Two Half-Size ATP-Binding Cassette Transporters Are Implicated in Aluminum Tolerance in Soybean
Abstract
:1. Introduction
2. Results
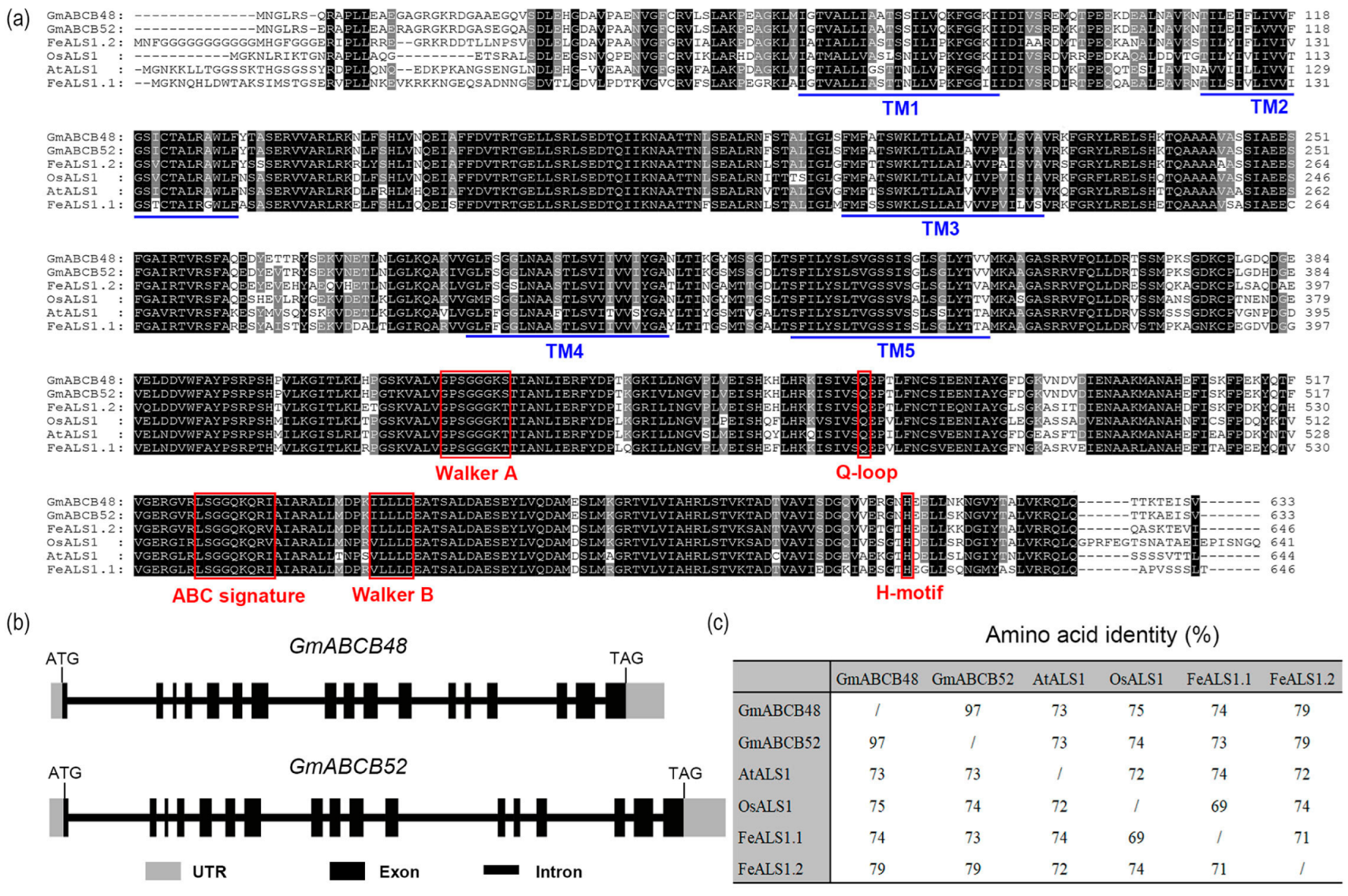
2.1. Sequence Analysis of GmABCB48 and GmABCB52 in Soybean
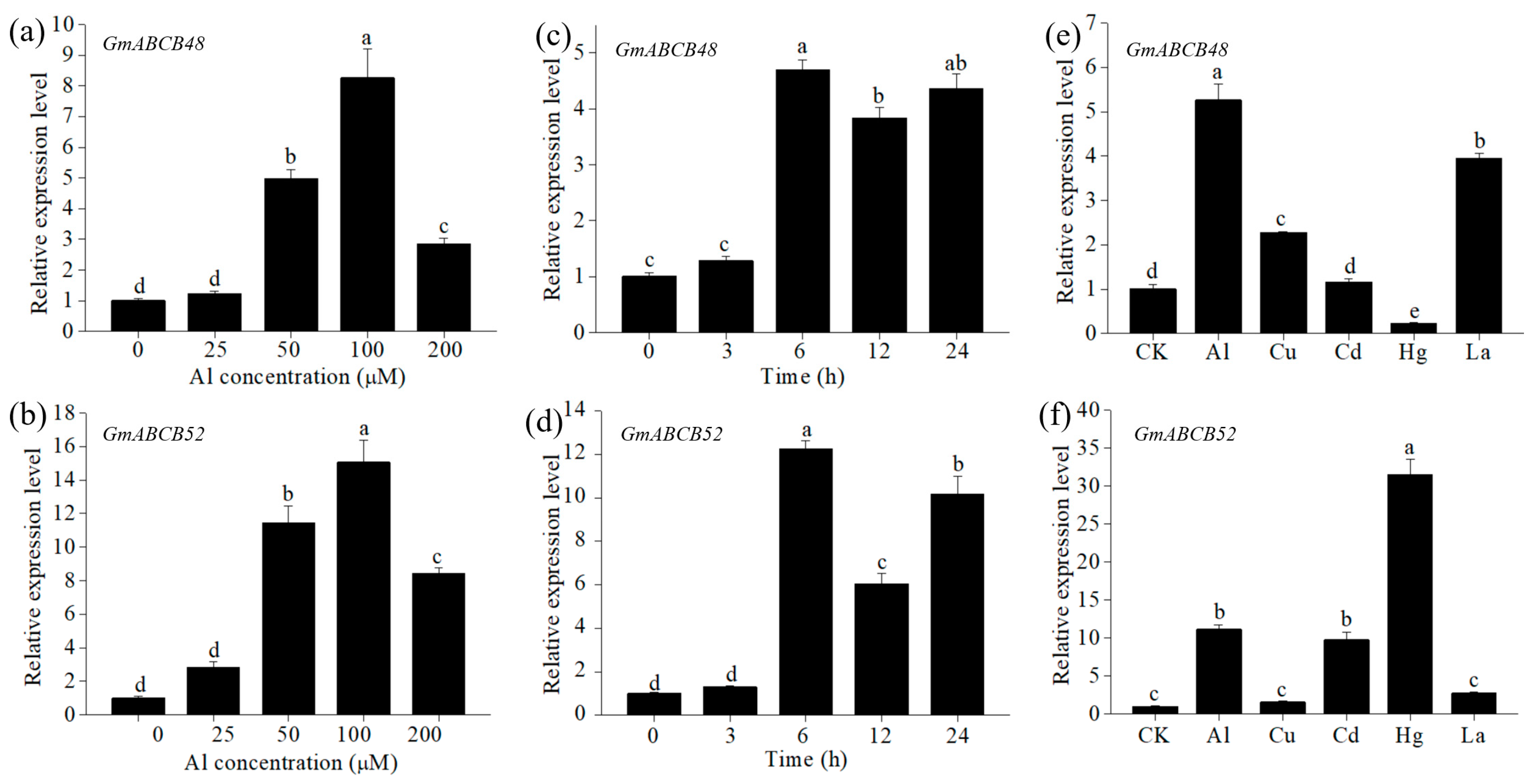
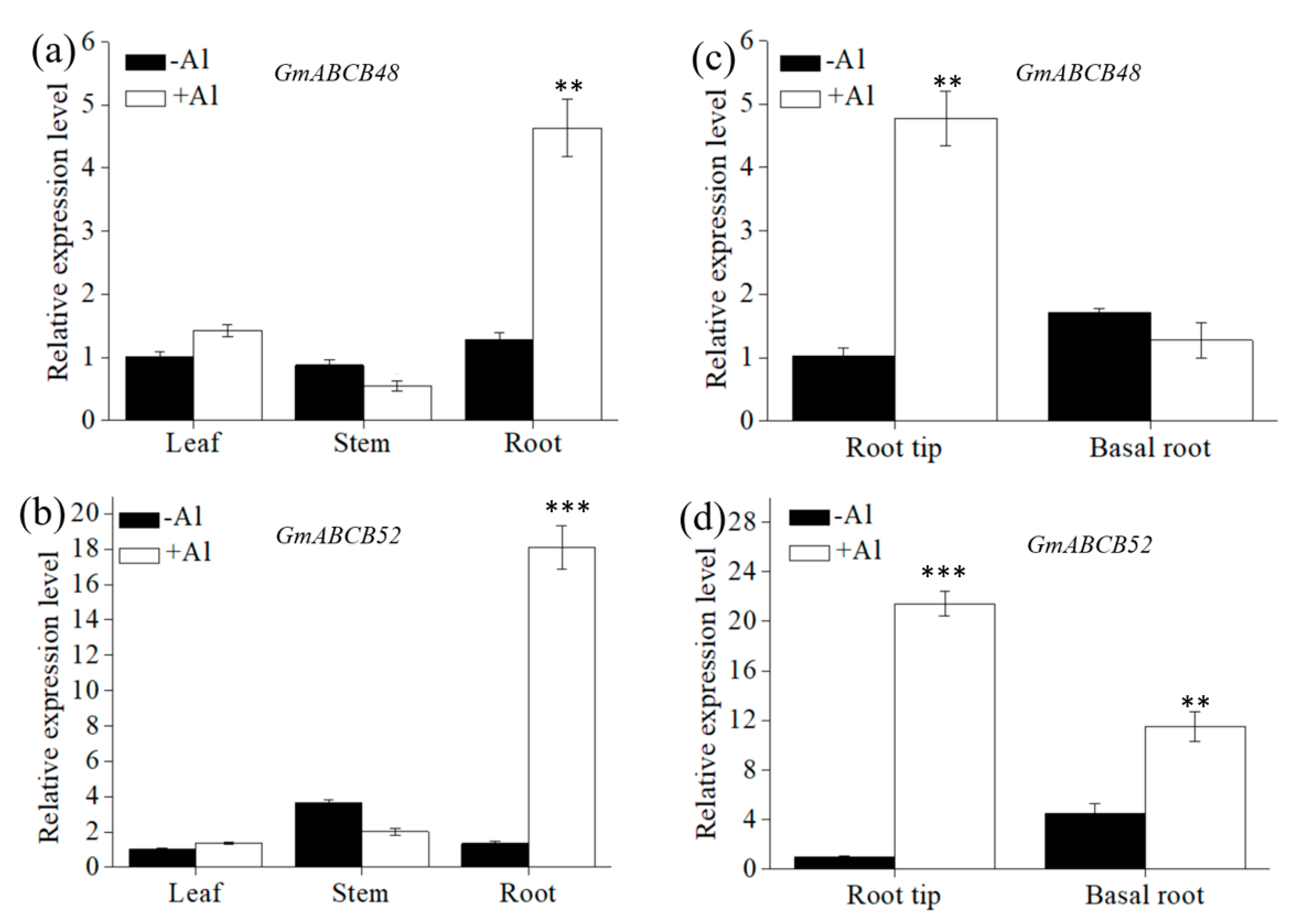
2.2. Expression Pattern of GmABCB48 and GmABCB52
2.3. Subcellular Localization of GmABCB48 and GmABCB52
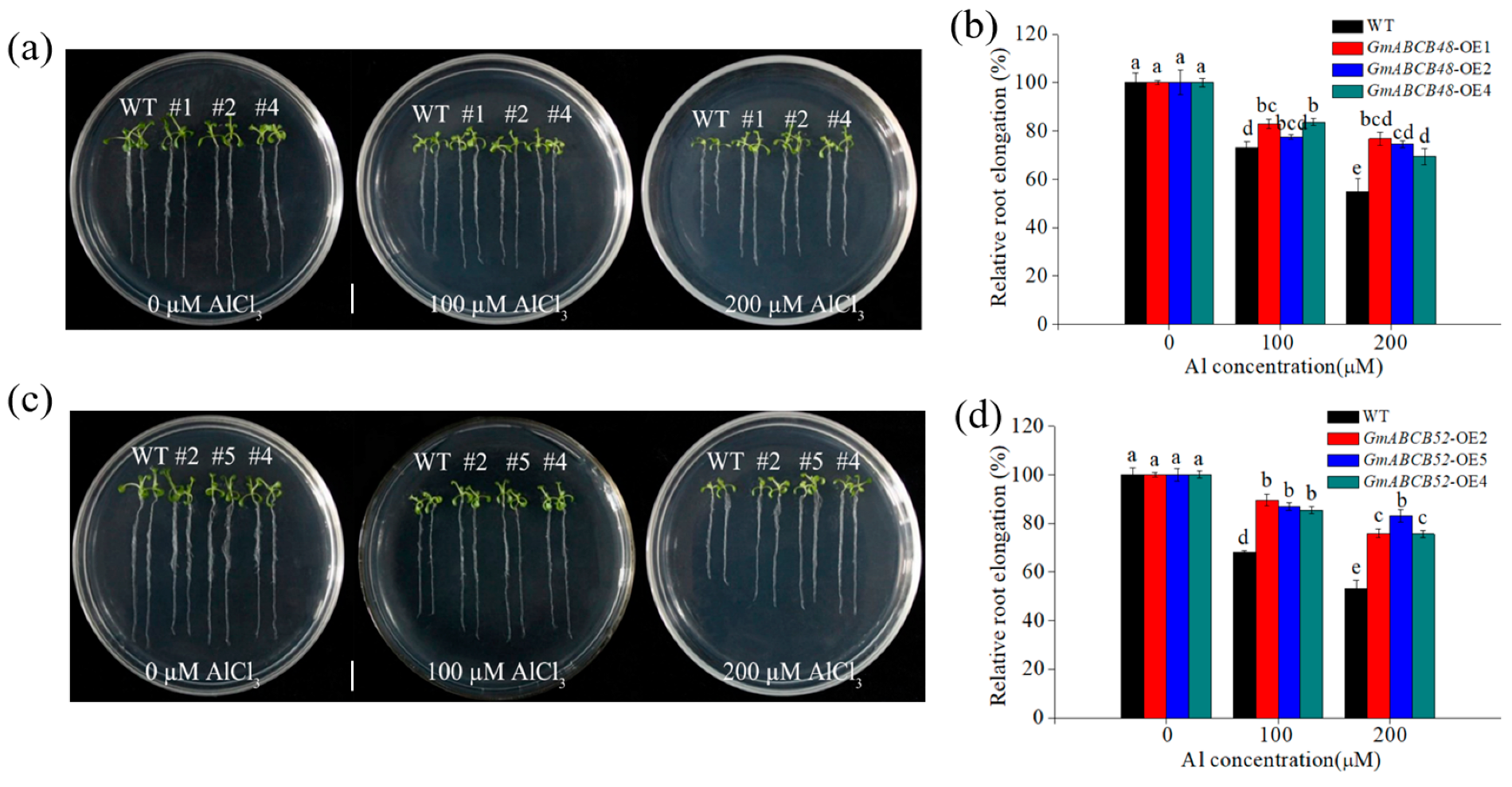
2.4. GmABCB48 and GmABCB52 Confer Al Tolerance
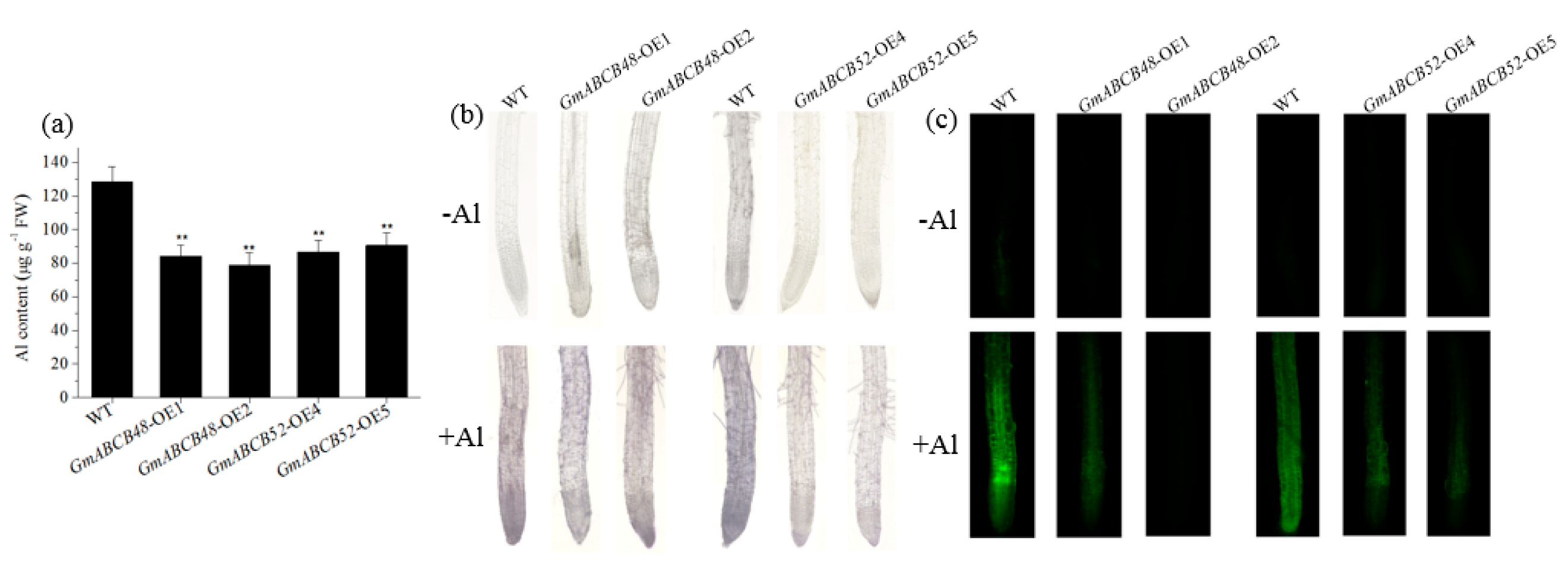
2.5. GmABCB48 and GmABCB52 Reduce Al Accumulation in Root Tips
2.6. Al Transport Activity of GmABCB48 and GmABCB52
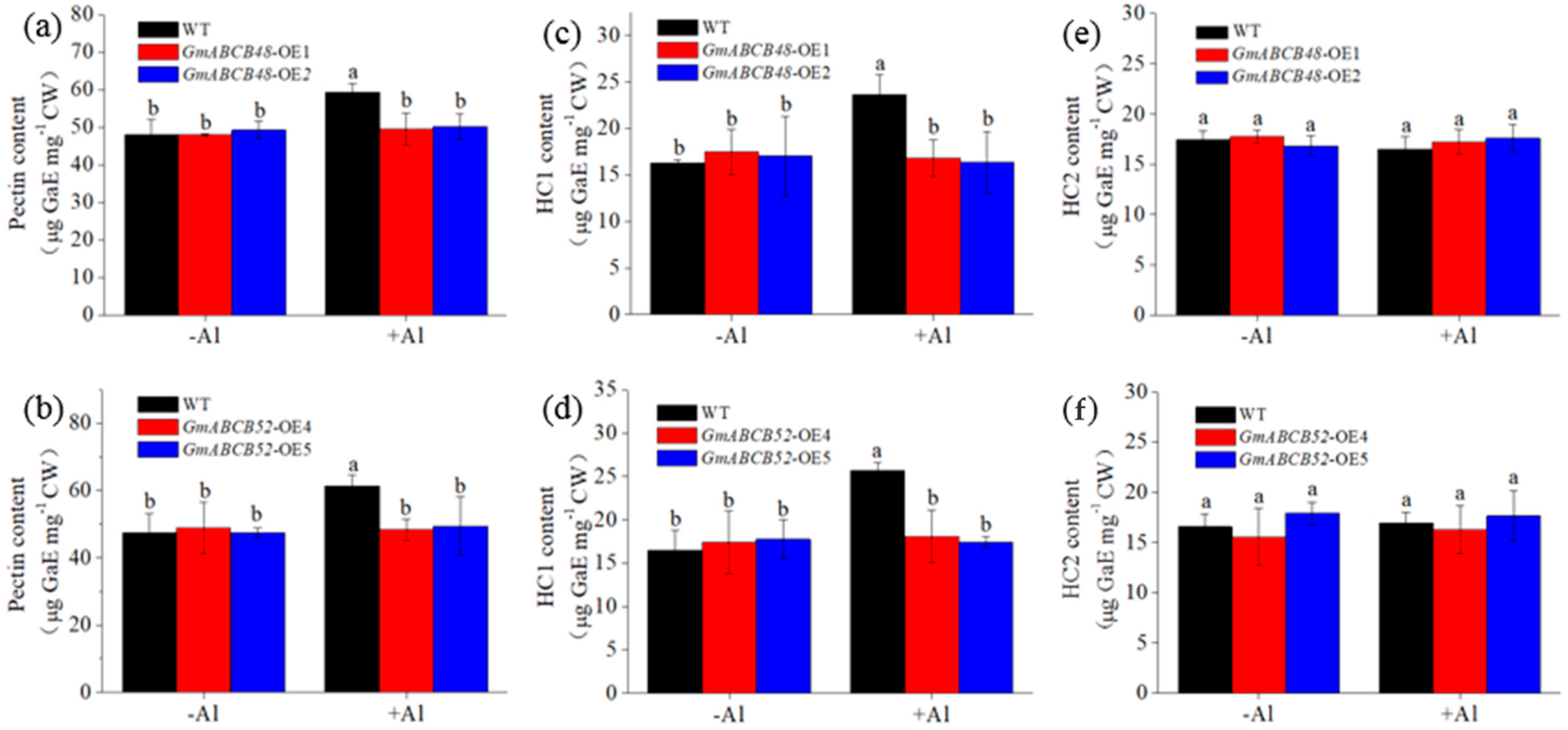
2.7. GmABCB48 and GmABCB52 Reduce Al Binding to Cell Wall
3. Discussion
4. Materials and Methods
4.1. Plant Growth and Treatment
4.2. Gene Expression Analysis
4.3. Subcellular Localization
4.4. Overexpression of GmABCBs in Arabidopsis
4.5. Assay of Al Transport Activity
4.6. Histochemical Staining
4.7. Isolation of Cell WALL Components
4.8. Analysis of Cell Wall Component Contents
4.9. Measurement of Al Content
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kochian, L.; Piñeros, M.A.; Liu, J.; Magalhaes, J.V. Plant adaptation to acid soils: The molecular basis for crop aluminum resistance. Annu. Rev. Plant Biol. 2015, 66, 571–598. [Google Scholar] [CrossRef] [PubMed]
- Kochian, L.V.; Hoekenga, O.A.; Pineros, M.A. How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu. Rev. Plant Biol. 2004, 55, 459–493. [Google Scholar] [CrossRef] [PubMed]
- Kochian, L.V. Cellular mechanisms of aluminum toxicity and resistance in plants. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1995, 46, 237–260. [Google Scholar] [CrossRef]
- Ma, J.F. Syndrome of aluminum toxicity and diversity of aluminum resistance in higher plants. Int. Rev. Cytol. 2007, 264, 225–252. [Google Scholar]
- Ma, J.F.; Shen, R.F.; Nagao, S.; Tanimoto, E. Aluminum targets elongating cells by reducing cell wall extensibility in wheat roots. Plant Cell Physiol. 2004, 45, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Horst, W.J.; Wang, Y.; Eticha, D. The role of root apoplast in aluminum-induced inhibition of root elongation and in aluminum resistance of plants: A review. Ann. Bot. 2010, 106, 185–197. [Google Scholar] [CrossRef]
- Kopittke, P.M.; Moore, K.L.; Lombi, E.; Gianoncelli, A.; Ferguson, B.J.; Blamey, F.P.C.; Menzies, N.W.; Nicholson, T.M.; McKenna, B.A.; Wang, P.; et al. Identification of the primary lesion of toxic aluminum in plant roots. Plant Physiol. 2015, 167, 1402–1411. [Google Scholar] [CrossRef]
- Yan, L.; Riaz, M.; Liu, J.Y.; Zeng, Y.; Yu, M.; Jiang, C.C. The aluminum tolerance and detoxification mechanisms in plants; recent advances and prospects. Crit. Rev. Environ. Sci. Technol. 2021, 4, 1–37. [Google Scholar] [CrossRef]
- Degenhardt, J.; Larsen, P.B.; Howell, S.H.; Kochian, L.V. Aluminum resistance in the arabidopsis mutant alr-104 is caused by an aluminum-induced increase in rhizosphere pH. Plant Physiol. 1998, 117, 19–27. [Google Scholar] [CrossRef]
- Ma, J.F. Role of organic acids in detoxification of aluminum in higher plants. Plant Cell Physiol. 2000, 41, 383–390. [Google Scholar] [CrossRef]
- Xiao, Z.; Yan, Z.; Yan, G.; Ye, M.; Liang, Y. Silicon relieves aluminum-induced inhibition of cell elongation in rice root apex by reducing the deposition of aluminum in the cell wall. Plant Soil. 2021, 462, 189–205. [Google Scholar] [CrossRef]
- Yan, L.; Li, S.; Cheng, J.; Liu, Y.; Liu, J.; Jiang, C. Boron contributes to excessive aluminum tolerance in trifoliate orange (Poncirus trifoliata (L.) Raf.) by inhibiting cell wall deposition and promoting vacuole compartmentation. J. Harzard. Mater. 2022, 437, 129275. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Y.; Mai, J.; Tao, L.; Qu, M.; Liu, J.; Shen, R.; Xu, G.; Feng, Y.; Xiao, H.; et al. Boron alleviates aluminium toxicity by promoting root alkalization in transition zone via polar auxin transport. Plant Physiol. 2018, 177, 1254–1266. [Google Scholar] [CrossRef] [PubMed]
- Li, X.W.; Liu, J.Y.; Fang, J.; Tao, L.; Shen, R.F.; Li, Y.L.; Xiao, H.D.; Feng, Y.M.; Wen, H.X.; Guan, J.H.; et al. Boron supply enhances aluminum tolerance in root border cells of pea (Pisum sativum) by interacting with cell wall pectins. Front. Plant Sci. 2017, 8, 742–753. [Google Scholar] [CrossRef] [PubMed]
- Riaz, M.; Yan, L.; Wu, X.W.; Hussain, S.; Aziz, O.; Imran, M.; Jiang, C.C. Boron deprivation induced inhibition of root elongation is provoked by oxidative damage, root injuries and changes in cell wall structure. Environ. Exp. Bot. 2018, 156, 74–85. [Google Scholar] [CrossRef]
- Ma, J.F.; Hiradate, S.; Nomoto, K.; Iwashita, T.; Matsumoto, H. Internal detoxification mechanism of Al in hydrangea: Identification of Al form in the leaves. Plant Physiol. 1997, 113, 1033–1039. [Google Scholar] [CrossRef]
- Shen, R.; Ma, J.; Kyo, M.; Iwashita, T. Compartmentation of aluminium in leaves of an Al-accumulator, Fagopyrum esculentum Moench. Planta 2002, 215, 394–398. [Google Scholar] [CrossRef]
- Larsen, P.B.; Cancel, J.; Rounds, M.; Ochoa, V. Arabidopsis ALS1 encodes a root tip and stele localized half type ABC transporter required for root growth in an aluminum toxic environment. Planta 2007, 225, 1447–1458. [Google Scholar] [CrossRef]
- Huang, C.; Yamaji, N.; Chen, Z.; Ma, J.F. A tonoplast-localized half-size ABC transporter is required for internal detoxification of aluminum in rice. Plant J. 2012, 69, 857–867. [Google Scholar] [CrossRef]
- Lefèvre, F.; Boutry, M. Towards identification of the substrates of ATP-binding cassette transporters. Plant Physiol. 2018, 178, 18–39. [Google Scholar] [CrossRef]
- Higgins, C.F.; Linton, K.J. The ATP switch model for ABC transporters. Nat. Struct. Mol. Biol. 2004, 11, 918–926. [Google Scholar] [CrossRef] [PubMed]
- Verrier, P.J.; Bird, D.; Burla, B.; Dassa, E.; Forestier, C.; Geisler, M.; Klein, M.; Kolukisaoglu, U.; Lee, Y.; Martinoia, E.; et al. Plant ABC proteins—A unified nomenclature and updated inventory. Trends Plant Sci. 2008, 13, 151–159. [Google Scholar] [CrossRef]
- Theodoulou, F.L.; Kerr, L.D. ABC transporter research: Going strong 40 years on. Biochem. Soc. Trans. 2015, 43, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Do, T.H.T.; Martinoia, E.; Lee, Y. Functions of ABC transporters in plant growth and development. Curr. Opin. Plant Biol. 2018, 41, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.F.; Yamaji, N.; Mitani, N.; Yano, M.; Nagamura, Y.; Ma, J.F. A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 2009, 21, 655–667. [Google Scholar] [CrossRef]
- Xu, J.M.; Wang, Z.Q.; Jin, J.F.; Chen, W.W.; Fan, W.; Zheng, S.J.; Yang, J.L. FeSTAR2 interacted by FeSTAR1 alters its subcellular location and regulates Al tolerance in buckwheat. Plant Soil. 2019, 436, 489–501. [Google Scholar] [CrossRef]
- Larsen, P.B.; Geisler, M.J.; Jones, C.A.; Williams, K.M.; Cancel, J.D. ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance in Arabidopsis. Plant J. 2005, 41, 353–363. [Google Scholar] [CrossRef]
- Huang, C.F.; Yamaji, N.; Ma, J.F. Knockout of a bacterial-type ATP-binding cassette transporter gene, AtSTAR1, results in increased aluminum sensitivity in Arabidopsis. Plant Physiol. 2010, 153, 1669–1677. [Google Scholar] [CrossRef]
- Lei, G.J.; Yokosho, K.; Yamaji, N.; Fujii-Kashino, M.; Ma, J.F. Functional characterization of two half-size ABC transporter genes in aluminium-accumulating buckwheat. New Phytol. 2017, 215, 1080–1089. [Google Scholar] [CrossRef]
- Huang, J.; Li, X.; Chen, X.; Guo, Y.; Liang, W.; Wang, H. Genome-wide identification of soybean ABC transporters relate to aluminum toxicity. Int. J. Mol. Sci. 2021, 22, 6556. [Google Scholar] [CrossRef]
- Kanga, J.; Park, J.; Choi, H.; Burla, B.; Kretzschmar, T.; Lee, Y.; Martinoia, E. Plant ABC transporters. Arabidopsis Book 2011, 9, e0153. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.Y.; Jin, J.Y.; Alejandro, S.; Martinoia, E.; Lee, Y. Overexpression of AtABCG36 improves drought and salt stress resistance in Arabidopsis. Physiol. Plant. 2010, 139, 170–180. [Google Scholar] [CrossRef]
- Liu, W.; Feng, X.; Cao, F.; Wu, D.; Zhang, G.; Vincze, E.; Wang, Y.; Chen, Z.H.; Wu, F. An ATP binding cassette transporter HvABCB25 confers aluminum detoxification in wild barley. J. Hazard. Mater. 2021, 401, 123371. [Google Scholar] [CrossRef]
- Ryan, P.R.; Delhaize, E.; Jones, D.L. Function and mechanism of organic anion exudation from plant roots. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 527–560. [Google Scholar] [CrossRef] [PubMed]
- Hoekenga, O.A.; Maron, L.G.; Pineros, M.A.; Cancado, G.M.A.; Shaff, J.; Kobayashi, Y.; Ryan, P.R.; Dong, B.; Delhaize, E.; Sasaki, T.; et al. AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 9738–9743. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ji, F.; Zhang, Y.; Hou, J.; Liu, W.; Huang, J.; Liang, W. Interactions between hydrogen sulphide and nitric oxide regulate two soybean citrate transporters during the alleviation of aluminium toxicity. Plant Cell Environ. 2009, 42, 2340–2356. [Google Scholar] [CrossRef]
- Gao, P.; Han, R.; Xu, H.; Wei, Y.; Yu, Y. Identification of MATE family and characterization of GmMATE13 and GmMATE75 in soybean’s response to aluminum stress. Int. J. Mol. Sci. 2024, 25, 3711. [Google Scholar] [CrossRef]
- Yan, L.; Riaz, M.; Liu, J.; Liu, Y.; Zeng, Y.; Jiang, C. Boron reduces aluminum deposition in alkali-soluble pectin and cytoplasm to release aluminum toxicity. J. Hazard. Mater. 2021, 401, 123388. [Google Scholar] [CrossRef]
- Horst, W.J. The role of the apoplast in aluminum toxicity and resistance of higher plants: A review. Z. Pflanzenernährung Bodenkd. 1995, 158, 419–428. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; Qu, M.; Xiao, H.; Feng, Y.; Liu, J.; Wu, L.; Yu, M. Cell wall pectin and its methyl-esterification in transition zone determine Al resistance in cultivars of pea (Pisum sativum). Front. Plant Sci. 2016, 7, 39–47. [Google Scholar] [CrossRef]
- Yang, J.L.; Zhu, X.F.; Peng, Y.X.; Zheng, C.; Li, G.X.; Liu, Y.; Shi, Y.Z.; Zheng, S.J. Cell wall hemicellulose contributes significantly to aluminum adsorption and root growth in Arabidopsis. Plant Physiol. 2011, 155, 1885–1892. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.L.; Li, Y.Y.; Zhang, Y.J.; Zhang, S.S.; Wu, Y.R.; Wu, P.; Zheng, S.J. Cell wall polysaccharides are specifically involved in the exclusion of aluminum from the rice root apex. Plant Physiol. 2008, 146, 602–611. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.L.; Zhu, X.F.; Zheng, C.; Zhang, Y.J.; Zheng, S.J. Genotypic differences in Al resistance and the role of cell-wall pectin in Al exclusion from the root apex in Fagopyrum tataricum. Ann. Bot. 2011, 107, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Horst, W.J.; Schmohl, N.; Kollmeier, M.; Baluska, F.; Sivaguru, M. Does aluminium affect root growth of maize through interaction with the cell wall-plasma membrane-cytoskeleton continuum? Plant Soil. 1999, 215, 163–174. [Google Scholar] [CrossRef]
- Sawaki, Y.; Iuchi, S.; Kobayashi, Y.; Kobayashi, Y.; Ikka, Y.; Sakural, N.; Fujita, M.; Shibata, D.; Kobayashi, M.; Koyama, H. STOP1 regulates multiple genes which protect Arabidopsis from proton and aluminum toxicities. Plant Physiol. 2009, 150, 281–294. [Google Scholar] [CrossRef]
- Huang, J.; Han, R.; Ji, F.; Yu, Y.; Wang, Y.; Hai, Z.; Liang, W.; Wang, H. Glucose-6-phosphate dehydrogenase and abscisic acid mediate programmed cell death induced by aluminum toxicity in soybean root tips. J. Hazard. Mater. 2022, 425, 127964. [Google Scholar] [CrossRef]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Y.; Hou, J.; Liu, W.; Huang, J.; Liang, W. Nitric oxide mediates aluminum-induced citrate secretion through regulating the metabolism and transport of citrate in soybean roots. Plant Soil. 2019, 435, 127–142. [Google Scholar] [CrossRef]
- Eticha, D.; Stass, A.; Horst, W.J. Localization of aluminium in the maize root apex: Can morin detect cell wall-bound aluminium? J. Exp. Bot. 2005, 56, 1351–1357. [Google Scholar] [CrossRef]
- Zhong, H.; Lauchli, A. Changes of cell wall composition and polymer size in primary roots of cotton seedlings under high salinity. J. Exp. Bot. 1993, 44, 773–778. [Google Scholar] [CrossRef]
- Taylor, K.; Buchanan-Smith, J. A colorimetric method for the quantitation of uronic acids and a specific assay for galacturonic acid. Anal. Biochem. 1992, 201, 190–196. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Li, H.; Chen, Y.; Li, X.; Jia, Z.; Cheng, K.; Wang, L.; Wang, H. Two Half-Size ATP-Binding Cassette Transporters Are Implicated in Aluminum Tolerance in Soybean. Int. J. Mol. Sci. 2024, 25, 10332. https://doi.org/10.3390/ijms251910332
Huang J, Li H, Chen Y, Li X, Jia Z, Cheng K, Wang L, Wang H. Two Half-Size ATP-Binding Cassette Transporters Are Implicated in Aluminum Tolerance in Soybean. International Journal of Molecular Sciences. 2024; 25(19):10332. https://doi.org/10.3390/ijms251910332
Chicago/Turabian StyleHuang, Junjun, Huanan Li, Yiwei Chen, Xiaoyu Li, Ziyu Jia, Kunxia Cheng, Luyu Wang, and Huahua Wang. 2024. "Two Half-Size ATP-Binding Cassette Transporters Are Implicated in Aluminum Tolerance in Soybean" International Journal of Molecular Sciences 25, no. 19: 10332. https://doi.org/10.3390/ijms251910332



