Selective Effect of DNA N6-Methyladenosine Modification on Transcriptional Genetic Variations in East Asian Samples
Abstract
:1. Introduction
2. Results
2.1. Identification of DNA 6mA Modification in East Asian Samples
2.2. Effect of 6mA Modification on DNA Variations
2.3. Effect of 6mA Modification on RNA Variations
2.4. Selective Effect of 6mA Modification on Transcriptional Variations from DNA to RNA
2.5. Validation of 6mA Methylation Effect on Variations in Imprinting Genes
2.6. Relationship between 6mA Modification and Variations in Coding and Regulated Regions
3. Discussion
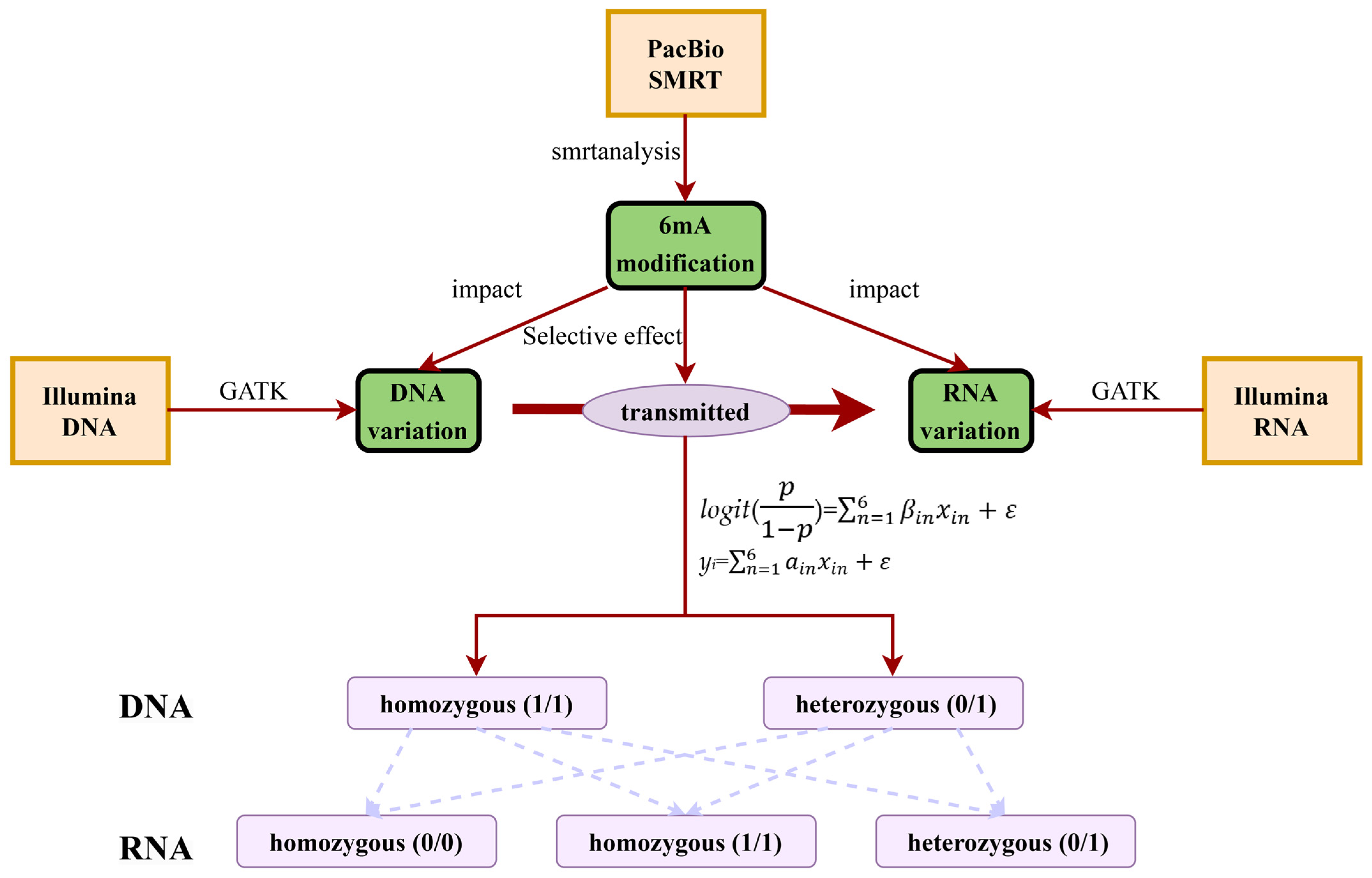
4. Materials and Methods
4.1. DNA Methylation Samples
4.2. Identification of 6mA Modifications in Genomic DNA
4.3. DNA and RNA Genetic Variation Samples
4.4. DNA and RNA Datasets for Genetic Variation Analysis
4.5. Statistical Analysis of Dynamic Transcriptional Genetic Variations
4.6. 6mA Methylation in Imprinting Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 6mA | DNA N6-methyladenosine |
| 5mC | 5-methylcytosine |
| SMRT | single-molecule, real-time sequencing |
| IPD | inter-pulse duration |
| KRT5 | keratin 5 |
| IDH2 | isocitrate dehydrogenase (NADP(+)) 2 |
| SRSF2 | serine- and arginine-rich splicing factor 2 |
| GPR15 | G protein-coupled receptor 15 |
| IGF2 | insulin-like growth factor 2 |
| SNRPN | small nuclear ribonucleoprotein polypeptide N |
| LC-MS/MS | liquid chromatography–tandem mass spectrometry |
| N6AMT1 | N-6 adenine-specific DNA methyltransferase 1 |
| ALKBH1 | alkB homolog 1, histone H2A dioxygenase |
| 6mACE-seq | 6mA cross-linking exonuclease sequencing |
| 6mA-IP-Seq | 6mA immunoprecipitation sequencing |
| SLE | systemic lupus erythematosus |
References
- Ebert, P.; Audano, P.A.; Zhu, Q.; Rodriguez-Martin, B.; Porubsky, D.; Bonder, M.J.; Sulovari, A.; Ebler, J.; Zhou, W.; Serra Mari, R.; et al. Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science 2021, 372, abf7117. [Google Scholar] [CrossRef] [PubMed]
- Sudmant, P.H.; Mallick, S.; Nelson, B.J.; Hormozdiari, F.; Krumm, N.; Huddleston, J.; Coe, B.P.; Baker, C.; Nordenfelt, S.; Bamshad, M.; et al. Global diversity, population stratification, and selection of human copy-number variation. Science 2015, 349, aab3761. [Google Scholar] [CrossRef]
- Moreno-Estrada, A.; Gignoux, C.R.; Fernandez-Lopez, J.C.; Zakharia, F.; Sikora, M.; Contreras, A.V.; Acuna-Alonzo, V.; Sandoval, K.; Eng, C.; Romero-Hidalgo, S.; et al. Human genetics. The genetics of Mexico recapitulates Native American substructure and affects biomedical traits. Science 2014, 344, 1280–1285. [Google Scholar] [CrossRef]
- Hill, M.S.; Vande Zande, P.; Wittkopp, P.J. Molecular and evolutionary processes generating variation in gene expression. Nat. Rev. Genet. 2021, 22, 203–215. [Google Scholar] [CrossRef]
- Ellegren, H.; Galtier, N. Determinants of genetic diversity. Nat. Rev. Genet. 2016, 17, 422–433. [Google Scholar] [CrossRef]
- Claussnitzer, M.; Cho, J.H.; Collins, R.; Cox, N.J.; Dermitzakis, E.T.; Hurles, M.E.; Kathiresan, S.; Kenny, E.E.; Lindgren, C.M.; MacArthur, D.G.; et al. A brief history of human disease genetics. Nature 2020, 577, 179–189. [Google Scholar] [CrossRef]
- Povysil, G.; Petrovski, S.; Hostyk, J.; Aggarwal, V.; Allen, A.S.; Goldstein, D.B. Rare-variant collapsing analyses for complex traits: Guidelines and applications. Nat. Rev. Genet. 2019, 20, 747–759. [Google Scholar] [CrossRef] [PubMed]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.; Lv, J.; Yang, M.; Wang, M.; Zhu, M.; Wang, T.; Yan, C.; Yu, C.; Ding, Y.; Li, G.; et al. Genetic risk, incident gastric cancer, and healthy lifestyle: A meta-analysis of genome-wide association studies and prospective cohort study. Lancet Oncol. 2020, 21, 1378–1386. [Google Scholar] [CrossRef] [PubMed]
- Oliva, M.; Munoz-Aguirre, M.; Kim-Hellmuth, S.; Wucher, V.; Gewirtz, A.D.H.; Cotter, D.J.; Parsana, P.; Kasela, S.; Balliu, B.; Vinuela, A.; et al. The impact of sex on gene expression across human tissues. Science 2020, 369, eaba3066. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, H.; Jiang, Z.; Wang, W.; Xu, R.; Wang, Q.; Zhang, Z.; Li, A.; Liang, Y.; Ou, S.; et al. Genomic basis of geographical adaptation to soil nitrogen in rice. Nature 2021, 590, 600–605. [Google Scholar] [CrossRef] [PubMed]
- Fournier-Level, A.; Korte, A.; Cooper, M.D.; Nordborg, M.; Schmitt, J.; Wilczek, A.M. A map of local adaptation in Arabidopsis thaliana. Science 2011, 334, 86–89. [Google Scholar] [CrossRef]
- Gusev, A.; Ko, A.; Shi, H.; Bhatia, G.; Chung, W.; Penninx, B.W.; Jansen, R.; de Geus, E.J.; Boomsma, D.I.; Wright, F.A.; et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016, 48, 245–252. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Tang, X.; Baheti, S.; Shameer, K.; Thompson, K.J.; Wills, Q.; Niu, N.; Holcomb, I.N.; Boutet, S.C.; Ramakrishnan, R.; Kachergus, J.M.; et al. The eSNV-detect: A computational system to identify expressed single nucleotide variants from transcriptome sequencing data. Nucleic Acids Res. 2014, 42, e172. [Google Scholar] [CrossRef] [PubMed]
- Brouard, J.S.; Schenkel, F.; Marete, A.; Bissonnette, N. The GATK joint genotyping workflow is appropriate for calling variants in RNA-seq experiments. J. Anim. Sci. Biotechnol. 2019, 10, 44. [Google Scholar] [CrossRef]
- Sahraeian, S.M.E.; Mohiyuddin, M.; Sebra, R.; Tilgner, H.; Afshar, P.T.; Au, K.F.; Bani Asadi, N.; Gerstein, M.B.; Wong, W.H.; Snyder, M.P.; et al. Gaining comprehensive biological insight into the transcriptome by performing a broad-spectrum RNA-seq analysis. Nat. Commun. 2017, 8, 59. [Google Scholar] [CrossRef]
- Saeidian, A.H.; Youssefian, L.; Vahidnezhad, H.; Uitto, J. Research Techniques Made Simple: Whole-Transcriptome Sequencing by RNA-Seq for Diagnosis of Monogenic Disorders. J. Investig. Dermatol. 2020, 140, 1117–1126.e1. [Google Scholar] [CrossRef]
- Yousefi, S.; Abbassi-Daloii, T.; Kraaijenbrink, T.; Vermaat, M.; Mei, H.; van ‘t Hof, P.; van Iterson, M.; Zhernakova, D.V.; Claringbould, A.; Franke, L.; et al. A SNP panel for identification of DNA and RNA specimens. BMC Genom. 2018, 19, 90. [Google Scholar] [CrossRef]
- Li, J.; Liu, C. Coding or Noncoding, the Converging Concepts of RNAs. Front. Genet. 2019, 10, 496. [Google Scholar] [CrossRef]
- Krasilnikova, M.M.; Kireeva, M.L.; Petrovic, V.; Knijnikova, N.; Kashlev, M.; Mirkin, S.M. Effects of Friedreich’s ataxia (GAA)n*(TTC)n repeats on RNA synthesis and stability. Nucleic Acids Res. 2007, 35, 1075–1084. [Google Scholar] [CrossRef] [PubMed]
- Eddy, J.; Maizels, N. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008, 36, 1321–1333. [Google Scholar] [CrossRef]
- Li, Q.; Seo, J.H.; Stranger, B.; McKenna, A.; Pe’er, I.; Laframboise, T.; Brown, M.; Tyekucheva, S.; Freedman, M.L. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell 2013, 152, 633–641. [Google Scholar] [CrossRef]
- Montgomery, S.B.; Dermitzakis, E.T. From expression QTLs to personalized transcriptomics. Nat. Rev. Genet. 2011, 12, 277–282. [Google Scholar] [CrossRef] [PubMed]
- St Hilaire, C.; Ziegler, S.G.; Markello, T.C.; Brusco, A.; Groden, C.; Gill, F.; Carlson-Donohoe, H.; Lederman, R.J.; Chen, M.Y.; Yang, D.; et al. NT5E mutations and arterial calcifications. N. Engl. J. Med. 2011, 364, 432–442. [Google Scholar] [CrossRef] [PubMed]
- Bulaj, Z.J.; Griffen, L.M.; Jorde, L.B.; Edwards, C.Q.; Kushner, J.P. Clinical and biochemical abnormalities in people heterozygous for hemochromatosis. N. Engl. J. Med. 1996, 335, 1799–1805. [Google Scholar] [CrossRef]
- Morin, T.; Martin, J.P.; Feldmann, G.; Rueff, B.; Benhamou, J.P.; Ropartz, C. Heterozygous alpha 1-antitrypsin deficiency and cirrhosis in adults, a fortuitous association. Lancet 1975, 1, 250–251. [Google Scholar] [CrossRef] [PubMed]
- Qu, H.Q.; Lawrence, S.G.; Guo, F.; Majewski, J.; Polychronakos, C. Strand bias in complementary single-nucleotide polymorphisms of transcribed human sequences: Evidence for functional effects of synonymous polymorphisms. BMC Genom. 2006, 7, 213. [Google Scholar] [CrossRef]
- Green, P.; Ewing, B.; Miller, W.; Thomas, P.J.; Program, N.C.S.; Green, E.D. Transcription-associated mutational asymmetry in mammalian evolution. Nat. Genet. 2003, 33, 514–517. [Google Scholar] [CrossRef]
- Polak, P.; Arndt, P.F. Transcription induces strand-specific mutations at the 5’ end of human genes. Genome Res. 2008, 18, 1216–1223. [Google Scholar] [CrossRef]
- Manning, K.S.; Cooper, T.A. The roles of RNA processing in translating genotype to phenotype. Nat. Rev. Mol. Cell Biol. 2017, 18, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Yoshimi, A.; Lin, K.T.; Wiseman, D.H.; Rahman, M.A.; Pastore, A.; Wang, B.; Lee, S.C.; Micol, J.B.; Zhang, X.J.; de Botton, S.; et al. Coordinated alterations in RNA splicing and epigenetic regulation drive leukaemogenesis. Nature 2019, 574, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Koks, G.; Uudelepp, M.L.; Limbach, M.; Peterson, P.; Reimann, E.; Koks, S. Smoking-induced expression of the GPR15 gene indicates its potential role in chronic inflammatory pathologies. Am. J. Pathol. 2015, 185, 2898–2906. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.R.; Busche, S.; Ge, B.; Kwan, T.; Pastinen, T.; Blanchette, M. The relationship between DNA methylation, genetic and expression inter-individual variation in untransformed human fibroblasts. Genome Biol. 2014, 15, R37. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Arcelus, M.; Lappalainen, T.; Montgomery, S.B.; Buil, A.; Ongen, H.; Yurovsky, A.; Bryois, J.; Giger, T.; Romano, L.; Planchon, A.; et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. eLife 2013, 2, e00523. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.T.; Pai, A.A.; Pickrell, J.K.; Gaffney, D.J.; Pique-Regi, R.; Degner, J.F.; Gilad, Y.; Pritchard, J.K. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011, 12, R10. [Google Scholar] [CrossRef]
- Luan, M.W.; Chen, W.; Xing, J.F.; Xiao, C.L.; Chen, Y.; Xie, S.Q. DNA N6-Methyladenosine modification role in transmitted variations from genomic DNA to RNA in Herrania umbratica. BMC Genom. 2019, 20, 508. [Google Scholar] [CrossRef] [PubMed]
- Metsalu, T.; Viltrop, T.; Tiirats, A.; Rajashekar, B.; Reimann, E.; Koks, S.; Rull, K.; Milani, L.; Acharya, G.; Basnet, P.; et al. Using RNA sequencing for identifying gene imprinting and random monoallelic expression in human placenta. Epigenetics 2014, 9, 1397–1409. [Google Scholar] [CrossRef]
- Vanyushin, B.F.; Belozersky, A.N.; Kokurina, N.A.; Kadirova, D.X. 5-methylcytosine and 6-methylamino-purine in bacterial DNA. Nature 1968, 218, 1066–1067. [Google Scholar] [CrossRef]
- Liang, Z.; Shen, L.; Cui, X.; Bao, S.; Geng, Y.; Yu, G.; Liang, F.; Xie, S.; Lu, T.; Gu, X.; et al. DNA N6-Adenine Methylation in Arabidopsis thaliana. Dev. Cell 2018, 45, 406–416.e3. [Google Scholar] [CrossRef]
- Vanyushin, B.F.; Tkacheva, S.G.; Belozersky, A.N. Rare bases in animal DNA. Nature 1970, 225, 948–949. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Cokus, S.J.; Zhang, X.; Chen, P.Y.; Bostick, M.; Goll, M.G.; Hetzel, J.; Jain, J.; Strauss, S.H.; Halpern, M.E.; et al. Conservation and divergence of methylation patterning in plants and animals. Proc. Natl. Acad. Sci. USA 2010, 107, 8689–8694. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Morgunova, E.; Jolma, A.; Kaasinen, E.; Sahu, B.; Khund-Sayeed, S.; Das, P.K.; Kivioja, T.; Dave, K.; Zhong, F.; et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 2017, 356, aaj2239. [Google Scholar] [CrossRef]
- Zhang, D.; Cheng, L.; Badner, J.A.; Chen, C.; Chen, Q.; Luo, W.; Craig, D.W.; Redman, M.; Gershon, E.S.; Liu, C. Genetic control of individual differences in gene-specific methylation in human brain. Am. J. Hum. Genet. 2010, 86, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Luo, G.Z.; Chen, K.; Deng, X.; Yu, M.; Han, D.; Hao, Z.; Liu, J.; Lu, X.; Dore, L.C.; et al. N6-methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell 2015, 161, 879–892. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.P.; Wang, T.; Seetin, M.G.; Lai, Y.; Zhu, S.; Lin, K.; Liu, Y.; Byrum, S.D.; Mackintosh, S.G.; Zhong, M.; et al. DNA methylation on N6-adenine in mammalian embryonic stem cells. Nature 2016, 532, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Liu, Y.; Li, X.; Zou, J.; Zou, S. DNA N6-methyladenine demethylase ALKBH1 enhances osteogenic differentiation of human MSCs. Bone Res. 2016, 4, 16033. [Google Scholar] [CrossRef] [PubMed]
- Mondo, S.J.; Dannebaum, R.O.; Kuo, R.C.; Louie, K.B.; Bewick, A.J.; LaButti, K.; Haridas, S.; Kuo, A.; Salamov, A.; Ahrendt, S.R.; et al. Widespread adenine N6-methylation of active genes in fungi. Nat. Genet. 2017, 49, 964–968. [Google Scholar] [CrossRef]
- Luo, G.Z.; Blanco, M.A.; Greer, E.L.; He, C.; Shi, Y. DNA N6-methyladenine: A new epigenetic mark in eukaryotes? Nat. Rev. Mol. Cell Biol. 2015, 16, 705–710. [Google Scholar] [CrossRef]
- Xiao, C.L.; Zhu, S.; He, M.; Chen, D.; Zhang, Q.; Chen, Y.; Yu, G.; Liu, J.; Xie, S.Q.; Luo, F.; et al. N6-Methyladenine DNA Modification in the Human Genome. Mol. Cell 2018, 71, 306–318.e7. [Google Scholar] [CrossRef]
- Yao, B.; Cheng, Y.; Wang, Z.; Li, Y.; Chen, L.; Huang, L.; Zhang, W.; Chen, D.; Wu, H.; Tang, B.; et al. DNA N6-methyladenine is dynamically regulated in the mouse brain following environmental stress. Nat. Commun. 2017, 8, 1122. [Google Scholar] [CrossRef]
- Zhang, G.; Huang, H.; Liu, D.; Cheng, Y.; Liu, X.; Zhang, W.; Yin, R.; Zhang, D.; Zhang, P.; Liu, J.; et al. N6-methyladenine DNA modification in Drosophila. Cell 2015, 161, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Blanco, M.A.; Gu, L.; Sendinc, E.; Liu, J.; Aristizabal-Corrales, D.; Hsu, C.H.; Aravind, L.; He, C.; Shi, Y. DNA Methylation on N6-Adenine in C. elegans. Cell 2015, 161, 868–878. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Wu, T.P.; Gimple, R.C.; Li, Z.; Prager, B.C.; Wu, Q.; Yu, Y.; Wang, P.; Wang, Y.; Gorkin, D.U.; et al. N6-Methyladenine DNA Modification in Glioblastoma. Cell 2018, 175, 1228–1243.e20. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Beaulaurier, J.; Deikus, G.; Wu, T.P.; Strahl, M.; Hao, Z.; Luo, G.; Gregory, J.A.; Chess, A.; He, C.; et al. Mapping and characterizing N6-methyladenine in eukaryotic genomes using single-molecule real-time sequencing. Genome Res. 2018, 28, 1067–1078. [Google Scholar] [CrossRef]
- Tian, L.F.; Liu, Y.P.; Chen, L.; Tang, Q.; Wu, W.; Sun, W.; Chen, Z.; Yan, X.X. Structural basis of nucleic acid recognition and 6mA demethylation by human ALKBH1. Cell Res. 2020, 30, 272–275. [Google Scholar] [CrossRef]
- Goh, W.S.S. Single-Nucleotide-Resolution Sequencing of N6-Methyldeoxyadenosine. Methods Mol. Biol. 2021, 2198, 369–377. [Google Scholar] [CrossRef]
- Flusberg, B.A.; Webster, D.R.; Lee, J.H.; Travers, K.J.; Olivares, E.C.; Clark, T.A.; Korlach, J.; Turner, S.W. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods 2010, 7, 461–465. [Google Scholar] [CrossRef]
- Zhou, C.; Wang, C.; Liu, H.; Zhou, Q.; Liu, Q.; Guo, Y.; Peng, T.; Song, J.; Zhang, J.; Chen, L.; et al. Identification and analysis of adenine N6-methylation sites in the rice genome. Nat. Plants 2018, 4, 554–563. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, Y.; Luo, G.Z.; Wang, X.; Yue, Y.; Wang, X.; Zong, X.; Chen, K.; Yin, H.; Fu, Y.; et al. Abundant DNA 6mA methylation during early embryogenesis of zebrafish and pig. Nat. Commun. 2016, 7, 13052. [Google Scholar] [CrossRef]
- Guo, Y.; Pei, Y.; Li, K.; Cui, W.; Zhang, D. DNA N6-methyladenine modification in hypertension. Aging 2020, 12, 6276–6291. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, M.; Guo, M. DNA N6-methyladenine increased in human esophageal squamous cell carcinoma. Discov. Med. 2020, 29, 85–90. [Google Scholar] [PubMed]
- Ma, C.; Niu, R.; Huang, T.; Shao, L.W.; Peng, Y.; Ding, W.; Wang, Y.; Jia, G.; He, C.; Li, C.Y.; et al. N6-methyldeoxyadenine is a transgenerational epigenetic signal for mitochondrial stress adaptation. Nat. Cell Biol. 2019, 21, 319–327. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Cobb, M. 60 years ago, Francis Crick changed the logic of biology. PLoS Biol. 2017, 15, e2003243. [Google Scholar] [CrossRef] [PubMed]
- Douvlataniotis, K.; Bensberg, M.; Lentini, A.; Gylemo, B.; Nestor, C.E. No evidence for DNA N6-methyladenine in mammals. Sci. Adv. 2020, 6, eaay3335. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, S.; Nelakanti, R.V.; Lin, K.; Wu, T.P.; Alderman, M.H., 3rd; Guo, C.; Wang, P.; Zhang, M.; Min, W.; et al. N6-Methyladenine in DNA antagonizes SATB1 in early development. Nature 2020, 583, 625–630. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.J. The epigenetic roles of DNA N6-Methyladenine (6mA) modification in eukaryotes. Cancer Lett. 2020, 494, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Karanthamalai, J.; Chodon, A.; Chauhan, S.; Pandi, G. DNA N6-Methyladenine Modification in Plant Genomes-A Glimpse into Emerging Epigenetic Code. Plants 2020, 9, 247. [Google Scholar] [CrossRef]
- Zhang, Q.; Liang, Z.; Cui, X.; Ji, C.; Li, Y.; Zhang, P.; Liu, J.; Riaz, A.; Yao, P.; Liu, M.; et al. N6-Methyladenine DNA Methylation in Japonica and Indica Rice Genomes and Its Association with Gene Expression, Plant Development, and Stress Responses. Mol. Plant 2018, 11, 1492–1508. [Google Scholar] [CrossRef]
- Iyer, L.M.; Zhang, D.; Aravind, L. Adenine methylation in eukaryotes: Apprehending the complex evolutionary history and functional potential of an epigenetic modification. Bioessays 2016, 38, 27–40. [Google Scholar] [CrossRef]
- Zheng, F.; Tang, D.; Xu, H.; Xu, Y.; Dai, W.; Zhang, X.; Hong, X.; Liu, D.; Dai, Y. Genomewide analysis of 6-methyladenine DNA in peripheral blood mononuclear cells of systemic lupus erythematosus. Lupus 2019, 28, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Koh, C.W.Q.; Goh, Y.T.; Toh, J.D.W.; Neo, S.P.; Ng, S.B.; Gunaratne, J.; Gao, Y.G.; Quake, S.R.; Burkholder, W.F.; Goh, W.S.S. Single-nucleotide-resolution sequencing of human N6-methyldeoxyadenosine reveals strand-asymmetric clusters associated with SSBP1 on the mitochondrial genome. Nucleic Acids Res. 2018, 46, 11659–11670. [Google Scholar] [CrossRef] [PubMed]
- Clark, T.A.; Murray, I.A.; Morgan, R.D.; Kislyuk, A.O.; Spittle, K.E.; Boitano, M.; Fomenkov, A.; Roberts, R.J.; Korlach, J. Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res. 2012, 40, e29. [Google Scholar] [CrossRef]
- Hu, H.; Dai, Y.; Liang, Z.; He, H.; Zhou, J.; Hu, Z.; Xu, Y.; Guo, H.; Tang, D. Analysis for 6-methyladenine modification of DNA in chorionic tissue from aborted fetuses with monosomy 21. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2020, 37, 747–750. [Google Scholar] [CrossRef]
- Zhang, X.; Blumenthal, R.M.; Cheng, X. A Role for N6-Methyladenine in DNA Damage Repair. Trends Biochem. Sci. 2021, 46, 175–183. [Google Scholar] [CrossRef]
- Joyce, J.A.; Lam, W.K.; Catchpoole, D.J.; Jenks, P.; Reik, W.; Maher, E.R.; Schofield, P.N. Imprinting of IGF2 and H19: Lack of reciprocity in sporadic Beckwith-Wiedemann syndrome. Hum. Mol. Genet. 1997, 6, 1543–1548. [Google Scholar] [CrossRef] [PubMed]
- Catchpoole, D.; Lam, W.W.; Valler, D.; Temple, I.K.; Joyce, J.A.; Reik, W.; Schofield, P.N.; Maher, E.R. Epigenetic modification and uniparental inheritance of H19 in Beckwith-Wiedemann syndrome. J. Med. Genet. 1997, 34, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Reik, W.; Brown, K.W.; Schneid, H.; Le Bouc, Y.; Bickmore, W.; Maher, E.R. Imprinting mutations in the Beckwith-Wiedemann syndrome suggested by altered imprinting pattern in the IGF2-H19 domain. Hum. Mol. Genet. 1995, 4, 2379–2385. [Google Scholar] [CrossRef] [PubMed]
- Weksberg, R.; Shen, D.R.; Fei, Y.L.; Song, Q.L.; Squire, J. Disruption of insulin-like growth factor 2 imprinting in Beckwith-Wiedemann syndrome. Nat. Genet. 1993, 5, 143–150. [Google Scholar] [CrossRef]
- Sutcliffe, J.S.; Nakao, M.; Christian, S.; Orstavik, K.H.; Tommerup, N.; Ledbetter, D.H.; Beaudet, A.L. Deletions of a differentially methylated CpG island at the SNRPN gene define a putative imprinting control region. Nat. Genet. 1994, 8, 52–58. [Google Scholar] [CrossRef]
- Reis, A.; Dittrich, B.; Greger, V.; Buiting, K.; Lalande, M.; Gillessen-Kaesbach, G.; Anvret, M.; Horsthemke, B. Imprinting mutations suggested by abnormal DNA methylation patterns in familial Angelman and Prader-Willi syndromes. Am. J. Hum. Genet. 1994, 54, 741–747. [Google Scholar] [PubMed]
- Glenn, C.C.; Nicholls, R.D.; Robinson, W.P.; Saitoh, S.; Niikawa, N.; Schinzel, A.; Horsthemke, B.; Driscoll, D.J. Modification of 15q11-q13 DNA methylation imprints in unique Angelman and Prader-Willi patients. Hum. Mol. Genet. 1993, 2, 1377–1382. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.E.; Jiang, Y.H. Novel epigenetic molecular therapies for imprinting disorders. Mol. Psychiatry 2023, 28, 3182–3193. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, N.; Sharma, A.R.; Baylin, S.B. Epigenetic Therapeutics: A New Weapon in the War Against Cancer. Annu. Rev. Med. 2016, 67, 73–89. [Google Scholar] [CrossRef] [PubMed]
- Dawson, M.A.; Kouzarides, T. Cancer epigenetics: From mechanism to therapy. Cell 2012, 150, 12–27. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.S.; Rhie, A.; Kim, J.; Lee, S.; Sohn, M.H.; Kim, C.U.; Hastie, A.; Cao, H.; Yun, J.Y.; Kim, J.; et al. De novo assembly and phasing of a Korean human genome. Nature 2016, 538, 243–247. [Google Scholar] [CrossRef]
- Shi, L.; Guo, Y.; Dong, C.; Huddleston, J.; Yang, H.; Han, X.; Fu, A.; Li, Q.; Li, N.; Gong, S.; et al. Long-read sequencing and de novo assembly of a Chinese genome. Nat. Commun. 2016, 7, 12065. [Google Scholar] [CrossRef]
- Chaisson, M.J.P.; Sanders, A.D.; Zhao, X.; Malhotra, A.; Porubsky, D.; Rausch, T.; Gardner, E.J.; Rodriguez, O.L.; Guo, L.; Collins, R.L.; et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun. 2019, 10, 1784. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Quaglieri, A.; Flensburg, C.; Speed, T.P.; Majewski, I.J. Finding a suitable library size to call variants in RNA-Seq. BMC Bioinform. 2020, 21, 553. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Reik, W.; Walter, J. Genomic imprinting: Parental influence on the genome. Nat. Rev. Genet. 2001, 2, 21–32. [Google Scholar] [CrossRef]
- Falls, J.G.; Pulford, D.J.; Wylie, A.A.; Jirtle, R.L. Genomic imprinting: Implications for human disease. Am. J. Pathol. 1999, 154, 635–647. [Google Scholar] [CrossRef]

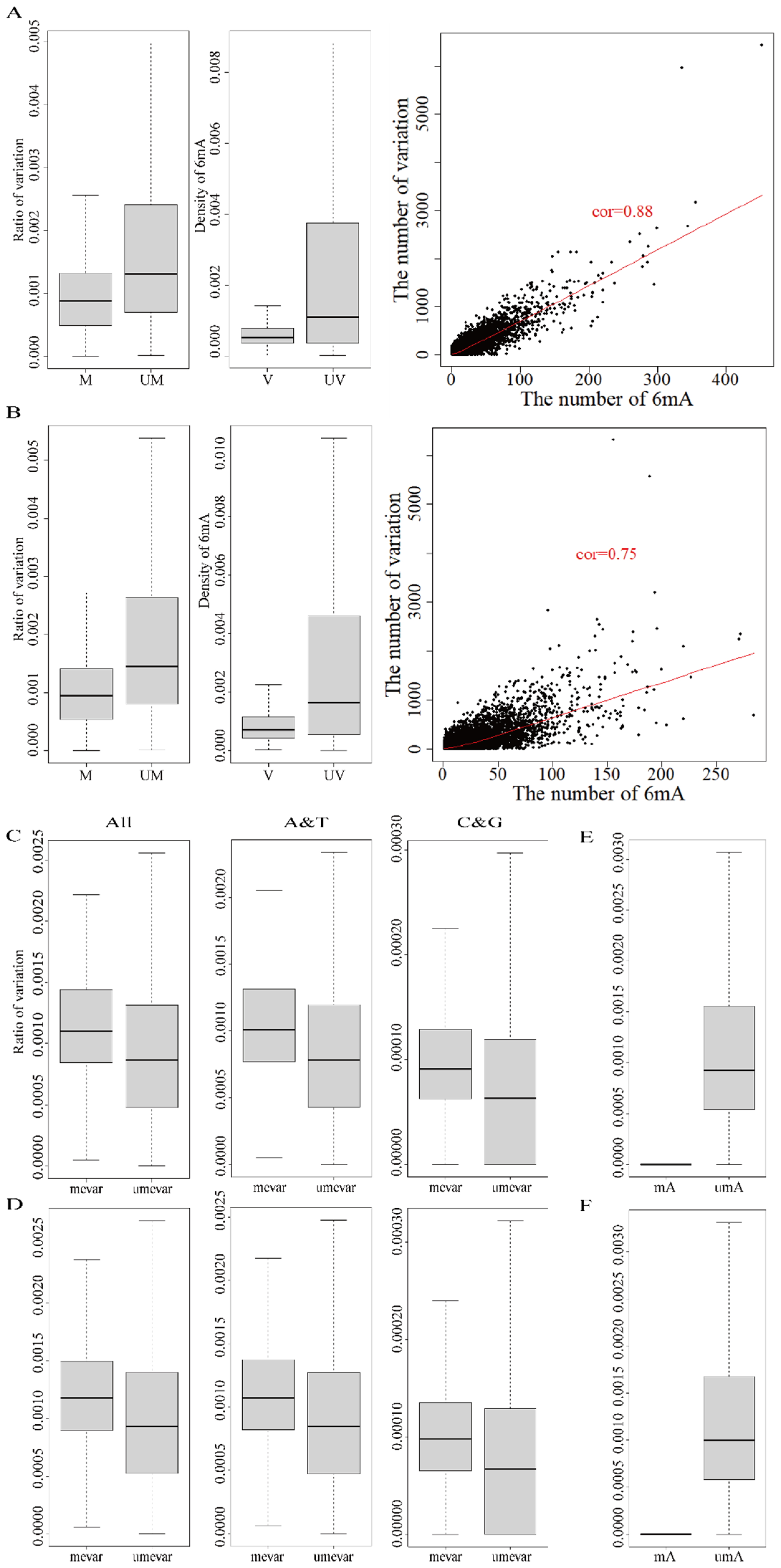

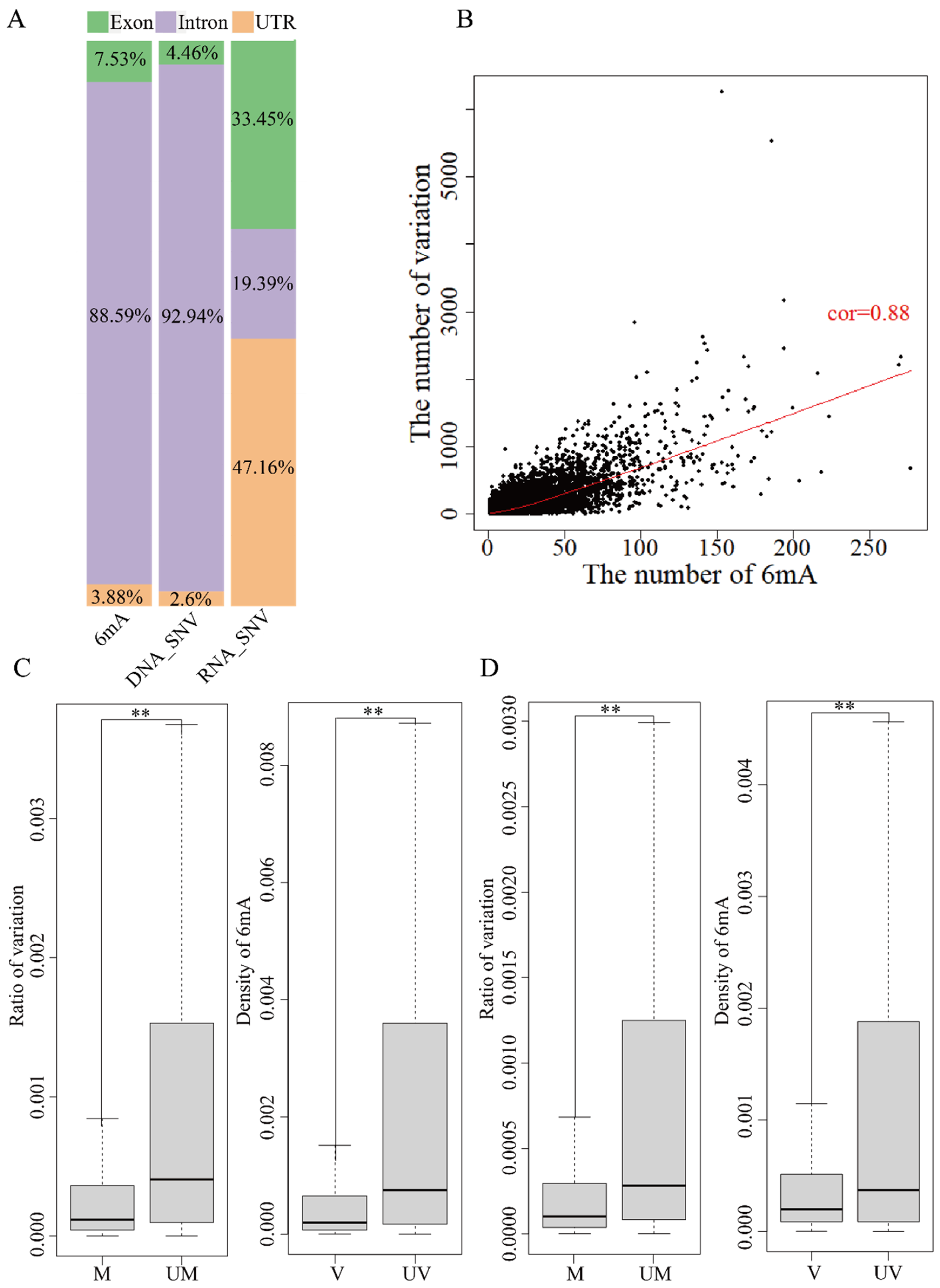
| Sample | No. of Methyloci 1 | No. of Methyloci 1 on Genes | Ratio of Methyloci 1 on Genes (%) | Strand-Methylated Genes 2 |
|---|---|---|---|---|
| HX1 | 753,242 | 238,597 | 31.68 | 25,926 |
| AK1 | 852,570 | 280,650 | 32.92 | 27,096 |
| HG00514 | 827,068 | 257,444 | 31.13 | 22,339 |
| Sample | Total Variations No. | No. of Variations in Genes | No. of Genes with Variations | No. of Methylated Variations | No. of Methylated Genes with Variations | No. of Genes with Methylated Variations |
|---|---|---|---|---|---|---|
| HX1 | 2,593,902 | 1,522,416 | 35,232 | 2270 | 22,280 | 707 |
| AK1 | 2,788,637 | 1,638,029 | 35,685 | 2308 | 23,293 | 809 |
| Type | East Asian Samples | HeLa | ||
|---|---|---|---|---|
| HX1 | AK1 | Consistency | ||
| Total RNA variations | 21,346 | 80,139 | 18,519 | 29,007 |
| No. of variations in genes | 21,044 | 77,791 | 18,345 | 28,632 |
| No. of genes with variations | 6630 | 13,129 | 6139 | 8481 |
| No. of transmitted variations | 7631 | 28,787 | 4219 | 6370 |
| No. of genes with transmitted variations | 3786 | 9309 | 2587 | 3841 |
| Transmit Type | HX1 | AK1 | ||
|---|---|---|---|---|
| Estimate | Pr (>|t|) | Estimate | Pr (>|t|) | |
| (Intercept) | 0.0684 | 0.280968 | 0.077022 | 0.22456 |
| 0/1_0/0 | 1.57906 | 0.144657 | −0.007638 | 0.99214 |
| 0/1_0/1 | 0.11667 | 0.000448 *** | 0.103532 | 0.00264 ** |
| 0/1_1/1 | −0.63351 | 0.174365 | −0.736716 | 0.10425 |
| 1/1_1/1 | 0.13076 | 3.89 × 10−5 *** | 0.136978 | 6.28 × 10−6 *** |
| Transmit Type | HX1 | AK1 | ||
|---|---|---|---|---|
| Estimate | Pr (>|t|) | Estimate | Pr (>|t|) | |
| (Intercept) | 1.27 × 10−4 | <2 × 10−16 *** | 1.30 × 10−4 | <2 × 10−16 *** |
| 0/1_0/0 | 4.69 × 10−4 | 2.93 × 10−7 *** | −6.50 × 10−5 | 0.6425 |
| 0/1_0/1 | 4.03 × 10−6 | 0.25 | −2.81 × 10−7 | 0.9396 |
| 0/1_1/1 | 3.07 × 10−5 | 0.553 | −9.57 × 10−5 | 0.0598 |
| 1/1_1/1 | 2.51 × 10−6 | 0.46 | 5.44 × 10−6 | 0.0892 |
| Transmit Type | Estimate | Pr (>|t|) |
|---|---|---|
| (Intercept) | −0.09068 | 0.2267 |
| 0/1_0/0 | 0.17215. | 0.0855 |
| 0/1_0/1 | 0.21859 | 7.35 × 10−6 *** |
| 0/1_1/1 | 0.01122 | 0.8107 |
| 1/1_0/0 | 0.28214 | 0.4466 |
| 1/1_0/1 | 0.10845 | 0.4791 |
| 1/1_1/1 | 0.08597 | 0.107 |
| Classification | HX1 | AK1 | ||
|---|---|---|---|---|
| Methylated- Non-Imprinted | Unmethylated- Non-Imprinted | Methylated- Non-Imprinted | Unmethylated- Non-Imprinted | |
| Methylated- imprinted | 0.01 | 3.91 × 10−7 | 4.28 × 10−3 | 1.11 × 10−7 |
| Unmethylated- imprinted | 2.81 × 10−292 | 0.22 | 2.05 × 10−238 | 0.04 |
| Classification | HX1 | AK1 | ||
|---|---|---|---|---|
| Unmethylated- Imprinted | Unmethylated- Non-Imprinted | Unmethylated- Imprinted | Unmethylated- Non-Imprinted | |
| Methylated- imprinted | 4.55 × 10−7 | 3.91 × 10−7 | 1.47 × 10−7 | 1.11 × 10−7 |
| Methylated- non-imprinted | 2.81 × 10−292 | 0.00 | 2.05 × 10−292 | 0.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luan, M.; Chen, K.; Zhao, W.; Tang, M.; Wang, L.; Liu, S.; Zhu, L.; Xie, S. Selective Effect of DNA N6-Methyladenosine Modification on Transcriptional Genetic Variations in East Asian Samples. Int. J. Mol. Sci. 2024, 25, 10400. https://doi.org/10.3390/ijms251910400
Luan M, Chen K, Zhao W, Tang M, Wang L, Liu S, Zhu L, Xie S. Selective Effect of DNA N6-Methyladenosine Modification on Transcriptional Genetic Variations in East Asian Samples. International Journal of Molecular Sciences. 2024; 25(19):10400. https://doi.org/10.3390/ijms251910400
Chicago/Turabian StyleLuan, Meiwei, Kaining Chen, Wenwen Zhao, Minqiang Tang, Lingxia Wang, Shoubai Liu, Linan Zhu, and Shangqian Xie. 2024. "Selective Effect of DNA N6-Methyladenosine Modification on Transcriptional Genetic Variations in East Asian Samples" International Journal of Molecular Sciences 25, no. 19: 10400. https://doi.org/10.3390/ijms251910400
APA StyleLuan, M., Chen, K., Zhao, W., Tang, M., Wang, L., Liu, S., Zhu, L., & Xie, S. (2024). Selective Effect of DNA N6-Methyladenosine Modification on Transcriptional Genetic Variations in East Asian Samples. International Journal of Molecular Sciences, 25(19), 10400. https://doi.org/10.3390/ijms251910400






