Molecular Epidemiology and Phyloevolutionary Analysis of Porcine Parvoviruses (PPV1 through PPV7) Detected in Replacement Gilts from Colombia
Abstract
1. Introduction
2. Results
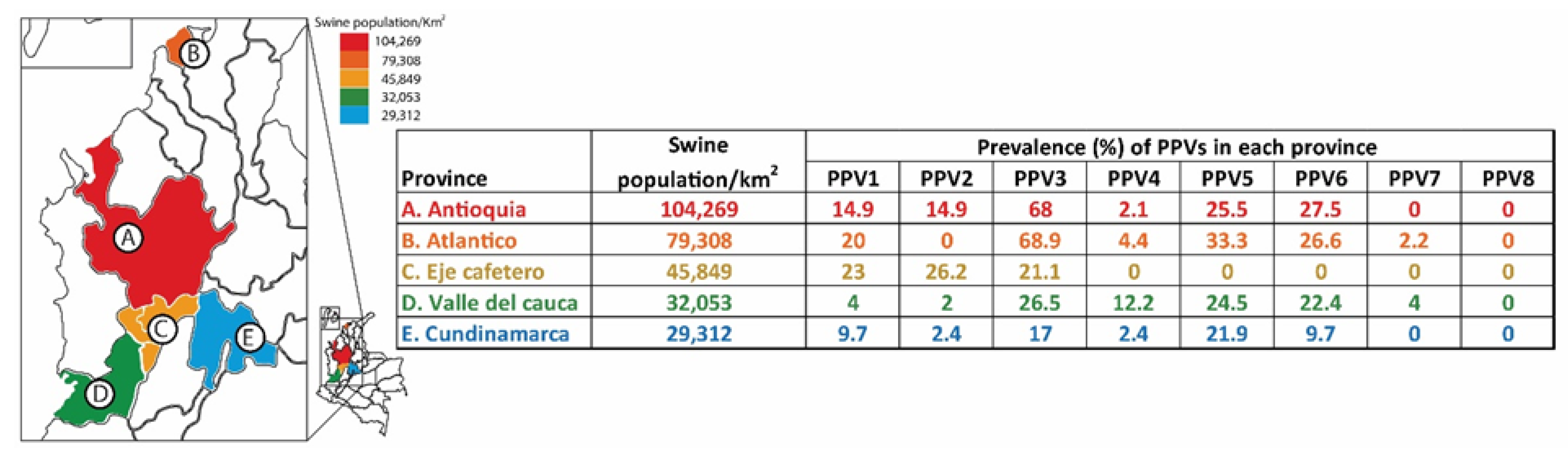
2.1. Detection Rates of PPVs in Replacement Gilts
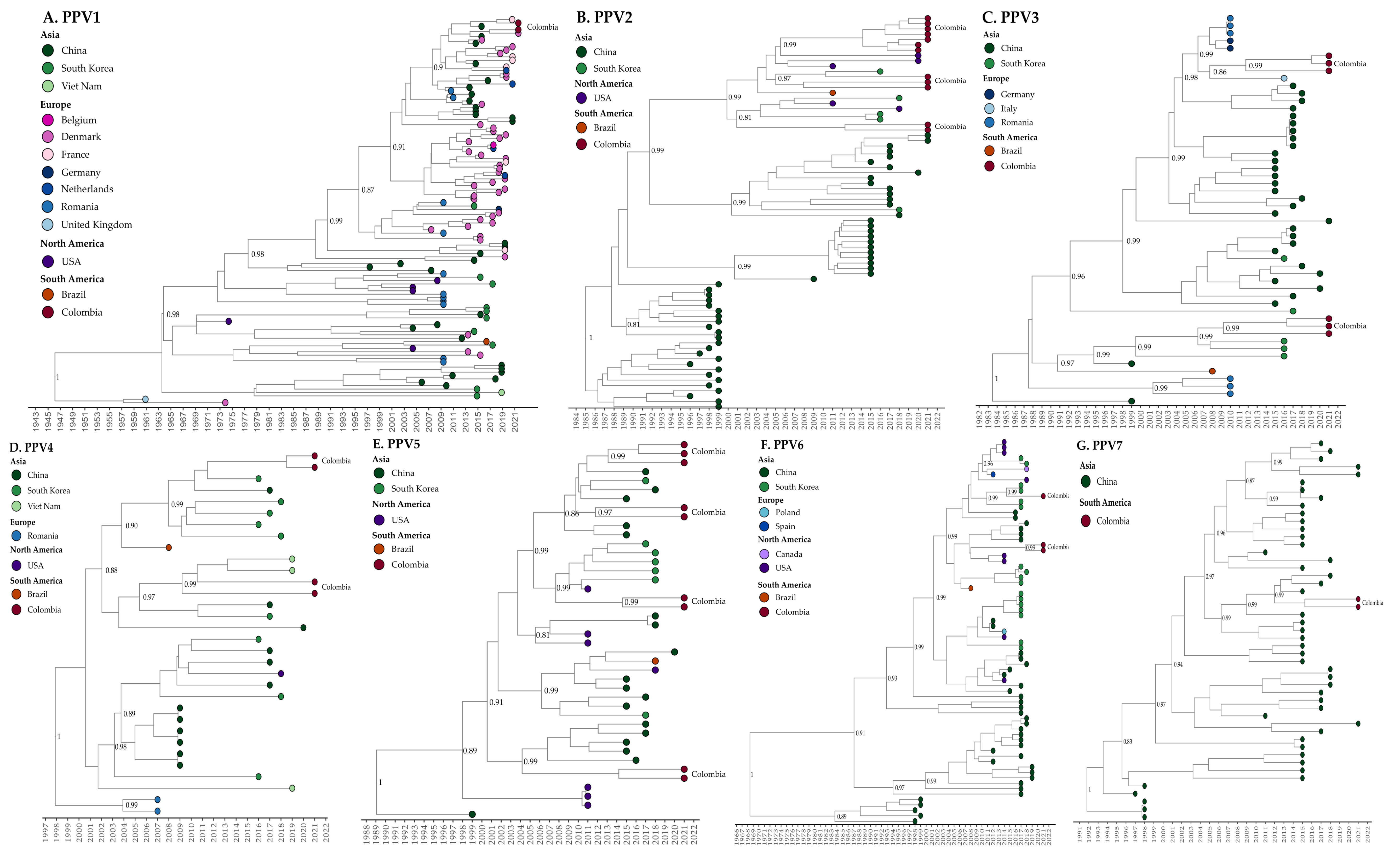
2.2. Phylogenetic and Evolutionary Rate Analysis of PPV1 and nPPVs
2.3. nPPVS Recombination Detection
3. Discussion
3.1. nPPVs as Etiologic Agents, Prevalence, and Coinfections
3.2. Phylogeny and Evolution of nPPVs
3.3. PPV1
4. Conclusions
5. Materials and Methods
5.1. Samples Selection
5.2. Detection of PPV1 and nPPVs
5.3. Sequencing of PPV1 and nPPVs
5.4. Quality Assurance (QA) Protocols
5.5. Phylogenetic Analysis of PPV1 and nPPVs
5.6. Bayesian Analyses of PPV1 through PPV7
5.7. Recombination Analysis in nPPVs
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pénzes, J.J.; de Souza, W.M.; Agbandje-McKenna, M.; Gifford, R.J. An Ancient Lineage of Highly Divergent Parvoviruses Infects both Vertebrate and Invertebrate Hosts. Viruses 2019, 11, 525. [Google Scholar] [CrossRef] [PubMed]
- Pénzes, J.J.; Söderlund-Venermo, M.; Canuti, M.; Eis-Hübinger, A.M.; Hughes, J.; Cotmore, S.F.; Harrach, B. Reorganizing the family Parvoviridae: A revised taxonomy independent of the canonical approach based on host association. Arch. Virol. 2020, 165, 2133–2146. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Bermudez, D.S.; Mogollon, J.D.; Franco-Rodriguez, C.; Jaime, J. The novel porcine parvoviruses: Current state of knowledge and their possible implications in clinical syndromes in pigs. Viruses 2023, 15, 2398. [Google Scholar] [CrossRef] [PubMed]
- Streck, A.F.; Truyen, U. Porcine Parvovirus. Curr. Issues Mol. Biol. 2020, 37, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, P.; Ritzmann, M.; Selbitz, H.J.; Heinritzi, K.; Truyen, U. VP1 sequences of German porcine parvovirus isolates define two genetic lineages. J. Gen. Virol. 2006, 87, 295–301. [Google Scholar] [CrossRef]
- Cadar, D.; Dán, Á.; Tombácz, K.; Lőrincz, M.; Kiss, T.; Becskei, Z.; Spînu, M.; Tuboly, T.; Cságola, A. Phylogeny and evolutionary genetics of porcine parvovirus in wild boars. Infect. Genet. Evol. 2012, 12, 1163–1171. [Google Scholar] [CrossRef]
- Oh, W.-T.; Kim, R.-Y.; Nguyen, V.-G.; Chung, H.-C.; Park, B.-K. Perspectives on the evolution of porcine parvovirus. Viruses 2017, 9, 196. [Google Scholar] [CrossRef]
- Vereecke, N.; Kvisgaard, L.K.; Baele, G.; Boone, C.; Kunze, M.; Larsen, L.E.; Theuns, S.; Nauwynck, H. Molecular epidemiology of Porcine Parvovirus Type 1 (PPV1) and the reactivity of vaccine-induced antisera against historical and current PPV1 strains. Virus Evol. 2022, 8, veac053. [Google Scholar] [CrossRef]
- Hijikata, M.; Abe, K.; Win, K.M.; Shimizu, Y.K.; Keicho, N.; Yoshikura, H. Identification of new parvovirus DNA sequence in swine sera from Myanmar. Jpn. J. Infect. Dis. 2001, 54, 244–245. [Google Scholar]
- Wang, F.; Wei, Y.; Zhu, C.; Huang, X.; Xu, Y.; Yu, L.; Yu, X. Novel parvovirus sublineage in the family of Parvoviridae. Virus Genes 2010, 41, 305–308. [Google Scholar] [CrossRef]
- Cadar, D.; Lőrincz, M.; Kiss, T.; Novosel, D.; Podgorska, K.; Becskei, Z.; Tuboly, T.; Cságola, A. Emerging novel porcine parvoviruses in Europe: Origin, evolution, phylodynamics and phylogeography. J. Gen. Virol. 2013, 94, 2330–2337. [Google Scholar] [CrossRef] [PubMed]
- Afolabi, K.O.; Iweriebor, B.C.; Obi, L.C.; Okoh, A.I. Prevalence of porcine parvoviruses in some South African swine herds with background of porcine circovirus type 2 infection. Acta Trop. 2019, 190, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.-T.; Gerber, P.F.; Giménez-Lirola, L.G.; Halbur, P.G.; Opriessnig, T. Characterization of porcine parvovirus type 2 (PPV2) which is highly prevalent in the USA. Vet. Microbiol. 2013, 161, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Novosel, D.; Cadar, D.; Tuboly, T.; Jungic, A.; Stadejek, T.; Ait-Ali, T.; Cságola, A. Investigating porcine parvoviruses genogroup 2 infection using in situ polymerase chain reaction. BMC Vet. Res. 2018, 14, 163. [Google Scholar] [CrossRef]
- Nelsen, A.; Lin, C.-M.; Hause, B.M. Porcine parvovirus 2 is predominantly associated with macrophages in porcine respiratory disease complex. Front. Vet. Sci. 2021, 8, 726884. [Google Scholar] [CrossRef]
- Vargas-Bermudez, D.S.; Mainenti, M.; Naranjo-Ortiz, M.F.; Mogollon, J.D.; Piñeyro, P.; Jaime, J. First Report of Porcine Parvovirus 2 (PPV2) in Pigs from Colombia Associated with Porcine Reproductive Failure (PRF) and Porcine Respiratory Disease Complex (PRDC). Transbound. Emerg. Dis. 2024, 2024, 1471536. [Google Scholar] [CrossRef]
- Saekhow, P.; Mawatari, T.; Ikeda, H. Coexistence of multiple strains of porcine parvovirus 2 in pig farms. Microbiol. Immunol. 2014, 58, 382–387. [Google Scholar] [CrossRef]
- Cadar, D.; Cságola, A.; Kiss, T.; Tuboly, T. Capsid protein evolution and comparative phylogeny of novel porcine parvoviruses. Mol. Phylogenet. Evol. 2013, 66, 243–253. [Google Scholar] [CrossRef]
- Sun, J.; Huang, L.; Wei, Y.; Wang, Y.; Chen, D.; Du, W.; Wu, H.; Liu, C. Prevalence of emerging porcine parvoviruses and their co-infections with porcine circovirus type 2 in China. Arch. Virol. 2015, 160, 1339–1344. [Google Scholar] [CrossRef]
- Lau, S.K.P.; Woo, P.C.Y.; Tse, H.; Fu, C.T.Y.; Au, W.-K.; Chen, X.-C.; Tsoi, H.-W.; Tsang, T.H.F.; Chan, J.S.Y.; Tsang, D.N.C.; et al. Identification of novel porcine and bovine parvoviruses closely related to human parvovirus 4. J. Gen. Virol. 2008, 89, 1840–1848. [Google Scholar] [CrossRef]
- Adlhoch, C.; Kaiser, M.; Ellerbrok, H.; Pauli, G. High prevalence of porcine Hokovirus in German wild boar populations. Virol. J. 2010, 7, 171. [Google Scholar] [CrossRef] [PubMed]
- Cságola, A.; Lőrincz, M.; Cadar, D.; Tombácz, K.; Biksi, I.; Tuboly, T. Detection, prevalence and analysis of emerging porcine parvovirus infections. Arch. Virol. 2012, 157, 1003–1010. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Biernacka, K.; Fan, J.; Gerber, P.F.; Stadejek, T.; Opriessnig, T. Circulation of Porcine Parvovirus Types 1 through 6 in Serum Samples Obtained from Six Commercial Polish Pig Farms. Transbound. Emerg. Dis. 2017, 64, 1945–1952. [Google Scholar] [CrossRef] [PubMed]
- Sliz, I.; Vlasakova, M.; Jackova, A.; Vilcek, S. Characterization of porcine parvovirus type 3 and porcine circovirus type 2 in wild boars (sus scrofa) in slovakia. J. Wildl. Dis. 2015, 51, 703–711. [Google Scholar] [CrossRef]
- Lagan Tregaskis, P.; Staines, A.; Gordon, A.; Sheridan, P.; McMenamy, M.; Duffy, C.; Collins, P.J.; Mooney, M.H.; Lemon, K. Co-infection status of novel parvovirus’s (PPV2 to 4) with porcine circovirus 2 in porcine respiratory disease complex and porcine circovirus-associated disease from 1997 to 2012. Transbound. Emerg. Dis. 2021, 68, 1979–1994. [Google Scholar] [CrossRef]
- Cadar, D.; Cságola, A.; Lorincz, M.; Tombácz, K.; Spînu, M.; Tuboly, T. Distribution and genetic diversity of porcine hokovirus in wild boars. Arch. Virol. 2011, 156, 2233–2239. [Google Scholar] [CrossRef]
- Cheung, A.K.; Wu, G.; Wang, D.; Bayles, D.O.; Lager, K.M.; Vincent, A.L. Identification and molecular cloning of a novel porcine parvovirus. Arch. Virol. 2010, 155, 801–806. [Google Scholar] [CrossRef]
- Amoroso, M.G.; Cerutti, F.; D’Alessio, N.; Lucibelli, M.G.; Cerrone, A.; Acutis, P.L.; Galiero, G.; Fusco, G.; Peletto, S. First identification of porcine parvovirus 3 in a wild boar in Italy by viral metagenomics Short communication. Acta Vet. Hung. 2019, 67, 135–139. [Google Scholar] [CrossRef]
- Kim, S.-C.; Kim, J.-H.; Kim, J.-Y.; Park, G.-S.; Jeong, C.-G.; Kim, W.-I. Prevalence of porcine parvovirus 1 through 7 (PPV1-PPV7) and co-factor association with PCV2 and PRRSV in Korea. BMC Vet. Res. 2022, 18, 133. [Google Scholar] [CrossRef]
- Blomström, A.-L.; Ståhl, K.; Masembe, C.; Okoth, E.; Okurut, A.R.; Atmnedi, P.; Kemp, S.; Bishop, R.; Belák, S.; Berg, M. Viral metagenomic analysis of bushpigs (Potamochoerus larvatus) in Uganda identifies novel variants of Porcine parvovirus 4 and Torque teno sus virus 1 and 2. Virol. J. 2012, 9, 192. [Google Scholar] [CrossRef]
- Gava, D.; Souza, C.K.; Schaefer, R.; Vincent, A.L.; Cantão, M.E.; Coldebella, A.; Ciacci-Zanella, J.R. A TaqMan-based real-time PCR for detection and quantification of porcine parvovirus 4. J. Virol. Methods 2015, 219, 14–17. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Camacho, L.A.; Vargas-Ruiz, A.; Marin-Flamand, E.; Ramírez-Álvarez, H.; Brown, C. A retrospective study of DNA prevalence of porcine parvoviruses in Mexico and its relationship with porcine circovirus associated disease. Microbiol. Immunol. 2020, 64, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.-T.; Halbur, P.G.; Opriessnig, T. Complete genome sequence of a novel porcine parvovirus (PPV) provisionally designated PPV5. Genome Announc. 2013, 1, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Wen, Y.; Huang, X.; Wen, X.; Yan, Q.; Huang, Y.; Ma, X.; Cao, S. First complete genomic characterization of a porcine parvovirus 5 isolate from China. Arch. Virol. 2014, 159, 1533–1536. [Google Scholar] [CrossRef]
- Cibulski, S.; Alves de Lima, D.; Fernandes Dos Santos, H.; Teixeira, T.F.; Tochetto, C.; Mayer, F.Q.; Roehe, P.M. A plate of viruses: Viral metagenomics of supermarket chicken, pork and beef from Brazil. Virology 2021, 552, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Wang, G.; Chen, J.; Zhang, W.; He, Y.; Qian, P. Molecular epidemiology of porcine circovirus type 2 and porcine parvoviruses in guangxi autonomous region, china. Microbiol. Res. 2023, 14, 1331–1342. [Google Scholar] [CrossRef]
- Ni, J.; Qiao, C.; Han, X.; Han, T.; Kang, W.; Zi, Z.; Cao, Z.; Zhai, X.; Cai, X. Identification and genomic characterization of a novel porcine parvovirus (PPV6) in China. Virol. J. 2014, 11, 203. [Google Scholar] [CrossRef]
- Schirtzinger, E.E.; Suddith, A.W.; Hause, B.M.; Hesse, R.A. First identification of porcine parvovirus 6 in North America by viral metagenomic sequencing of serum from pigs infected with porcine reproductive and respiratory syndrome virus. Virol. J. 2015, 12, 170. [Google Scholar] [CrossRef]
- Franzo, G.; Kekarainen, T.; Llorens, A.; Correa-Fiz, F.; Segalés, J. Exploratory metagenomic analyses of periweaning failure-to-thrive syndrome-affected pigs. Vet. Rec. 2019, 184, 25. [Google Scholar] [CrossRef]
- Cui, J.; Fan, J.; Gerber, P.F.; Biernacka, K.; Stadejek, T.; Xiao, C.-T.; Opriessnig, T. First identification of porcine parvovirus 6 in Poland. Virus Genes 2017, 53, 100–104. [Google Scholar] [CrossRef]
- Palinski, R.M.; Mitra, N.; Hause, B.M. Discovery of a novel Parvovirinae virus, porcine parvovirus 7, by metagenomic sequencing of porcine rectal swabs. Virus Genes 2016, 52, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Xing, X.; Zhou, H.; Tong, L.; Chen, Y.; Sun, Y.; Wang, H.; Zhang, G. First identification of porcine parvovirus 7 in China. Arch. Virol. 2018, 163, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Miłek, D.; Woźniak, A.; Stadejek, T. The detection and genetic diversity of novel porcine parvovirus 7 (PPV7) on Polish pig farms. Res. Vet. Sci. 2018, 120, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Bermudez, D.S.; Rendon-Marin, S.; Ruiz-Saenz, J.; Mogollón, D.; Jaime, J. The first report of porcine parvovirus 7 (PPV7) in Colombia demonstrates the presence of variants associated with modifications at the level of the VP2-capsid protein. PLoS ONE 2021, 16, e0258311. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zheng, C.; Lv, Z.; Xue, S.; Chen, Y.; Liu, Y.; Huang, X.; Luo, G.; Yang, X.; Dai, A. Genetic and epidemic characteristics of porcine parvovirus 7 in the Fujian and Guangdong regions of southern China. Front. Vet. Sci. 2022, 9, 949764. [Google Scholar] [CrossRef]
- Guo, Y.; Yan, G.; Chen, S.; Han, H.; Li, J.; Zhang, H.; Luo, S.; Liu, M.; Wu, Q.; Li, Q.; et al. Identification and genomic characterization of a novel porcine parvovirus in China. Front. Vet. Sci. 2022, 9, 1009103. [Google Scholar] [CrossRef]
- Igriczi, B.; Dénes, L.; Schönhardt, K.; Balka, G. First report of porcine parvovirus 8 in europe: Widespread detection and genetic characterization on commercial pig farms in hungary and slovakia. Animals 2024, 14, 1974. [Google Scholar] [CrossRef]
- Vargas-Bermudez, D.S.; Jaime, J. The first report of porcine parvovirus 8 (PPV8) on the American continent is associated with pigs in Colombia with porcine respiratory disease. Arch. Virol. 2024, 169, 179. [Google Scholar] [CrossRef]
- Li, J.; Xiao, Y.; Qiu, M.; Li, X.; Li, S.; Lin, H.; Li, X.; Zhu, J.; Chen, N. A Systematic Investigation Unveils High Coinfection Status of Porcine Parvovirus Types 1 through 7 in China from 2016 to 2020. Microbiol. Spectr. 2021, 9, e0129421. [Google Scholar] [CrossRef]
- Park, G.-N.; Song, S.; Cha, R.M.; Choe, S.; Shin, J.; Kim, S.-Y.; Hyun, B.-H.; Park, B.-K.; An, D.-J. Genetic analysis of porcine parvoviruses detected in Republic of Korean wild boars. Arch. Virol. 2021, 166, 2249–2254. [Google Scholar] [CrossRef]
- Shackelton, L.A.; Parrish, C.R.; Truyen, U.; Holmes, E.C. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc. Natl. Acad. Sci. USA 2005, 102, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Shackelton, L.A.; Hoelzer, K.; Parrish, C.R.; Holmes, E.C. Comparative analysis reveals frequent recombination in the parvoviruses. J. Gen. Virol. 2007, 88, 3294–3301. [Google Scholar] [CrossRef] [PubMed]
- Cartwright, S.F.; Lucas, M.; Huck, R.A. A small haemagglutinating porcine DNA virus. I. Isolation and properties. J. Comp. Pathol. 1969, 79, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Fu, P.-F.; Wang, Y.-H.; Liu, G.; Wang, D.-M.; Huang, W.-W.; Guo, D.-Q.; Li, X.-Y.; Liu, P.; Wei, M.-X.; Lu, M.; et al. First molecular detection and genetic characterization of porcine circovirus 4 in the Gansu Province of China. PLoS ONE 2024, 19, e0293135. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, G.; Piscopo, N.; Pagnini, U.; Esposito, L.; Montagnaro, S. Detection of selected pathogens in reproductive tissues of wild boars in the Campania region, southern Italy. Acta Vet. Scand. 2024, 66, 9. [Google Scholar] [CrossRef]
- Qin, S.; Ruan, W.; Yue, H.; Tang, C.; Zhou, K.; Zhang, B. Viral communities associated with porcine respiratory disease complex in intensive commercial farms in Sichuan province, China. Sci. Rep. 2018, 8, 13341. [Google Scholar] [CrossRef]
- Cságola, A.; Zádori, Z.; Mészáros, I.; Tuboly, T. Detection of porcine parvovirus 2 (ungulate tetraparvovirus 3) specific antibodies and examination of the serological profile of an infected swine herd. PLoS ONE 2016, 11, e0151036. [Google Scholar] [CrossRef]
- Faustini, G.; Tucciarone, C.M.; Franzo, G.; Donneschi, A.; Boniotti, M.B.; Alborali, G.L.; Drigo, M. Molecular Survey on Porcine Parvoviruses (PPV1-7) and Their Association with Major Pathogens in Reproductive Failure Outbreaks in Northern Italy. Viruses 2024, 16, 157. [Google Scholar] [CrossRef]
- Vargas-Bermudez, D.S.; Diaz, A.; Polo, G.; Mogollon, J.D.; Jaime, J. Infection and Coinfection of Porcine-Selected Viruses (PPV1 to PPV8, PCV2 to PCV4, and PRRSV) in Gilts and Their Associations with Reproductive Performance. Vet. Sci. 2024, 11, 185. [Google Scholar] [CrossRef]
- Ouh, I.-O.; Lee, J.-Y.; Choi, H.; Moon, S.Y.; Song, J.Y.; Hyun, B.-H.; Kwak, D.; Lee, Y.-H.; Park, C.-K. Prevalence of Porcine Parvoviruses 1 to 6 and Porcine Circovirus 3 Infections and of Their Co-infections with Porcine Circovirus 2 in the Republic of Korea. Preprints 2023, 2023051112. [Google Scholar] [CrossRef]
- Mai, J.; Wang, D.; Zou, Y.; Zhang, S.; Meng, C.; Wang, A.; Wang, N. High Co-infection Status of Novel Porcine Parvovirus 7 With Porcine Circovirus 3 in Sows That Experienced Reproductive Failure. Front. Vet. Sci. 2021, 8, 695553. [Google Scholar] [CrossRef] [PubMed]
- Miłek, D.; Woźniak, A.; Guzowska, M.; Stadejek, T. Detection Patterns of Porcine Parvovirus (PPV) and Novel Porcine Parvoviruses 2 through 6 (PPV2-PPV6) in Polish Swine Farms. Viruses 2019, 11, 474. [Google Scholar] [CrossRef] [PubMed]
- Bisimwa, P.N.; Wasso, D.S.; Bantuzeko, F.; Aksanti, C.B.; Tonui, R.; Birindwa, A.B.; Bisimwa, E.B. First investigation on the presence of porcine parvovirus type 3 in domestic pig farms without reproductive failure in the Democratic Republic of Congo. Vet. Anim. Sci. 2021, 13, 100187. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Sharma, S.; Barua, S.; Tripathi, B.N.; Rouse, B.T. Virological and immunological outcomes of coinfections. Clin. Microbiol. Rev. 2018, 31, 1–39. [Google Scholar] [CrossRef] [PubMed]
- Ndze, V.N.; Cadar, D.; Cságola, A.; Kisfali, P.; Kovács, E.; Farkas, S.; Ngu, A.F.; Esona, M.D.; Dán, Á.; Tuboly, T.; et al. Detection of novel porcine bocaviruses in fecal samples of asymptomatic pigs in Cameroon. Infect. Genet. Evol. 2013, 17, 277–282. [Google Scholar] [CrossRef]
- Parsyan, A.; Szmaragd, C.; Allain, J.-P.; Candotti, D. Identification and genetic diversity of two human parvovirus B19 genotype 3 subtypes. J. Gen. Virol. 2007, 88, 428–431. [Google Scholar] [CrossRef]
- Opriessnig, T.; Xiao, C.-T.; Gerber, P.F.; Halbur, P.G. Identification of recently described porcine parvoviruses in archived North American samples from 1996 and association with porcine circovirus associated disease. Vet. Microbiol. 2014, 173, 9–16. [Google Scholar] [CrossRef]
- Chung, H.-C.; Nguyen, V.-G.; Huynh, T.-M.-L.; Park, Y.-H.; Park, K.-T.; Park, B.-K. PCR-based detection and genetic characterization of porcine parvoviruses in Republic of Korea in 2018. BMC Vet. Res. 2020, 16, 113. [Google Scholar] [CrossRef]
- Lukashov, V.V.; Goudsmit, J. Evolutionary relationships among parvoviruses: Virus-host coevolution among autonomous primate parvoviruses and links between adeno-associated and avian parvoviruses. J. Virol. 2001, 75, 2729–2740. [Google Scholar] [CrossRef]
- Wang, D.; Mai, J.; Yang, Y.; Wang, N. Porcine Parvovirus 7: Evolutionary Dynamics and Identification of Epitopes toward Vaccine Design. Vaccines 2020, 8, 359. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, Y.; Sun, H.; Lan, D. Stability of reference gene expression after porcine sapelovirus infection in porcine intestinal epithelial cells. Viral Immunol. 2016, 29, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Pallier, C.; Greco, A.; Le Junter, J.; Saib, A.; Vassias, I.; Morinet, F. The 3’ untranslated region of the B19 parvovirus capsid protein mRNAs inhibits its own mRNA translation in nonpermissive cells. J. Virol. 1997, 71, 9482–9489. [Google Scholar] [CrossRef] [PubMed]
- Thuy, N.T.D.; Trung, N.T.; Dung, T.Q.; Khoa, D.V.A.; Thuy, D.T.N.; Opriessnig, T. First investigation of the prevalence of parvoviruses in slaughterhouse pigs and genomic characterization of ungulate copiparvovirus 2 in Vietnam. Arch. Virol. 2021, 166, 779–788. [Google Scholar] [CrossRef] [PubMed]
- Beltran-Alcrudo, D.; Falco, J.R.; Raizman, E.; Dietze, K. Transboundary spread of pig diseases: The role of international trade and travel. BMC Vet. Res. 2019, 15, 64. [Google Scholar] [CrossRef]
- Opriessnig, T.; Fenaux, M.; Yu, S.; Evans, R.B.; Cavanaugh, D.; Gallup, J.M.; Pallares, F.J.; Thacker, E.L.; Lager, K.M.; Meng, X.J.; et al. Effect of porcine parvovirus vaccination on the development of PMWS in segregated early weaned pigs coinfected with type 2 porcine circovirus and porcine parvovirus. Vet. Microbiol. 2004, 98, 209–220. [Google Scholar] [CrossRef]
- Franzo, G.; Zerbo, H.L.; Ouoba, B.L.; Dji-Tombo, A.D.; Kindo, M.G.; Sawadogo, R.; Chang’a, J.; Bitanyi, S.; Kamigwe, A.; Mayenga, C.; et al. A phylogeographic analysis of porcine parvovirus 1 in africa. Viruses 2023, 15, 207. [Google Scholar] [CrossRef]
- Mészáros, I.; Olasz, F.; Cságola, A.; Tijssen, P.; Zádori, Z. Biology of Porcine Parvovirus (Ungulate parvovirus 1). Viruses 2017, 9, 393. [Google Scholar] [CrossRef] [PubMed]
- Gava, D.; Souza, C.K.; Mores, T.J.; Argenti, L.E.; Streck, A.F.; Canal, C.W.; Bortolozzo, F.P.; Wentz, I. Dynamics of vanishing of maternally derived antibodies of Ungulate protoparvovirus 1 suggests an optimal age for gilts vaccination. Trop. Anim. Health Prod. 2017, 49, 1085–1088. [Google Scholar] [CrossRef]
- Joo, H.S.; Donaldson-Wood, C.R.; Johnson, R.H. Observations on the pathogenesis of porcine parvovirus infection. Arch. Virol. 1976, 51, 123–129. [Google Scholar] [CrossRef]
- Mengeling, W.L.; Lager, K.M.; Vorwald, A.C. The effect of porcine parvovirus and porcine reproductive and respiratory syndrome virus on porcine reproductive performance. Anim. Reprod. Sci. 2000, 60–61, 199–210. [Google Scholar] [CrossRef]
- Choi, C.S.; Molitor, T.W.; Joo, H.S.; Gunther, R. Pathogenicity of a skin isolate of porcine parvovirus in swine fetuses. Vet. Microbiol. 1987, 15, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Joo, H.S.; Donaldson-Wood, C.R.; Johnson, R.H. A standardised haemagglutination inhibition test for porcine parvovirus antibody. Aust. Vet. J. 1976, 52, 422–424. [Google Scholar] [CrossRef] [PubMed]
- Zeeuw, E.J.L.; Leinecker, N.; Herwig, V.; Selbitz, H.J.; Truyen, U. Study of the virulence and cross-neutralization capability of recent porcine parvovirus field isolates and vaccine viruses in experimentally infected pregnant gilts. J. Gen. Virol. 2007, 88, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Streck, A.F.; Canal, C.W.; Truyen, U. Molecular epidemiology and evolution of porcine parvoviruses. Infect. Genet. Evol. 2015, 36, 300–306. [Google Scholar] [CrossRef]
- Streck, A.F.; Bonatto, S.L.; Homeier, T.; Souza, C.K.; Gonçalves, K.R.; Gava, D.; Canal, C.W.; Truyen, U. High rate of viral evolution in the capsid protein of porcine parvovirus. J. Gen. Virol. 2011, 92, 2628–2636. [Google Scholar] [CrossRef]
- Stevenson, M.; Nunes, T.; Heuer, C.; Marshall, J.; Sanchez, J. Tools for the Analysis of Epidemiological Data. In Package EpiR: CRAN. 2017. Available online: http://cran.nexr.com/web/packages/epiR/epiR.pdf (accessed on 23 April 2024).
- Serena, M.S.; Cappuccio, J.A.; Metz, G.E.; Aspitia, C.G.; Dibárbora, M.; Calderón, M.G.; Echeverría, M.G. Detection and molecular characterization of porcine parvovirus in fetal tissues from sows without reproductive failure in Argentina. Heliyon 2019, 5, e02874. [Google Scholar] [CrossRef]
- Huang, L.; Zhai, S.-L.; Cheung, A.K.; Zhang, H.-B.; Long, J.-X.; Yuan, S.-S. Detection of a novel porcine parvovirus, PPV4, in Chinese swine herds. Virol. J. 2010, 7, 333. [Google Scholar] [CrossRef]
- Xiao, C.-T.; Giménez-Lirola, L.G.; Jiang, Y.-H.; Halbur, P.G.; Opriessnig, T. Characterization of a novel porcine parvovirus tentatively designated PPV5. PLoS ONE 2013, 8, e65312. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, bbx108. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Lam, T.T.; Max Carvalho, L.; Pybus, O.G. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2016, 2, vew007. [Google Scholar] [CrossRef] [PubMed]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef] [PubMed]
- Baele, G.; Li, W.L.S.; Drummond, A.J.; Suchard, M.A.; Lemey, P. Accurate model selection of relaxed molecular clocks in bayesian phylogenetics. Mol. Biol. Evol. 2013, 30, 239–243. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A.; Shapiro, B.; Pybus, O.G. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 2005, 22, 1185–1192. [Google Scholar] [CrossRef]
- Drummond, A.J.; Ho, S.Y.W.; Phillips, M.J.; Rambaut, A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006, 4, e88. [Google Scholar] [CrossRef]
- Gill, M.S.; Lemey, P.; Faria, N.R.; Rambaut, A.; Shapiro, B.; Suchard, M.A. Improving Bayesian population dynamics inference: A coalescent-based model for multiple loci. Mol. Biol. Evol. 2013, 30, 713–724. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Yu, G.; Lam, T.T.-Y.; Zhu, H.; Guan, Y. Two methods for mapping and visualizing associated data on phylogeny using ggtree. Mol. Biol. Evol. 2018, 35, 3041–3043. [Google Scholar] [CrossRef]
- Yu, G. Using ggtree to Visualize Data on Tree-Like Structures. Curr. Protoc. Bioinform. 2020, 69, e96. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Bruen, T.C.; Philippe, H.; Bryant, D. A simple and robust statistical test for detecting the presence of recombination. Genetics 2006, 172, 2665–2681. [Google Scholar] [CrossRef] [PubMed]


| PPVs | Reporting Year | Latest Proposed Classification |
|---|---|---|
| PPV1 | 1969 [53] | PPV1a to PPV1d [8] |
| PPV2 | 2001 [9] | Clades A and B [18,19] |
| PPV3 | 2008 [20] | Group 1, 2, and 3 [25] |
| PPV4 | 2010 [27] | Group 1 and 2 [11,19,25] |
| PPV5 | 2013 [33] | Group A and B [36] |
| PPV6 | 2014 [37] | Clades I to III [29] |
| PPV7 | 2016 [41] | Clades a to f [45] |
| PPV8 | 2022 [54] | Not established |
| Virus | % Positive Herds (Positive/All Tested) | % Positive Gilt Sera (Positive/All Tested) |
|---|---|---|
| PPV1 | 17.5 (7/40) | 14.5 (34/234) |
| PPV2 | 22.5 (9/40) | 9.8 (23/234) |
| PPV3 | 67.5 (27/40) | 40.1 (94/234) |
| PPV4 | 15 (6/40) | 4.2 (10/234) |
| PPV5 | 40 (16/40) | 20.5 (48/234) |
| PPV6 | 32.5 (13/40) | 17 (40/234) |
| PPV7 | 5 (2/40) | 1.28 (3/234) |
| PPV8 | 0 (0/40) | 0 (0/234) |
| PPVs | PPV1 | PPV2 | PPV3 | PPV4 | PPV5 | PPV6 | PPV7 |
|---|---|---|---|---|---|---|---|
| PPV1 | 17 | ||||||
| PPV2 | 1 | 10 | |||||
| PPV3 | 4 | 7 | 36 | ||||
| PPV4 | 0 | 0 | 1 | 1 | |||
| PPV5 | 2 | 1 | 14 | 0 | 22 | ||
| PPV6 | 1 | 0 | 16 | 1 | 1 | 11 | |
| PPV7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Study from Germany [5] | Study from Romania, Hungary, and Serbia [6] | Study from China [7] | Study from Belgium and Denmark [8] | This Study from Colombia 2024 | |
|---|---|---|---|---|---|
| Classification | Cluster I and II | Cluster A to F | Four lineages: 1–4 | Four genotypes (PPV1a-PPV1d) | Two clades and two subclades in each clade |
| Region of PPV1 analyzed | VP1 gene (1740 nt) | VP1/VP2 gene (2190 nt) | VP1/VP2 gene (2190) | VP1/2/3 (2190nt) | VP1/2/3 gene (2190 nt) |
| Number of sequences analyzed | 15 sequences | 50 sequences | 75 sequences | 170 sequences | 96 sequences |
| Nucleotide substitution rates | Unrealized | 2.74 × 10−4 | 3.27 × 10−5 | 4.71 × 10−5 | 1.34 × 10−4 |
| PPV1-IDT DEU 1964 (AY684872) | Cluster I | Cluster A | Lineage 2 | PPV1a | Clade I Subclade II |
| PPV1-NALD2 USA (NC001718) | Cluster II | Cluster F | Lineage 3 | PPV1c | Clade I Subclade I |
| PPV1 NADL8 USA (ON014603) | Cluster II | Cluster C-E | Lineage 1, 3, and 4 | PPV1d | Clade I Subclade II |
| PPV1 27a like (AY684871) | Cluster II | B | Lineage 1 | PPV1b | Clade II Subclade I |
| PPV1 Colombian (PP921711) | Cluster II | B | Lineage 1 | PPV1b | Clade II Subclade II |
| nPPV | Genetic Identity | %NCG * Identity | % NS # Identity | % VP ¶ Identity | ||
|---|---|---|---|---|---|---|
| nt § | aa ¤ | nt | aa | |||
| PPV1 | Within Colombian sequences Within worldwide sequences | Unrealized | Unrealized | 99.8 96.4–100 | 100 95.2–100 | |
| PPV2 | Within Colombian sequences | 97.9–99.7 | 97.8–99.7 | 97.5–99.7 | 97.9–99 | 98–99.7 |
| Within worldwide sequences | 93.5–98.9 | 93.3–99.1 | 95.5–98.6 | 92.5–99.2 | 94.9–99.1 | |
| PPV3 | Within Colombian sequences | 97.7–99.8 | 97–99.8 | 98.1–100 | 98.1–99.9 | 99.2–99.9 |
| Within worldwide sequences | 97.3–99.4 | 96.2–99.2 | 96–100 | 97.6–99.4 | 98.6–100 | |
| PPV4 | Within Colombian sequences | 98.9–99.8 | 99.2–99.9 | 99.3–99.8 | 98.7–99.9 | 98.7–100 |
| Within worldwide sequences | 96.1–99.6 | 96.3–99.9 | 97.2–100 | 94.5–100 | 97.9–100 | |
| PPV5 | Within Colombian sequences | 98–99.8 | 98.9–100 | 99.7–100 | 98.9–99.8 | 97.6–99.4 |
| Within worldwide sequences | 97.8–99.9 | 97.1–100 | 97.6–100 | 97.8–99.8 | 96.3–99.8 | |
| PPV6 | Within Colombian sequences | 98.7–99.7 | 99.7–99.9 | 99.5–99.8 | 98.5–99.7 | 99.3–99.9 |
| Within worldwide sequences | 96.6–99.4 | 98.9–99.9 | 99.1–99.8 | 95.9–99.2 | 96.5–99.3 | |
| PPV7 | Within Colombian sequences | 95.4–97.4 | 95–98.2 | 94.3–98.6 | 92.4–98.2 | 91.7–98.9 |
| Within worldwide sequences | 93.5–99.8 | 93.8–99.9 | 91.6–99.7 | 90.4–99.6 | 87.8–99.8 | |
| Period Range of Sequences | Mean Substitution Rate | HPD Substitution Rate | TMRCA (Years) | |
|---|---|---|---|---|
| PPV1 | 1963–2021 | 1.34 × 10−4 | 5.30 × 10−5–1.08 × 10−4 | 78 |
| PPV2 | 1996–2021 | 5.74 × 10−4 | 4.74 × 10−4–6.71 × 10−4 | 40 |
| PPV3 | 1999–2021 | 5.57 × 10−4 | 3.63 × 10−4–6.50 × 10−4 | 41 |
| PPV4 | 2007–2021 | 3.03 × 10−4 | 1.11 × 10−4–3.73 × 10−4 | 27 |
| PPV5 | 1999–2021 | 2.88 × 10−4 | 1.14 × 10−4–3.48 × 10−4 | 35 |
| PPV6 | 1998–2021 | 4.22 × 10−4 | 2.18 × 10−4–5.36 × 10−4 | 56 |
| PPV7 | 1997–2021 | 3.90 × 10−3 | 2.81 × 10−3–4.97 × 10−3 | 33 |
| Study from Romania (Cadar et al., 2013) [18] | Study from China (Sun et al., 2015) [19] | Study from Europe (Tregaskis et al., 2021) [25] | Study from Korea (Kim et al., 2022) [29] | Study from China (Chen et al., 2023) [36] | Current study from Colombia (2024) Statistical Support Values (≥%) | |
|---|---|---|---|---|---|---|
| PPV2 | Clusters A and B 2 subclusters in A 2 subclusters in B 15 sequences analyzed | Clusters A and B 2 subclusters in A 2 subclusters in B 12 sequences analyzed | Clades 1 and 2 114 sequences analyzed (*partial variable region) | Clades 1 and 2 20 sequences analyzed | Clusters A and B (15 sequences) | Clades I (≥81) and II (≥70) 4 subclades in clade II (I ≥ 73, II ≥ 70, III ≥ 79, and IV ≥75) 206 sequences analyzed |
| PPV3 | Clusters A, B, C, and D 2 subclusters in B 2 subclusters in C 3 subclusters in D 38 sequences analyzed | Cluster A and B 2 subclusters in B 2 subclusters in C 3 subclusters in D 14 sequences analyzed | Clades 1, 2, and 3 69 sequences | Not divided into clusters 30 sequences analyzed | Clusters A and B (19 sequences) | Clades I (≥74) and II (≥70) 5 subclades in clade II (I ≥ 86, II ≥ 70, III ≥ 76, IV ≥ 73, and V ≥ 71) 122 sequences analyzed |
| PPV4 | Clusters A and B 20 sequences analyzed | Clusters A and B 11 sequences analyzed | Clades 1 and 2 49 sequences | Not divided into clusters 18 sequences analyzed | No data | Clades I (≥78), II (≥78), and III (≥73) 2 subclades in clade II (I ≥ 78 and II ≥ 80) 2 subclades in clade III (I ≥ 73 and II ≥ 82) 87 sequences analyzed |
| PPV5 | No data | No data | No data | Not divided into clusters 17 sequences analyzed | Clusters A and B (8 sequences) | Clades I (≥80), II (≥71), and III (≥72) 2 subclades in clade II (I ≥ 72 and II ≥ 77) 2 subclades in clade III (I ≥ 77 and II ≥ 72) 100 sequences analyzed |
| PPV6 | No data | No data | No data | Clades 1, 2, and 3 27 sequences analyzed | No data | Clades I (≥86) and II (≥72) 2 subclades in clade I (I ≥ 90 and II ≥ 86) 3 subclades in clade II (I ≥ 72, II ≥ 73, and III ≥ 72) 130 sequences analyzed |
| PPV7 | No data | No data | No data | Clades 1, 2, and 3 21 sequences analyzed | Clades I (≥72) and II (≥70) 2 subclades in clade I (I ≥ 65 and II ≥ 72) 2 subclades in clade II (I ≥ 71 and II ≥ 70) 185 sequences analyzed |
| Virus | Primer Name | Sequence | Annealing Temp | Bp Size | Reference |
|---|---|---|---|---|---|
| PPV1 | PPV1F | CTTGGAGCCGTGGAGCGAGC | 60 | 147 | [87] |
| PPV1 | PPV1R | TGCACAGTTTTCACCAAAGCAGGC | |||
| PPV2 | PPV-2-F | AGCTCTGCGACAAGTGGG | 58 | 563 | [56] |
| PPV2 | PPV-2-R | GTCTACGGCCTGCAAGAA | |||
| PPV3 | PPV-3-F | CCACGCCAAATCAAAGTC | 58 | 514 | |
| PPV3 | PPV-3-R | CTCCCACTCCCATCCACT | |||
| PPV4 | PPV-4-F | TATGTGGGCTGGGCAAGGAATGTC | 58 | 416 | |
| PPV4 | PPV-4-R | GTTGCGGAATGCTATCAGGCTCTT | |||
| PPV5 | PPV-5-F | ACACCTCCTGCGGCTTAT | 58 | 959 | |
| PPV5 | PPV-5-R | GTGTAGCGATGTCCTGGC | |||
| PPV6 | PPV-6-F | CTTTGGTGTAGAGGGCTTGA | 58 | 650 | |
| PPV6 | PPV-6-R | CGGTTGTAGCAGGTCCAA | |||
| PPV7 | PPV7F | CCTCCATCAGCAGCGACCAGT | 58 | 241 | [42] |
| PPV7 | PPV7R | ACCAGGGTTCCGTTTTCGTCT | |||
| PPV8 | PPV8-outF | TGTTGGTTTGCACCTAGCG | 58 | [46] | |
| PPV8 | PPV8-outR | TGATGAGATGGTGGAACGC | |||
| PPV8 | PPV8-inF | TCCAAGTTGCCCTAGACAGC | 58 | 554 | |
| PPV8 | PPV8-inR | GCCTCGTACATGTGGACCTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vargas-Bermudez, D.S.; Prandi, B.A.; Souza, U.J.B.d.; Durães-Carvalho, R.; Mogollón, J.D.; Campos, F.S.; Roehe, P.M.; Jaime, J. Molecular Epidemiology and Phyloevolutionary Analysis of Porcine Parvoviruses (PPV1 through PPV7) Detected in Replacement Gilts from Colombia. Int. J. Mol. Sci. 2024, 25, 10354. https://doi.org/10.3390/ijms251910354
Vargas-Bermudez DS, Prandi BA, Souza UJBd, Durães-Carvalho R, Mogollón JD, Campos FS, Roehe PM, Jaime J. Molecular Epidemiology and Phyloevolutionary Analysis of Porcine Parvoviruses (PPV1 through PPV7) Detected in Replacement Gilts from Colombia. International Journal of Molecular Sciences. 2024; 25(19):10354. https://doi.org/10.3390/ijms251910354
Chicago/Turabian StyleVargas-Bermudez, Diana S., Bruno Aschidamini Prandi, Ueric José Borges de Souza, Ricardo Durães-Carvalho, José Darío Mogollón, Fabrício Souza Campos, Paulo Michel Roehe, and Jairo Jaime. 2024. "Molecular Epidemiology and Phyloevolutionary Analysis of Porcine Parvoviruses (PPV1 through PPV7) Detected in Replacement Gilts from Colombia" International Journal of Molecular Sciences 25, no. 19: 10354. https://doi.org/10.3390/ijms251910354
APA StyleVargas-Bermudez, D. S., Prandi, B. A., Souza, U. J. B. d., Durães-Carvalho, R., Mogollón, J. D., Campos, F. S., Roehe, P. M., & Jaime, J. (2024). Molecular Epidemiology and Phyloevolutionary Analysis of Porcine Parvoviruses (PPV1 through PPV7) Detected in Replacement Gilts from Colombia. International Journal of Molecular Sciences, 25(19), 10354. https://doi.org/10.3390/ijms251910354








