Proteomic Characterization of Corneal Epithelial and Stromal Cell-Derived Extracellular Vesicles
Abstract
1. Introduction
2. Results
2.1. Characterization of HCE-, HCK-, HCF- and HCM-EVs
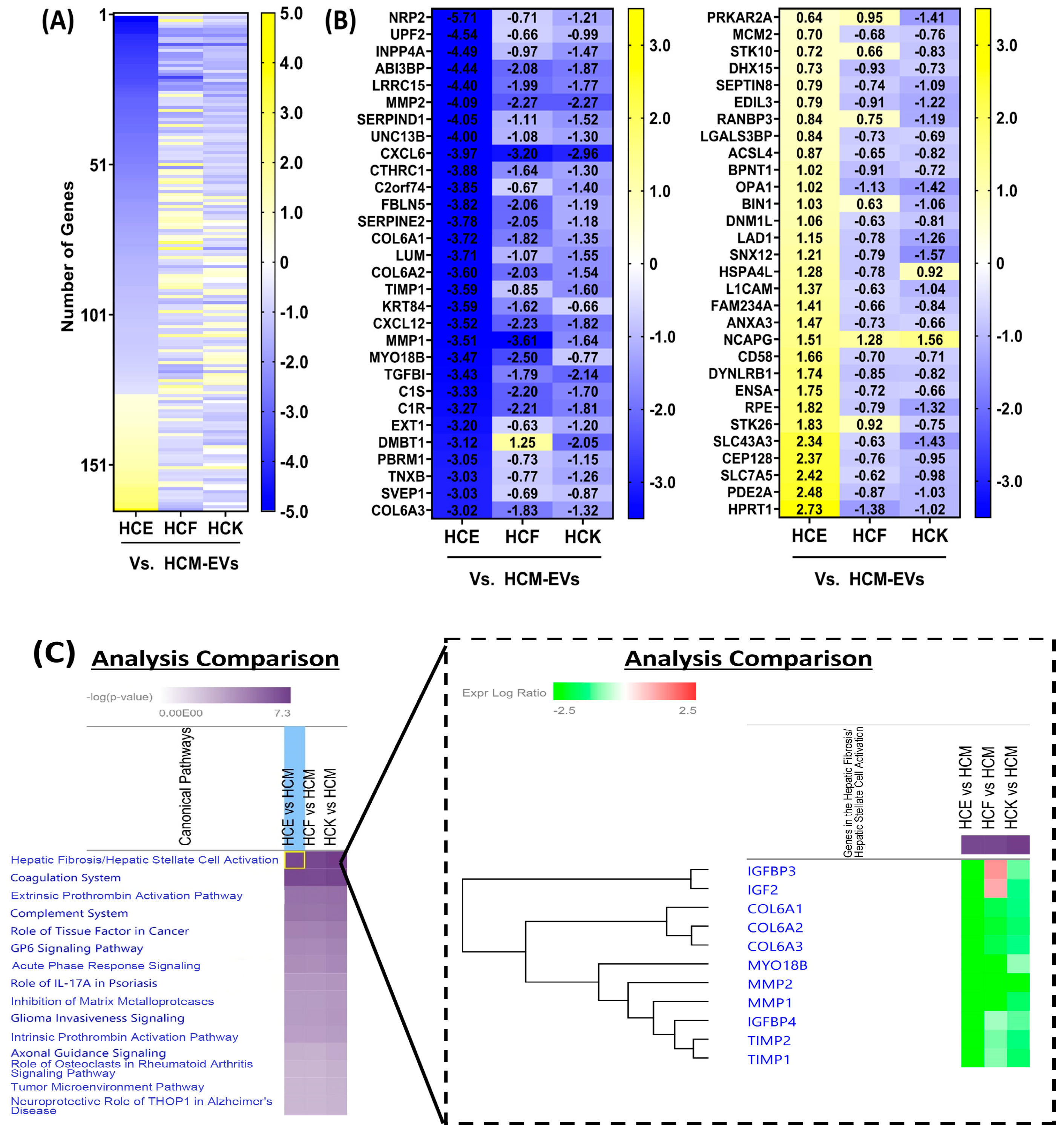
2.2. Proteomic Characterization of HCE-EVs
2.3. Proteomic Characterization of HCK-EVs
2.4. Proteomic Characterization of HCF-EVs
2.5. Proteomic Characterization of HCM-EVs
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Extracellular Vesicle (EV) Isolation
4.3. Western Blot
4.4. Nanoparticle Tracking Analysis (NTA) and Zeta Potential Measurements
4.5. Transmission Electron Microscopy (TEM)
4.6. Tandem Mass Tag (TMT) Mass Spectrometry
4.6.1. Sample Protein Digestion and Labeling
4.6.2. Sample Labeling and High pH Fractionation
4.6.3. Mass Spectrometry Analysis
4.7. Qiagen™ Ingenuity Pathway Analysis (IPA)
4.8. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Willoughby, C.E.; Ponzin, D.; Ferrari, S.; Lobo, A.; Landau, K.; Omidi, Y. Anatomy and physiology of the human eye: Effects of mucopolysaccharidoses disease on structure and function—A review. Clin. Exp. Ophthalmol. 2010, 38, 2–11. [Google Scholar] [CrossRef]
- Yeung, V.; Zhang, T.C.; Yuan, L.; Parekh, M.; Cortinas, J.A.; Delavogia, E.; Hutcheon, A.E.K.; Guo, X.; Ciolino, J.B. Extracellular Vesicles Secreted by Corneal Myofibroblasts Promote Corneal Epithelial Cell Migration. Int. J. Mol. Sci. 2022, 23, 3136. [Google Scholar] [CrossRef]
- Ljubimov, A.V.; Saghizadeh, M. Progress in corneal wound healing. Prog. Retin. Eye Res. 2015, 49, 17–45. [Google Scholar] [CrossRef]
- Kowtharapu, B.S.; Murín, R.; Jünemann, A.G.M.; Stachs, O. Role of Corneal Stromal Cells on Epithelial Cell Function during Wound Healing. Int. J. Mol. Sci. 2018, 19, 464. [Google Scholar] [CrossRef]
- McKay, T.B.; Hutcheon, A.E.K.; Zieske, J.D.; Ciolino, J.B. Extracellular Vesicles Secreted by Corneal Epithelial Cells Promote Myofibroblast Differentiation. Cells 2020, 9, 1080. [Google Scholar] [CrossRef]
- Zieske, J.D.; E Hutcheon, A.; Guo, X.; Chung, E.H.; Joyce, N.C. TGF-beta receptor types I and II are differentially expressed during corneal epithelial wound repair. Investig. Ophthalmol. Vis. Sci. 2001, 42, 1465–1471. [Google Scholar]
- Yeung, V.; Sriram, S.; Tran, J.A.; Guo, X.; Hutcheon, A.E.K.; Zieske, J.D.; Karamichos, D.; Ciolino, J.B. FAK Inhibition Attenuates Corneal Fibroblast Differentiation In Vitro. Biomolecules 2021, 11, 1682. [Google Scholar] [CrossRef]
- Bomb, R.; Heckle, M.R.; Sun, Y.; Mancarella, S.; Guntaka, R.V.; Gerling, I.C.; Weber, K.T. Myofibroblast secretome and its auto-/paracrine signaling. Expert. Rev. Cardiovasc. Ther. 2016, 14, 591–598. [Google Scholar] [CrossRef]
- Gartz, M.; Strande, J.L. Examining the Paracrine Effects of Exosomes in Cardiovascular Disease and Repair. J. Am. Heart Assoc. 2018, 7, e007954. [Google Scholar] [CrossRef] [PubMed]
- Yeung, V.; Willis, G.R.; Taglauer, E.; Mitsialis, S.A.; Kourembanas, S. Paving the Road for Mesenchymal Stem Cell-Derived Exosome Therapy in Bronchopulmonary Dysplasia and Pulmonary Hypertension. In Stem Cell-Based Therapy for Lung Disease; Springer: Cham, Switzerland, 2019; pp. 131–152. [Google Scholar] [CrossRef]
- Habibi, A.; Zarei-Behjani, Z.; Falamarzi, K.; Malekpour, M.; Ebrahimi, F.; Soleimani, M.; Nejabat, M.; Khosravi, A.; Moayedfard, Z.; Pakbaz, S.; et al. Extracellular vesicles as a new horizon in the diagnosis and treatment of inflammatory eye diseases: A narrative review of the literature. Front. Immunol. 2023, 14, 1097456. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Chen, T.; Li, C.; Xu, H.; Feng, Q.; Su, Y.; Cai, J.; Zhu, Q.; Liu, F.; Hu, L. Metabolic signatures of tear extracellular vesicles caused by herpes simplex keratitis. Ocul. Surf. 2024, 31, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Angourani, K.J.; Mazhari, S.; Farivar, S.; Mahini, D.S.; Rouintan, A.; Baghaei, K. Fibroblast-myofibroblast crosstalk after exposure to mesenchymal stem cells secretome. Gastroenterol. Hepatol. Bed Bench 2018, 11, S73–S79. [Google Scholar]
- Han, K.-Y.; Tran, J.A.; Chang, J.-H.; Azar, D.T.; Zieske, J.D. Potential role of corneal epithelial cell-derived exosomes in corneal wound healing and neovascularization. Sci. Rep. 2017, 7, srep40548. [Google Scholar] [CrossRef]
- Welsh, J.A.; Goberdhan, D.C.I.; O’DRiscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Di Vizio, D.; Driedonks, T.A.P.; Erdbrügger, U.; et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J. Extracell. Vesicles 2024, 13, e12404. [Google Scholar] [CrossRef]
- Harding, H.P.; Novoa, I.; Zhang, Y.; Zeng, H.; Wek, R.; Schapira, M.; Ron, D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 2000, 6, 1099–1108. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-H.; I, C.-C.; Kuo, C.-H.; Hsu, Y.-Y.; Lee, F.-T.; Shi, G.-Y.; Tseng, S.-H.; Wu, H.-L. Thrombomodulin promotes corneal epithelial wound healing. PLoS ONE 2015, 10, e0122491. [Google Scholar] [CrossRef]
- Wilson, S.E. Corneal myofibroblasts and fibrosis. Exp. Eye Res. 2020, 201, 108272. [Google Scholar] [CrossRef] [PubMed]
- Araki-Sasaki, K.; Ohashi, Y.; Sasabe, T.; Hayashi, K.; Watanabe, H.; Tano, Y.; Handa, H. An SV40-immortalized human corneal epithelial cell line and its characterization. Investig. Ophthalmol. Vis. Sci. 1995, 36, 614–621. [Google Scholar]
- Rubelowski, A.-K.; Latta, L.; Katiyar, P.; Stachon, T.; Käsmann-Kellner, B.; Seitz, B.; Szentmáry, N. HCE-T cell line lacks cornea-specific differentiation markers compared to primary limbal epithelial cells and differentiated corneal epithelium. Graefe’s Arch. Clin. Exp. Ophthalmol. 2020, 258, 565–575. [Google Scholar] [CrossRef]
- Torricelli, A.A.M.; Singh, V.; Santhiago, M.R.; Wilson, S.E. The corneal epithelial basement membrane: Structure, function, and disease. Investig. Opthalmol. Vis. Sci. 2013, 54, 6390–6400. [Google Scholar] [CrossRef] [PubMed]
- McKay, T.B.; Yeung, V.; Hutcheon, A.E.K.; Guo, X.; Zieske, J.D.; Ciolino, J.B. Extracellular Vesicles in the Cornea: Insights from Other Tissues. Anal. Cell. Pathol. 2021, 2021, e9983900. [Google Scholar] [CrossRef] [PubMed]
- Nieto-Miguel, T.; Calonge, M.; De La Mata, A.; López-Paniagua, M.; Galindo, S.; De La Paz, M.F.; Corrales, R.M. A comparison of stem cell-related gene expression in the progenitor-rich limbal epithelium and the differentiating central corneal epithelium. Mol. Vis. 2011, 17, 2102–2117. [Google Scholar]
- Albert, R.; Veréb, Z.; Csomós, K.; Moe, M.C.; Johnsen, E.O.; Olstad, O.K.; Nicolaissen, B.; Rajnavölgyi, E.; Fésüs, L.; Berta, A.; et al. Cultivation and characterization of cornea limbal epithelial stem cells on lens capsule in animal material-free medium. PLoS ONE 2012, 7, e47187. [Google Scholar] [CrossRef]
- Lupasco, T.; He, Z.; Cassagne, M.; Sagnial, T.; Brion, L.; Fournié, P.; Gain, P.; Thuret, G.; Allouche, M.; Malecaze, F.; et al. Corneal epithelium in keratoconus underexpresses active NRF2 and a subset of oxidative stress-related genes. PLoS ONE 2022, 17, e0273807. [Google Scholar] [CrossRef]
- Kao, W.W.-Y. Keratin expression by corneal and limbal stem cells during development. Exp. Eye Res. 2020, 200, 108206. [Google Scholar] [CrossRef] [PubMed]
- Collin, J.; Queen, R.; Zerti, D.; Bojic, S.; Dorgau, B.; Moyse, N.; Molina, M.M.; Yang, C.; Dey, S.; Reynolds, G.; et al. A single cell atlas of human cornea that defines its development, limbal progenitor cells and their interactions with the immune cells. Ocul. Surf. 2021, 21, 279–298. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.B.; Shen, X.; Pfeifer, C.W.; Shiau, F.; Santeford, A.; Ruzycki, P.A.; Clark, B.S.; Liu, Q.; Huang, A.J.W.; Apte, R.S. Dry eye disease in mice activates adaptive corneal epithelial regeneration distinct from constitutive renewal in homeostasis. Proc. Natl. Acad. Sci. USA 2023, 120, e2204134120. [Google Scholar] [CrossRef]
- Bulanova, D.R.; Akimov, Y.A.; Rokka, A.; Laajala, T.D.; Aittokallio, T.; Kouvonen, P.; Pellinen, T.; Kuznetsov, S.G. Orphan G protein-coupled receptor GPRC5A modulates integrin β1-mediated epithelial cell adhesion. Cell Adhes. Migr. 2017, 11, 434–446. [Google Scholar] [CrossRef]
- Liu, B.; Yang, H.; Pilarsky, C.; Weber, G.F. The Effect of GPRC5a on the Proliferation, Migration Ability, Chemotherapy Resistance, and Phosphorylation of GSK-3β in Pancreatic Cancer. Int. J. Mol. Sci. 2018, 19, 1870. [Google Scholar] [CrossRef]
- Sawada, Y.; Kikugawa, T.; Iio, H.; Sakakibara, I.; Yoshida, S.; Ikedo, A.; Yanagihara, Y.; Saeki, N.; Győrffy, B.; Kishida, T.; et al. GPRC5A facilitates cell proliferation through cell cycle regulation and correlates with bone metastasis in prostate cancer. Int. J. Cancer 2020, 146, 1369–1382. [Google Scholar] [CrossRef]
- Dougherty, G.W.; E Ostrowski, L.; Nöthe-Menchen, T.; Raidt, J.; Schramm, A.; Olbrich, H.; Yin, W.; Sears, P.R.; Dang, H.; Smith, A.J.; et al. Recessively Inherited Deficiency of Secreted WFDC2 (HE4) Causes Nasal Polyposis and Bronchiectasis. Am. J. Respir. Crit. Care Med. 2024, 210, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Lamandé, S.R.; Bateman, J.F. Collagen VI disorders: Insights on form and function in the extracellular matrix and beyond. Matrix Biol. 2018, 71, 348–367. [Google Scholar] [CrossRef]
- Zanotti, S.; Magri, F.; Salani, S.; Napoli, L.; Ripolone, M.; Ronchi, D.; Fortunato, F.; Ciscato, P.; Velardo, D.; D’aNgelo, M.G.; et al. Extracellular Matrix Disorganization and Sarcolemmal Alterations in COL6-Related Myopathy Patients with New Variants of COL6 Genes. Int. J. Mol. Sci. 2023, 24, 5551. [Google Scholar] [CrossRef] [PubMed]
- Zeichen, J.; van Griensven, M.; Albers, I.; Lobenhoffer, P.; Bosch, U. Immunohistochemical localization of collagen VI in arthrofibrosis. Arch. Orthop. Trauma Surg. 1999, 119, 315–318. [Google Scholar] [CrossRef]
- Fontana, R.J.; Goodman, Z.D.; Dienstag, J.L.; Bonkovsky, H.L.; Naishadham, D.; Sterling, R.K.; Su, G.L.; Ghosh, M.; Wright, E.C.; HALT-C Trial Group. Relationship of serum fibrosis markers with liver fibrosis stage and collagen content in patients with advanced chronic hepatitis C. Hepatology 2008, 47, 789–798. [Google Scholar] [CrossRef]
- Williams, L.M.; McCann, F.E.; Cabrita, M.A.; Layton, T.; Cribbs, A.; Knezevic, B.; Fang, H.; Knight, J.; Zhang, M.; Fischer, R.; et al. Identifying collagen VI as a target of fibrotic diseases regulated by CREBBP/EP300. Proc. Natl. Acad. Sci. USA 2020, 117, 20753–20763. [Google Scholar] [CrossRef] [PubMed]
- Jessen, H.; Hoyer, N.; Prior, T.S.; Frederiksen, P.; Rønnow, S.R.; Karsdal, M.A.; Leeming, D.J.; Bendstrup, E.; Sand, J.M.B.; Shaker, S.B. Longitudinal serological assessment of type VI collagen turnover is related to progression in a real-world cohort of idiopathic pulmonary fibrosis. BMC Pulm. Med. 2021, 21, 382. [Google Scholar] [CrossRef]
- Cai, X.; Li, Z.; Zhang, Q.; Qu, Y.; Xu, M.; Wan, X.; Lu, L. CXCL6-EGFR-induced Kupffer cells secrete TGF-β1 promoting hepatic stellate cell activation via the SMAD2/BRD4/C-MYC/EZH2 pathway in liver fibrosis. J. Cell. Mol. Med. 2018, 22, 5050–5061. [Google Scholar] [CrossRef]
- Kersey, A.L.; Cheng, D.Y.; Deo, K.A.; Dubell, C.R.; Wang, T.-C.; Jaiswal, M.K.; Kim, M.H.; Murali, A.; Hargett, S.E.; Mallick, S.; et al. Stiffness assisted cell-matrix remodeling trigger 3D mechanotransduction regulatory programs. Biomaterials 2024, 306, 122473. [Google Scholar] [CrossRef]
- Prakash, J.; Shaked, Y. The Interplay between Extracellular Matrix Remodeling and Cancer Therapeutics. Cancer Discov. 2024, 14, 1375–1388. [Google Scholar] [CrossRef]
- Holmberg, C.; Ghesquière, B.; Impens, F.; Gevaert, K.; Kumar, J.D.; Cash, N.; Kandola, S.; Hegyi, P.; Wang, T.C.; Dockray, G.J.; et al. Mapping proteolytic processing in the secretome of gastric cancer-associated myofibroblasts reveals activation of MMP-1, MMP-2, and MMP-3. J. Proteome Res. 2013, 12, 3413–3422. [Google Scholar] [CrossRef]
- Roeb, E. Matrix metalloproteinases and liver fibrosis (translational aspects). Matrix Biol. 2018, 68, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Skotland, T.; Sagini, K.; Sandvig, K.; Llorente, A. An emerging focus on lipids in extracellular vesicles. Adv. Drug Deliv. Rev. 2020, 159, 308–321. [Google Scholar] [CrossRef] [PubMed]
- Hallal, S.; Tűzesi, Á.; Grau, G.E.; Buckland, M.E.; Alexander, K.L. Understanding the extracellular vesicle surface for clinical molecular biology. J. Extracell. Vesicles 2022, 11, e12260. [Google Scholar] [CrossRef] [PubMed]
- Chitti, S.V.; Gummadi, S.; Kang, T.; Shahi, S.; Marzan, A.L.; Nedeva, C.; Sanwlani, R.; Bramich, K.; Stewart, S.; Petrovska, M.; et al. Vesiclepedia 2024: An extracellular vesicles and extracellular particles repository. Nucleic Acids Res. 2024, 52, D1694–D1698. [Google Scholar] [CrossRef]
- Dogra, N.; Chen, T.; Gonzalez-Kozlova, E.; Miceli, R.; Cordon-Cardo, C.; Tewari, A.K.; Losic, B.; Stolovitzky, G. Extracellular vesicles carry transcriptional ‘dark matter’ revealing tissue-specific information. J. Extracell. Vesicles 2024, 13, e12481. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Wu, Z.; Zhao, M.; Murtazina, R.; Cai, J.; Zhang, A.; Li, R.; Sun, D.; Li, W.; Zhao, L.; et al. Rab5-dependent autophagosome closure by ESCRT. J. Cell Biol. 2019, 218, 1908–1927. [Google Scholar] [CrossRef]
- Hattori, T.; Takahashi, Y.; Chen, L.; Tang, Z.; Wills, C.A.; Liang, X.; Wang, H.-G. Targeting the ESCRT-III component CHMP2A for noncanonical Caspase-8 activation on autophagosomal membranes. Cell Death Differ. 2021, 28, 657–670. [Google Scholar] [CrossRef]
- Long, C.J.; Roth, M.R.; Tasheva, E.S.; Funderburgh, M.; Smit, R.; Conrad, G.W.; Funderburgh, J.L. Fibroblast growth factor-2 promotes keratan sulfate proteoglycan expression by keratocytes in vitro. J. Biol. Chem. 2000, 275, 13918–13923. [Google Scholar] [CrossRef]
- Foster, J.W.; Gouveia, R.M.; Connon, C.J. Low-glucose enhances keratocyte-characteristic phenotype from corneal stromal cells in serum-free conditions. Sci. Rep. 2015, 5, 10839. [Google Scholar] [CrossRef]
- Yusoff, N.Z.B.M.; Riau, A.K.; Yam, G.H.F.; Halim, N.S.H.B.; Mehta, J.S. Isolation and Propagation of Human Corneal Stromal Keratocytes for Tissue Engineering and Cell Therapy. Cells 2022, 11, 178. [Google Scholar] [CrossRef] [PubMed]
- Beales, M.P.; Funderburgh, J.L.; Jester, J.V.; Hassell, J.R. Proteoglycan synthesis by bovine keratocytes and corneal fibroblasts: Maintenance of the keratocyte phenotype in culture. Investig. Ophthalmol. Vis. Sci. 1999, 40, 1658–1663. [Google Scholar]
- Karamichos, D.; Rich, C.B.; Zareian, R.; Hutcheon, A.E.K.; Ruberti, J.W.; Trinkaus-Randall, V.; Zieske, J.D. TGF-β3 Stimulates Stromal Matrix Assembly by Human Corneal Keratocyte-Like Cells. Investig. Opthalmol. Vis. Sci. 2013, 54, 6612–6619. [Google Scholar] [CrossRef] [PubMed]
- Ren, R.; Hutcheon, A.; Guo, X.; Saeidi, N.; Melotti, S.; Ruberti, J.; Zieske, J.; Trinkaus-Randall, V. Human Primary Corneal Fibroblasts Synthesize and Deposit Proteoglycans in Long-Term 3-D Cultures. Dev. Dyn. 2008, 237, 2705–2715. [Google Scholar] [CrossRef] [PubMed]
- Yeung, V.; Webber, J.P.; Dunlop, E.A.; Morgan, H.; Hutton, J.; Gurney, M.; Jones, E.; Falcon-Perez, J.; Tabi, Z.; Errington, R.; et al. Rab35-dependent extracellular nanovesicles are required for induction of tumour supporting stroma. Nanoscale 2018, 10, 8547–8559. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, C.; Ferreira, Y.J.; Dragovic, R.A.; Redman, C.W.G.; Sargent, I.L. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. J. Extracell. Vesicles 2013, 2, 19671. [Google Scholar] [CrossRef]
- Thery, C.; Clayton, A.; Amigorena, S.; Raposo, G. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3–22. [Google Scholar] [CrossRef]
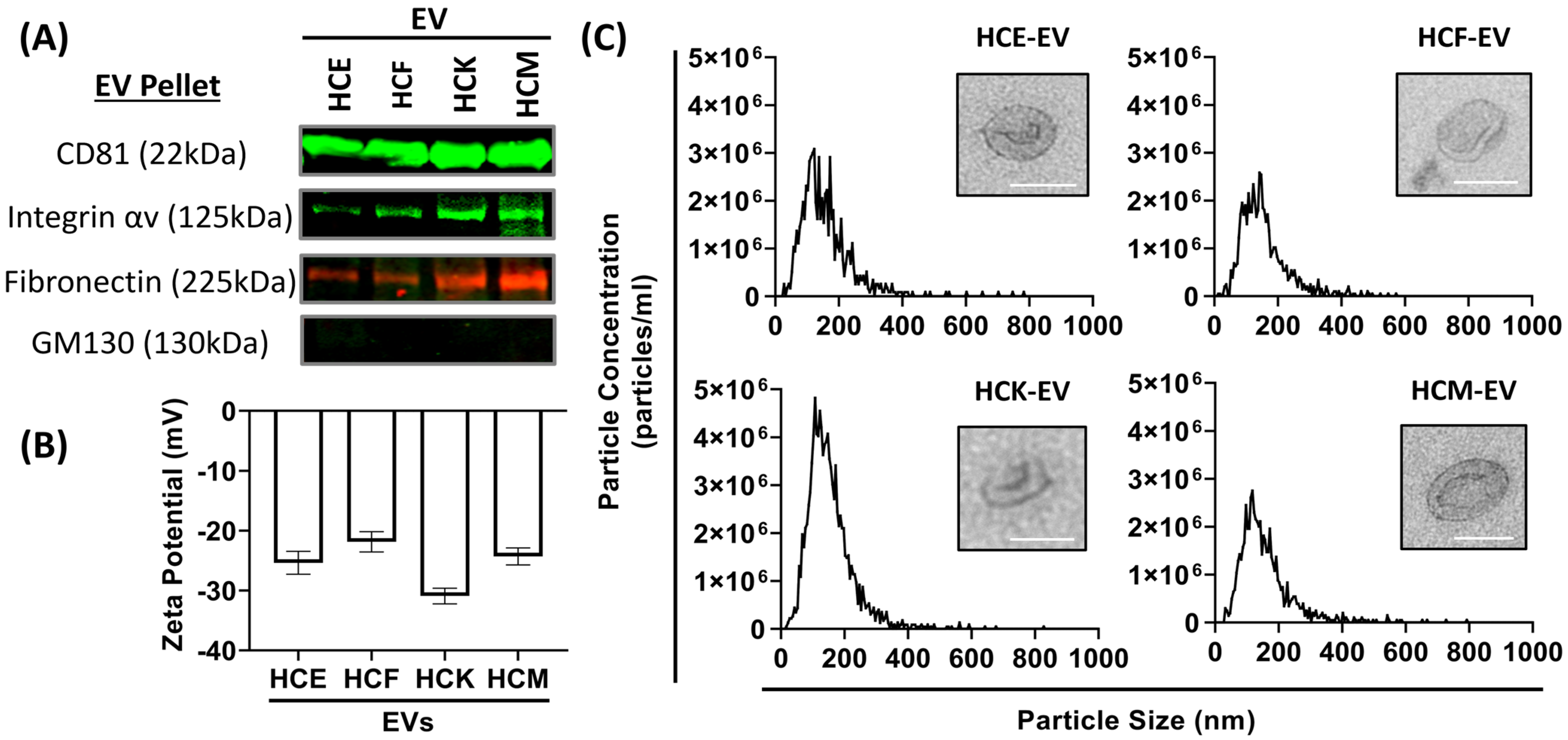



| EV Type | Enriched Protein Markers | IPA |
|---|---|---|
| HCE-EV | GPRC5A, KRT7, KRT8, KRT18, KRT19, WFDC2, FLOT2 | EIF Signaling Pathway |
| HCK-EV | APOA1, APOM, DMKN, IAH1, AGBL5, KRT13 | Acute-Phase Response Signaling |
| HCF-EV | FDXR, APEX1, LAMP1, PARP1, VAMP8 | Extrinsic Prothrombic Activation, Coagulation Pathway |
| HCM-EV | COL6A1, COL6A2, COL6A3, MMP1, MMP2, TIMP1, TIMP2 CXCL6, CXCL12 | Fibrosis Pathway |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yeung, V.; Boychev, N.; Kanu, L.N.; Ng, V.; Ross, A.E.; Hutcheon, A.E.K.; Ciolino, J.B. Proteomic Characterization of Corneal Epithelial and Stromal Cell-Derived Extracellular Vesicles. Int. J. Mol. Sci. 2024, 25, 10338. https://doi.org/10.3390/ijms251910338
Yeung V, Boychev N, Kanu LN, Ng V, Ross AE, Hutcheon AEK, Ciolino JB. Proteomic Characterization of Corneal Epithelial and Stromal Cell-Derived Extracellular Vesicles. International Journal of Molecular Sciences. 2024; 25(19):10338. https://doi.org/10.3390/ijms251910338
Chicago/Turabian StyleYeung, Vincent, Nikolay Boychev, Levi N. Kanu, Veronica Ng, Amy E. Ross, Audrey E. K. Hutcheon, and Joseph B. Ciolino. 2024. "Proteomic Characterization of Corneal Epithelial and Stromal Cell-Derived Extracellular Vesicles" International Journal of Molecular Sciences 25, no. 19: 10338. https://doi.org/10.3390/ijms251910338
APA StyleYeung, V., Boychev, N., Kanu, L. N., Ng, V., Ross, A. E., Hutcheon, A. E. K., & Ciolino, J. B. (2024). Proteomic Characterization of Corneal Epithelial and Stromal Cell-Derived Extracellular Vesicles. International Journal of Molecular Sciences, 25(19), 10338. https://doi.org/10.3390/ijms251910338






