C.; Bol, J.G.; Van Dam, A.M.; Hoozemans, J.J.; Rozemuller, A.J.; Drukarch, B. Transglutaminases and transglutaminase-catalyzed cross-links colocalize with the pathological lesions in Alzheimer’s disease brain. Brain Pathol. 2009, 19, 612–622. [Google Scholar] [CrossRef]
- Kim, S.-Y.; Grant, P.; Lee, J.-H.; Pant, H.C.; Steinert, P.M. Differential expression of multiple transglutaminases in human brain: Increased expression and cross-linking by transglutaminases 1 and 2 in Alzheimer’s disease. J. Biol. Chem. 1999, 274, 30715–30721. [Google Scholar] [CrossRef]
- Muma, N.A. Transglutaminase is linked to neurodegenerative diseases. J. Neuropathol. Exp. Neurol. 2007, 66, 258–263. [Google Scholar] [CrossRef]
- Wilhelmus, M.M.; de Jager, M.; Rozemuller, A.J.; Brevé, J.; Bol, J.G.; Eckert, R.L.; Drukarch, B. Transglutaminase 1 and its regulator tazarotene-induced gene 3 localize to neuronal tau inclusions in tauopathies. J. Pathol. 2012, 226, 132–142. [Google Scholar] [CrossRef]
- Cavanagh, J. Corpora-amylacea and the family of polyglucosan diseases. Brain Res. Rev. 1999, 29, 265–295. [Google Scholar] [CrossRef]
- Wilhelmus, M.M.; Verhaar, R.; Bol, J.G.; van Dam, A.-M.; Hoozemans, J.J.; Rozemuller, A.J.; Drukarch, B. Novel role of transglutaminase 1 in corpora amylacea formation? Neurobiol. Aging 2011, 32, 845–856. [Google Scholar] [CrossRef]
- Pai, P.; Rachagani, S.; Dhawan, P.; Batra, S.K. Mucins and Wnt/β-catenin signaling in gastrointestinal cancers: An unholy nexus. Carcinogenesis 2016, 37, 223–232. [Google Scholar] [CrossRef]
- Ding, Y.; Labitzky, V.; Legler, K.; Qi, M.; Schumacher, U.; Schmalfeldt, B.; Stürken, C.; Oliveira-Ferrer, L. Molecular characteristics and tumorigenicity of ascites-derived tumor cells: Mitochondrial oxidative phosphorylation as a novel therapy target in ovarian cancer. Mol. Oncol. 2021, 15, 3578–3595. [Google Scholar] [CrossRef]
- Zhang, S.; Yao, H.-F.; Li, H.; Su, T.; Jiang, S.-H.; Wang, H.; Zhang, Z.-G.; Dong, F.-Y.; Yang, Q.; Yang, X.-M. Transglutaminases are oncogenic biomarkers in human cancers and therapeutic targeting of TGM2 blocks chemoresistance and macrophage infiltration in pancreatic cancer. Cell. Oncol. 2023, 46, 1473–1492. [Google Scholar] [CrossRef]
- Wu, R.; Li, D.; Zhang, S.; Wang, J.; Chen, K.; Tuo, Z.; Miyamoto, A.; Yoo, K.H.; Wei, W.; Zhang, C. A pan-cancer analysis of the oncogenic and immunological roles of transglutaminase 1 (TGM1) in human cancer. J. Cancer Res. Clin. Oncol. 2024, 150, 123. [Google Scholar] [CrossRef]
- Luan, H.; He, Y.; Jian, L.; Zhang, T.; Zhou, L. Identification of Metastasis-Associated Gene and Its Correlation with Immune Infiltrates For Skin Cutaneous Melanoma. Research Square. 2021. [Google Scholar] [CrossRef]
- Hou, J.; Mei, K.; Wang, D.; Ke, S.; Chen, X.; Shang, J.; Li, G.; Gao, Y.; Xiong, H.; Zhang, H. TGM1/3-mediated transamidation of Exo70 promotes tumor metastasis upon LKB1 inactivation. Cell Rep. 2024, 43, 114604. [Google Scholar] [CrossRef]
- Boor, P.; Ostendorf, T.; Floege, J. Renal fibrosis: Novel insights into mechanisms and therapeutic targets. Nat. Rev. Nephrol. 2010, 6, 643–656. [Google Scholar] [CrossRef]
- Zhang, Z.; Xing, J.; Ma, L.; Gong, R.; Chin, Y.E.; Zhuang, S. Transglutaminase-1 regulates renal epithelial cell proliferation through activation of Stat-3. J. Biol. Chem. 2009, 284, 3345–3353. [Google Scholar] [CrossRef]
- Murtha, L.A.; Schuliga, M.J.; Mabotuwana, N.S.; Hardy, S.A.; Waters, D.W.; Burgess, J.K.; Knight, D.A.; Boyle, A.J. The processes and mechanisms of cardiac and pulmonary fibrosis. Front. Physiol. 2017, 8, 777. [Google Scholar] [CrossRef]
- Al-U’datt, D.G.F.; Tranchant, C.C.; Alu’datt, M.; Abusara, S.; Al-Dwairi, A.; AlQudah, M.; Al-Shboul, O.; Hiram, R.; Altuntas, Y.; Jaradat, S. Inhibition of transglutaminase 2 (TG2) ameliorates ventricular fibrosis in isoproterenol-induced heart failure in rats. Life Sci. 2023, 321, 121564. [Google Scholar]
- Tatsukawa, H.; Takeuchi, T.; Shinoda, Y.; Hitomi, K. Identification and characterization of substrates crosslinked by transglutaminases in liver and kidney fibrosis. Anal. Biochem. 2020, 604, 113629. [Google Scholar] [CrossRef]
- Pires, R.H.; Felix, S.B.; Delcea, M. The architecture of neutrophil extracellular traps investigated by atomic force microscopy. Nanoscale 2016, 8, 14193–14202. [Google Scholar] [CrossRef]
- Csomós, K.; Kristóf, E.; Jakob, B.; Csomós, I.; Kovács, G.; Rotem, O.; Hodrea, J.; Bagoly, Z.; Muszbek, L.; Balajthy, Z. Protein cross-linking by chlorinated polyamines and transglutamylation stabilizes neutrophil extracellular traps. Cell Death Dis. 2016, 7, e2332. [Google Scholar] [CrossRef]
- Mousa, A.; Cui, C.; Song, A.; Myneni, V.D.; Sun, H.; Li, J.J.; Murshed, M.; Melino, G.; Kaartinen, M.T. Transglutaminases factor XIII-A and TG2 regulate resorption, adipogenesis and plasma fibronectin homeostasis in bone and bone marrow. Cell Death Differ. 2017, 24, 844–854. [Google Scholar] [CrossRef]
- Sun, H.; Kaartinen, M. Assessment of expression and specific activities of transglutaminases TG1, TG2, and FXIII-A during osteoclastogenesis. Anal. Biochem. 2020, 591, 113512. [Google Scholar] [CrossRef]
- Sun, H.; Kaartinen, M.T. Transglutaminases in monocytes and macrophages. Med. Sci. 2018, 6, 115. [Google Scholar] [CrossRef]
- Sun, H.; Kaartinen, M.T. Transglutaminase activity regulates differentiation, migration and fusion of osteoclasts via affecting actin dynamics. J. Cell. Physiol. 2018, 233, 7497–7513. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.-S.; Kim, H.; Jeong, E.M.; Kim, H.J.; Lee, Z.H.; Kim, I.-G.; Kim, H.-H. Transglutaminase 2 regulates osteoclast differentiation via a Blimp1-dependent pathway. Sci. Rep. 2017, 7, 10626. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi Samani, S.; Kaartinen, M.T. Increased Osteoclastogenesis in Absence of TG2 Is Reversed by Transglutaminase Inhibition—Evidence for the Role for TG1 in Osteoclast Formation. Cells 2023, 12, 2139. [Google Scholar] [CrossRef] [PubMed]
- Oliver, V.; van Bysterveldt, K.; Merbs, S. Epigenetics in ocular medicine. In Medical Epigenetics; Elsevier: Amsterdam, The Netherlands, 2016; pp. 391–412. [Google Scholar]
- Priglinger, S.G.; Alge, C.S.; Kreutzer, T.C.; Neubauer, A.S.; Haritoglou, C.; Kampik, A.; Welge-Luessen, U. Keratinocyte transglutaminase in proliferative vitreoretinopathy. Investig. Ophthalmol. Vis. Sci. 2006, 47, 4990–4997. [Google Scholar] [CrossRef]
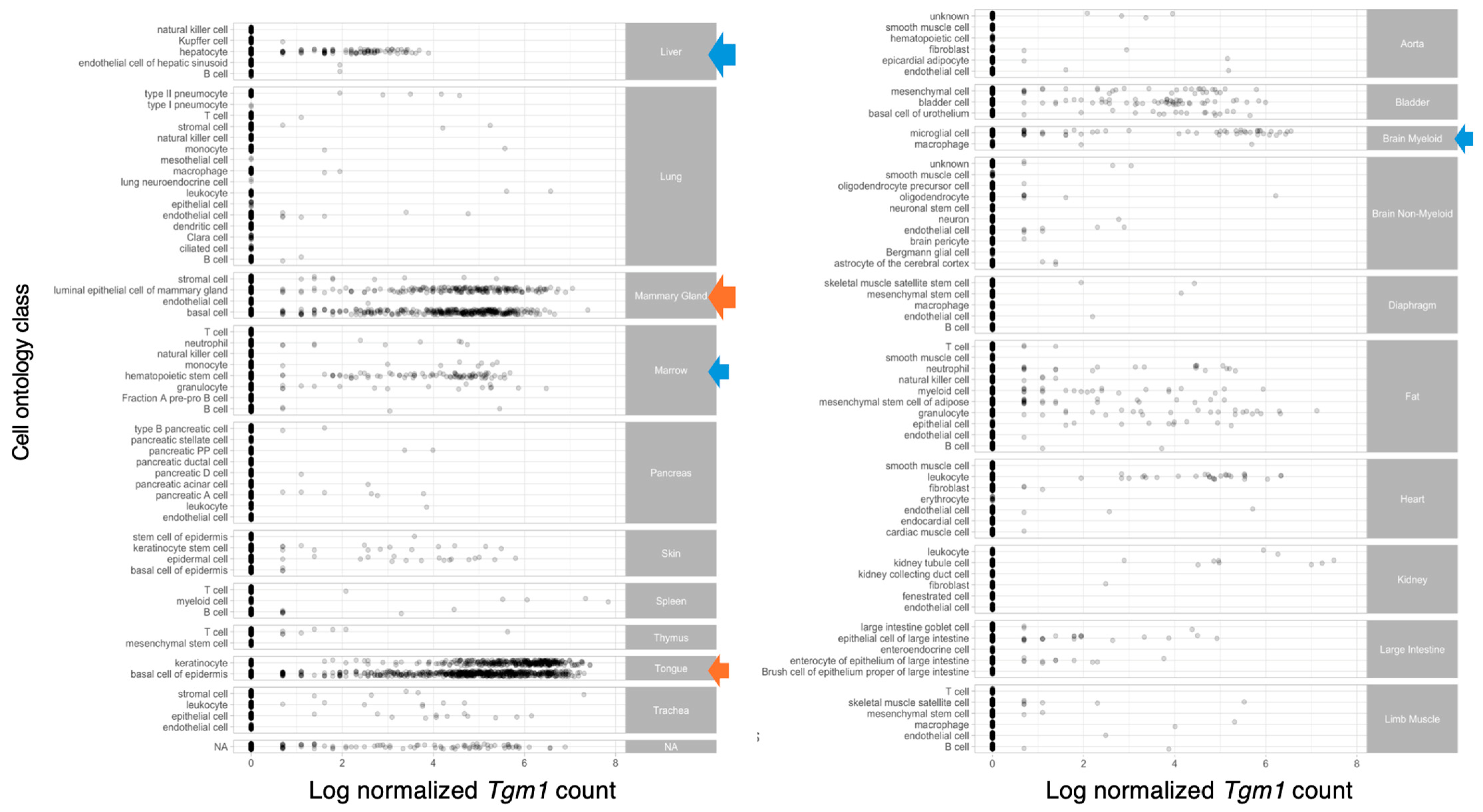
- A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature 2020, 583, 590–595. [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ebrahimi Samani, S.; Tatsukawa, H.; Hitomi, K.; Kaartinen, M.T. Transglutaminase 1: Emerging Functions beyond Skin. Int. J. Mol. Sci. 2024, 25, 10306. https://doi.org/10.3390/ijms251910306
Ebrahimi Samani S, Tatsukawa H, Hitomi K, Kaartinen MT. Transglutaminase 1: Emerging Functions beyond Skin. International Journal of Molecular Sciences. 2024; 25(19):10306. https://doi.org/10.3390/ijms251910306
Chicago/Turabian StyleEbrahimi Samani, Sahar, Hideki Tatsukawa, Kiyotaka Hitomi, and Mari T. Kaartinen. 2024. "Transglutaminase 1: Emerging Functions beyond Skin" International Journal of Molecular Sciences 25, no. 19: 10306. https://doi.org/10.3390/ijms251910306
APA StyleEbrahimi Samani, S., Tatsukawa, H., Hitomi, K., & Kaartinen, M. T. (2024). Transglutaminase 1: Emerging Functions beyond Skin. International Journal of Molecular Sciences, 25(19), 10306. https://doi.org/10.3390/ijms251910306






