RNA-Binding Proteins as Novel Effectors in Osteoblasts and Osteoclasts: A Systems Biology Approach to Dissect the Transcriptional Landscape
Abstract
:1. Introduction
2. Results
2.1. The Transcriptional Landscape of Genes Encoding 180 RBPs in Osteoblasts and Osteoclasts and Follow-Up Comparative Analysis
2.2. Regulation on the Gene Expression Level of 180 RBPs upon Various Treatment Conditions
2.3. Difference in the Expression Pattern of the 180 Genes Encoding RBPs in Osteoblasts and Osteoclast Dissected by Hierarchical Clustering
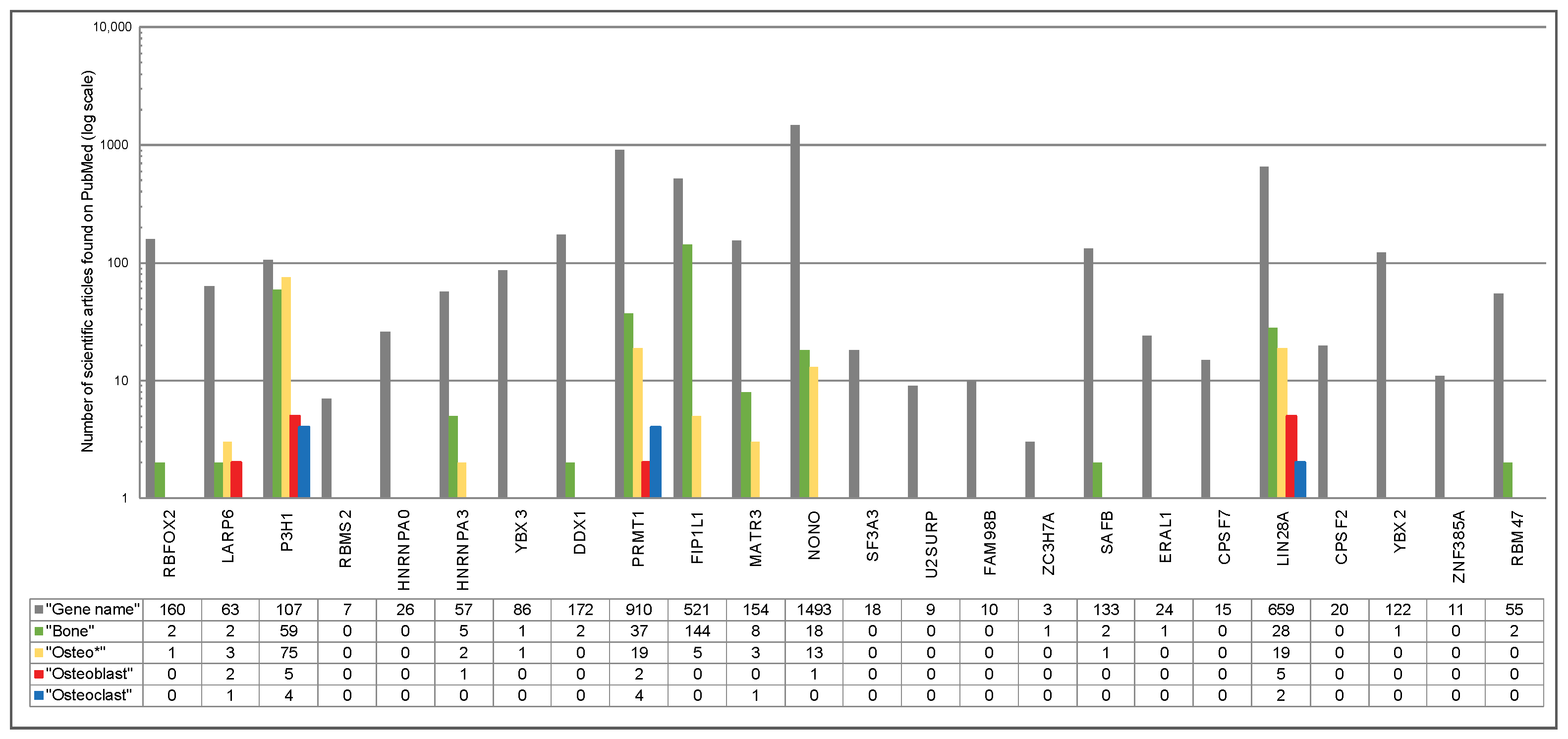
2.4. Osteoblast- and Osteoclast-Specific Gene Signatures
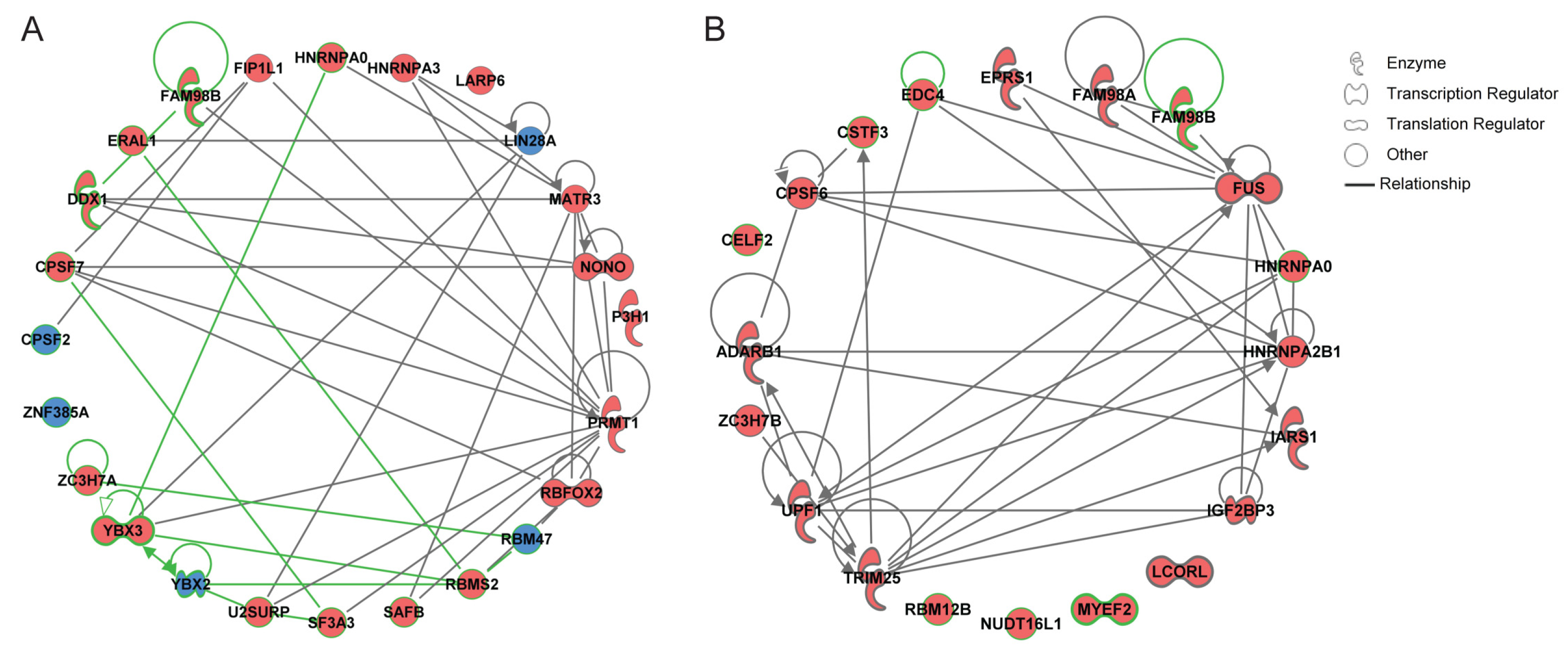
2.5. Gene Network Reconstruction and the Nomination of Genes Encoding RBPs as Promising Candidates in Osteoblasts and Osteoclasts
3. Discussion
4. Materials and Methods
4.1. Comprehensive Analysis of Transcriptomic Data
4.2. Statistical Analysis and Data Visualization
4.3. Gene Network Reconstruction
4.4. Comprehensive Literature Search
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Raggatt, L.J.; Partridge, N.C. Cellular and molecular mechanisms of bone remodeling. J. Biol. Chem. 2010, 285, 25103–25108. [Google Scholar] [CrossRef]
- Suzuki, A.; Minamide, M.; Iwaya, C.; Ogata, K.; Iwata, J. Role of Metabolism in Bone Development and Homeostasis. Int. J. Mol. Sci. 2020, 21, 8992. [Google Scholar] [CrossRef] [PubMed]
- Pietschmann, P.; Mechtcheriakova, D.; Meshcheryakova, A.; Föger-Samwald, U.; Ellinger, I. Immunology of Osteoporosis: A Mini-Review. Gerontology 2016, 62, 128–137. [Google Scholar] [CrossRef]
- Creecy, A.; Damrath, J.G.; Wallace, J.M. Control of Bone Matrix Properties by Osteocytes. Front. Endocrinol. 2020, 11, 578477. [Google Scholar] [CrossRef] [PubMed]
- Kenkre, J.S.; Bassett, J. The bone remodelling cycle. Ann. Clin. Biochem. 2018, 55, 308–327. [Google Scholar] [CrossRef] [PubMed]
- Hofbauer, L.C.; Schoppet, M. Clinical implications of the osteoprotegerin/RANKL/RANK system for bone and vascular diseases. JAMA 2004, 292, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Meshcheryakova, A.; Mechtcheriakova, D.; Pietschmann, P. Sphingosine 1-phosphate signaling in bone remodeling: Multifaceted roles and therapeutic potential. Expert Opin. Ther. Targets 2017, 21, 725–737. [Google Scholar] [CrossRef]
- Black, D.M.; Rosen, C.J. Clinical Practice. Postmenopausal Osteoporosis. N. Engl. J. Med. 2016, 374, 254–262. [Google Scholar] [CrossRef]
- Kanis, J.A. Diagnosis of osteoporosis and assessment of fracture risk. Lancet 2002, 359, 1929–1936. [Google Scholar] [CrossRef]
- Khosla, S.; Hofbauer, L.C. Osteoporosis treatment: Recent developments and ongoing challenges. Lancet Diabetes Endocrinol. 2017, 5, 898–907. [Google Scholar] [CrossRef]
- Tafaro, L.; Napoli, N. Current and Emerging Treatment of Osteoporosis. In Orthogeriatrics: The Management of Older Patients with Fragility Fractures; Falaschi, P., Marsh, D., Eds.; Springer: Cham, Switzerland, 2021; pp. 257–272. [Google Scholar]
- Fabre, S.; Funck-Brentano, T.; Cohen-Solal, M. Anti-Sclerostin Antibodies in Osteoporosis and Other Bone Diseases. J. Clin. Med. 2020, 9, 3439. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell. Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell. Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef]
- Matsuyama, H.; Suzuki, H.I. Systems and Synthetic microRNA Biology: From Biogenesis to Disease Pathogenesis. Int. J. Mol. Sci. 2019, 21, 132. [Google Scholar] [CrossRef] [PubMed]
- Annese, T.; Tamma, R.; De Giorgis, M.; Ribatti, D. microRNAs Biogenesis, Functions and Role in Tumor Angiogenesis. Front. Oncol. 2020, 10, 581007. [Google Scholar] [CrossRef] [PubMed]
- Romero-Cordoba, S.L.; Salido-Guadarrama, I.; Rodriguez-Dorantes, M.; Hidalgo-Miranda, A. miRNA biogenesis: Biological impact in the development of cancer. Cancer Biol. Ther. 2014, 15, 1444–1455. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Shoorei, H.; Anamag, F.T.; Taheri, M. The Role of Non-Coding RNAs in Controlling Cell Cycle Related Proteins in Cancer Cells. Front. Oncol. 2020, 10, 608975. [Google Scholar] [CrossRef]
- Scherm, M.G.; Daniel, C. miRNA-Mediated Immune Regulation in Islet Autoimmunity and Type 1 Diabetes. Front. Endocrinol. 2020, 11, 606322. [Google Scholar] [CrossRef]
- Domingo, S.; Solé, C.; Moliné, T.; Ferrer, B.; Cortés-Hernández, J. MicroRNAs in Several Cutaneous Autoimmune Diseases: Psoriasis, Cutaneous Lupus Erythematosus and Atopic Dermatitis. Cells 2020, 9, 2656. [Google Scholar] [CrossRef]
- Ciuffi, S.; Donati, S.; Marini, F.; Palmini, G.; Luzi, E.; Brandi, M.L. Circulating MicroRNAs as Novel Biomarkers for Osteoporosis and Fragility Fracture Risk: Is There a Use in Assessment Risk? Int. J. Mol. Sci. 2020, 21, 6927. [Google Scholar] [CrossRef]
- Yang, Y.; Yujiao, W.; Fang, W.; Linhui, Y.; Ziqi, G.; Zhichen, W.; Zirui, W.; Shengwang, W. The roles of miRNA, lncRNA and circRNA in the development of osteoporosis. Biol. Res. 2020, 53, 40. [Google Scholar] [CrossRef] [PubMed]
- Kerschan-Schindl, K.; Hackl, M.; Boschitsch, E.; Föger-Samwald, U.; Nägele, O.; Skalicky, S.; Weigl, M.; Grillari, J.; Pietschmann, P. Diagnostic Performance of a Panel of miRNAs (OsteomiR) for Osteoporosis in a Cohort of Postmenopausal Women. Calcif. Tissue Int. 2021, 108, 725–737. [Google Scholar] [CrossRef] [PubMed]
- Treiber, T.; Treiber, N.; Plessmann, U.; Harlander, S.; Daiß, J.L.; Eichner, N.; Lehmann, G.; Schall, K.; Urlaub, H.; Meister, G. A Compendium of RNA-Binding Proteins that Regulate MicroRNA Biogenesis. Mol. Cell 2017, 66, 270–284.e13. [Google Scholar] [CrossRef] [PubMed]
- Gebauer, F.; Schwarzl, T.; Valcárcel, J.; Hentze, M.W. RNA-binding proteins in human genetic disease. Nat. Rev. Genet. 2021, 22, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Granchi, D.; Ochoa, G.; Leonardi, E.; Devescovi, V.; Baglìo, S.R.; Osaba, L.; Baldini, N.; Ciapetti, G. Gene expression patterns related to osteogenic differentiation of bone marrow-derived mesenchymal stem cells during ex vivo expansion. Tissue Eng. Part C Methods 2010, 16, 511–524. [Google Scholar] [CrossRef]
- Yuen, T.; Stachnik, A.; Iqbal, J.; Sgobba, M.; Gupta, Y.; Lu, P.; Colaianni, G.; Ji, Y.; Zhu, L.L.; Kim, S.M.; et al. Bisphosphonates inactivate human EGFRs to exert antitumor actions. Proc. Natl. Acad. Sci. USA 2014, 111, 17989–17994. [Google Scholar] [CrossRef]
- Grundberg, E.; Brändström, H.; Lam, K.C.; Gurd, S.; Ge, B.; Harmsen, E.; Kindmark, A.; Ljunggren, O.; Mallmin, H.; Nilsson, O.; et al. Systematic assessment of the human osteoblast transcriptome in resting and induced primary cells. Physiol. Genom. 2008, 33, 301–311. [Google Scholar] [CrossRef]
- Ogasawara, T.; Kawaguchi, H.; Jinno, S.; Hoshi, K.; Itaka, K.; Takato, T.; Nakamura, K.; Okayama, H. Bone morphogenetic protein 2-induced osteoblast differentiation requires Smad-mediated down-regulation of Cdk6. Mol. Cell. Biol. 2004, 24, 6560–6568. [Google Scholar] [CrossRef]
- Zhou, D.; Couture, S.; Scott, M.S.; Abou Elela, S. RBFOX2 alters splicing outcome in distinct binding modes with multiple protein partners. Nucleic Acids Res. 2021, 49, 8370–8383. [Google Scholar] [CrossRef]
- Cibi, D.M.; Mia, M.M.; Guna Shekeran, S.; Yun, L.S.; Sandireddy, R.; Gupta, P.; Hota, M.; Sun, L.; Ghosh, S.; Singh, M.K. Neural crest-specific deletion of Rbfox2 in mice leads to craniofacial abnormalities including cleft palate. Elife 2019, 8, e45418. [Google Scholar] [CrossRef]
- Cho, N.; Kim, J.O.; Lee, S.; Choi, S.; Kim, J.; Ko, M.S.; Park, S.J.; Ji, J.H.; Kim, K.K. Alternative splicing induces cytoplasmic localization of RBFOX2 protein in calcific tendinopathy. Exp. Mol. Pathol. 2019, 109, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Stefanovic, B. LARP6 Meets Collagen mRNA: Specific Regulation of Type I Collagen Expression. Int. J. Mol. Sci. 2016, 17, 419. [Google Scholar] [CrossRef] [PubMed]
- Blair, H.C.; Larrouture, Q.C.; Li, Y.; Lin, H.; Beer-Stoltz, D.; Liu, L.; Tuan, R.S.; Robinson, L.J.; Schlesinger, P.H.; Nelson, D.J. Osteoblast Differentiation and Bone Matrix Formation In Vivo and In Vitro. Tissue Eng. Part B Rev. 2017, 23, 268–280. [Google Scholar] [CrossRef] [PubMed]
- Oxlund, H.; Mosekilde, L.; Ortoft, G. Reduced concentration of collagen reducible cross links in human trabecular bone with respect to age and osteoporosis. Bone 1996, 19, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Stavraka, C.; Blagden, S. The La-Related Proteins, a Family with Connections to Cancer. Biomolecules 2015, 5, 2701–2722. [Google Scholar] [CrossRef]
- Marini, J.C.; Blissett, A.R. New genes in bone development: What’s new in osteogenesis imperfecta. J. Clin. Endocrinol. Metab. 2013, 98, 3095–3103. [Google Scholar] [CrossRef]
- Sun, X.; Hu, Y.; Wu, J.; Shi, L.; Zhu, L.; Xi, P.W.; Wei, J.F.; Ding, Q. RBMS2 inhibits the proliferation by stabilizing P21 mRNA in breast cancer. J. Exp. Clin. Cancer Res. 2018, 37, 298. [Google Scholar] [CrossRef]
- Premnath, P.; Jorgenson, B.; Hess, R.; Tailor, P.; Louie, D.; Taiani, J.; Boyd, S.; Krawetz, R. p21(-/-) mice exhibit enhanced bone regeneration after injury. BMC Musculoskelet Disord. 2017, 18, 435. [Google Scholar] [CrossRef]
- Premnath, P.; Ferrie, L.; Louie, D.; Boyd, S.; Krawetz, R. Absence of p21(WAF1/CIP1/SDI1) protects against osteopenia and minimizes bone loss after ovariectomy in a mouse model. PLoS ONE 2019, 14, e0215018. [Google Scholar] [CrossRef]
- Guan, R.; El-Rass, S.; Spillane, D.; Lam, S.; Wang, Y.; Wu, J.; Chen, Z.; Wang, A.; Jia, Z.; Keating, A.; et al. rbm47, a novel RNA binding protein, regulates zebrafish head development. Dev. Dyn. 2013, 242, 1395–1404. [Google Scholar] [CrossRef]
- Peng, Q.; Li, J.; Wu, Q.; Wang, P.; Kang, Z.; Deng, Y.; Xiao, Y.; Zheng, P.; Ge, F.; Chen, Y. ZNF385A and ZNF346 Serve as Prognostic Biomarkers Associated with an Inflamed Immunosuppressive Tumor Microenvironment in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2023, 24, 3155. [Google Scholar] [CrossRef] [PubMed]
- Jen, J.; Wang, Y.C. Zinc finger proteins in cancer progression. J. Biomed. Sci. 2016, 23, 53. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Kawagishi, H.; Watanabe, A.; Sugimoto, K.; Maruyama, M.; Sugimoto, M. Cooperative role of the RNA-binding proteins Hzf and HuR in p53 activation. Mol. Cell. Biol. 2011, 31, 1997–2009. [Google Scholar] [CrossRef] [PubMed]
- Kadmiel, M.; Cidlowski, J.A. Glucocorticoid receptor signaling in health and disease. Trends Pharmacol. Sci. 2013, 34, 518–530. [Google Scholar] [CrossRef]
- Licht, K.; Jantsch, M.F. Rapid and dynamic transcriptome regulation by RNA editing and RNA modifications. J. Cell. Biol. 2016, 213, 15–22. [Google Scholar] [CrossRef]
- Heraud-Farlow, J.E.; Walkley, C.R. What do editors do? Understanding the physiological functions of A-to-I RNA editing by adenosine deaminase acting on RNAs. Open Biol. 2020, 10, 200085. [Google Scholar] [CrossRef]
- Broni, E.; Striegel, A.; Ashley, C.; Sakyi, P.O.; Peracha, S.; Velazquez, M.; Bebla, K.; Sodhi, M.; Kwofie, S.K.; Ademokunwa, A.; et al. Molecular Docking and Dynamics Simulation Studies Predict Potential Anti-ADAR2 Inhibitors: Implications for the Treatment of Cancer, Neurological, Immunological and Infectious Diseases. Int. J. Mol. Sci. 2023, 24, 6795. [Google Scholar] [CrossRef]
- Yu, S.; Sharma, R.; Nie, D.; Jiao, H.; Im, H.J.; Lai, Y.; Zhao, Z.; Zhu, K.; Fan, J.; Chen, D.; et al. ADAR1 ablation decreases bone mass by impairing osteoblast function in mice. Gene 2013, 513, 101–110. [Google Scholar] [CrossRef]
- Yanaizu, M.; Washizu, C.; Nukina, N.; Satoh, J.I.; Kino, Y. CELF2 regulates the species-specific alternative splicing of TREM2. Sci. Rep. 2020, 10, 17995. [Google Scholar] [CrossRef]
- Yeung, Y.T.; Fan, S.; Lu, B.; Yin, S.; Yang, S.; Nie, W.; Wang, M.; Zhou, L.; Li, T.; Li, X.; et al. CELF2 suppresses non-small cell lung carcinoma growth by inhibiting the PREX2-PTEN interaction. Carcinogenesis 2020, 41, 377–389. [Google Scholar] [CrossRef]
- Ramalingam, S.; Ramamoorthy, P.; Subramaniam, D.; Anant, S. Reduced Expression of RNA Binding Protein CELF2, a Putative Tumor Suppressor Gene in Colon Cancer. Immunogastroenterology 2012, 1, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Januszewski, J.; Forma, A.; Zembala, J.; Flieger, M.; Tyczyńska, M.; Dring, J.C.; Dudek, I.; Świątek, K.; Baj, J. Nutritional Supplements for Skin Health-A Review of What Should Be Chosen and Why. Medicina 2023, 60, 68. [Google Scholar] [CrossRef] [PubMed]
- Voltan, G.; Cannito, M.; Ferrarese, M.; Ceccato, F.; Camozzi, V. Vitamin D: An Overview of Gene Regulation, Ranging from Metabolism to Genomic Effects. Genes 2023, 14, 1691. [Google Scholar] [CrossRef] [PubMed]
- Ganss, B.; Kim, R.H.; Sodek, J. Bone sialoprotein. Crit. Rev. Oral Biol. Med. 1999, 10, 79–98. [Google Scholar] [CrossRef] [PubMed]
- Hafez, A.; Squires, R.; Pedracini, A.; Joshi, A.; Seegmiller, R.E.; Oxford, J.T. Col11a1 Regulates Bone Microarchitecture during Embryonic Development. J. Dev. Biol. 2015, 3, 158–176. [Google Scholar] [CrossRef]
- Izu, Y.; Sun, M.; Zwolanek, D.; Veit, G.; Williams, V.; Cha, B.; Jepsen, K.J.; Koch, M.; Birk, D.E. Type XII collagen regulates osteoblast polarity and communication during bone formation. J. Cell. Biol. 2011, 193, 1115–1130. [Google Scholar] [CrossRef]
- Tanaka, Y.; Morimoto, I.; Nakano, Y.; Okada, Y.; Hirota, S.; Nomura, S.; Nakamura, T.; Eto, S. Osteoblasts are regulated by the cellular adhesion through ICAM-1 and VCAM-1. J. Bone Min. Res. 1995, 10, 1462–1469. [Google Scholar] [CrossRef]
- Di Cesare, P.E.; Fang, C.; Leslie, M.P.; Tulli, H.; Perris, R.; Carlson, C.S. Expression of cartilage oligomeric matrix protein (COMP) by embryonic and adult osteoblasts. J. Orthop. Res. 2000, 18, 713–720. [Google Scholar] [CrossRef]
- Di Benedetto, A.; Watkins, M.; Grimston, S.; Salazar, V.; Donsante, C.; Mbalaviele, G.; Radice, G.L.; Civitelli, R. N-cadherin and cadherin 11 modulate postnatal bone growth and osteoblast differentiation by distinct mechanisms. J. Cell. Sci. 2010, 123, 2640–2648. [Google Scholar] [CrossRef]
- Samee, N.; Geoffroy, V.; Marty, C.; Schiltz, C.; Vieux-Rochas, M.; Levi, G.; de Vernejoul, M.C. Dlx5, a positive regulator of osteoblastogenesis, is essential for osteoblast-osteoclast coupling. Am. J. Pathol. 2008, 173, 773–780. [Google Scholar] [CrossRef]
- Udagawa, N.; Koide, M.; Nakamura, M.; Nakamichi, Y.; Yamashita, T.; Uehara, S.; Kobayashi, Y.; Furuya, Y.; Yasuda, H.; Fukuda, C.; et al. Osteoclast differentiation by RANKL and OPG signaling pathways. J. Bone Min. Metab. 2021, 39, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Geissmann, F.; Manz, M.G.; Jung, S.; Sieweke, M.H.; Merad, M.; Ley, K. Development of monocytes, macrophages, and dendritic cells. Science 2010, 327, 656–661. [Google Scholar] [CrossRef] [PubMed]
- Otero, K.; Shinohara, M.; Zhao, H.; Cella, M.; Gilfillan, S.; Colucci, A.; Faccio, R.; Ross, F.P.; Teitelbaum, S.L.; Takayanagi, H.; et al. TREM2 and beta-catenin regulate bone homeostasis by controlling the rate of osteoclastogenesis. J. Immunol. 2012, 188, 2612–2621. [Google Scholar] [CrossRef] [PubMed]
- Burstone, M.S. Histochemical demonstration of acid phosphatase activity in osteoclasts. J. Histochem. Cytochem. 1959, 7, 39–41. [Google Scholar] [CrossRef] [PubMed]
- Paiva, K.B.S.; Granjeiro, J.M. Chapter Six—Matrix Metalloproteinases in Bone Resorption, Remodeling, and Repair. In Progress in Molecular Biology and Translational Science; Khalil, R.A., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 203–303. [Google Scholar]
- Tokuhara, C.K.; Santesso, M.R.; Oliveira, G.S.N.; Ventura, T.M.D.S.; Doyama, J.T.; Zambuzzi, W.F.; Oliveira, R.C. Updating the role of matrix metalloproteinases in mineralized tissue and related diseases. J. Appl. Oral Sci. 2019, 27, e20180596. [Google Scholar] [CrossRef]
- Meshcheryakova, A.; Zimmermann, P.; Ecker, R.; Mungenast, F.; Heinze, G.; Mechtcheriakova, D. An Integrative MuSiCO Algorithm: From the Patient-Specific Transcriptional Profiles to Novel Checkpoints in disease pathobiology. In Systems Biology; Rajewsky, N., Jurga, S., Barciszewski, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 351–372. [Google Scholar]
- Meshcheryakova, A.; Zimmermann, P.; Salzmann, M.; Pietschmann, P.; Mechtcheriakova, D. Singularity and Commonality in Response to SARS-CoV-2 in Lung and Colon Cell Models. Int. J. Mol. Sci. 2022, 23, 10451. [Google Scholar] [CrossRef]
- Eisen, M.B.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [Google Scholar] [CrossRef]
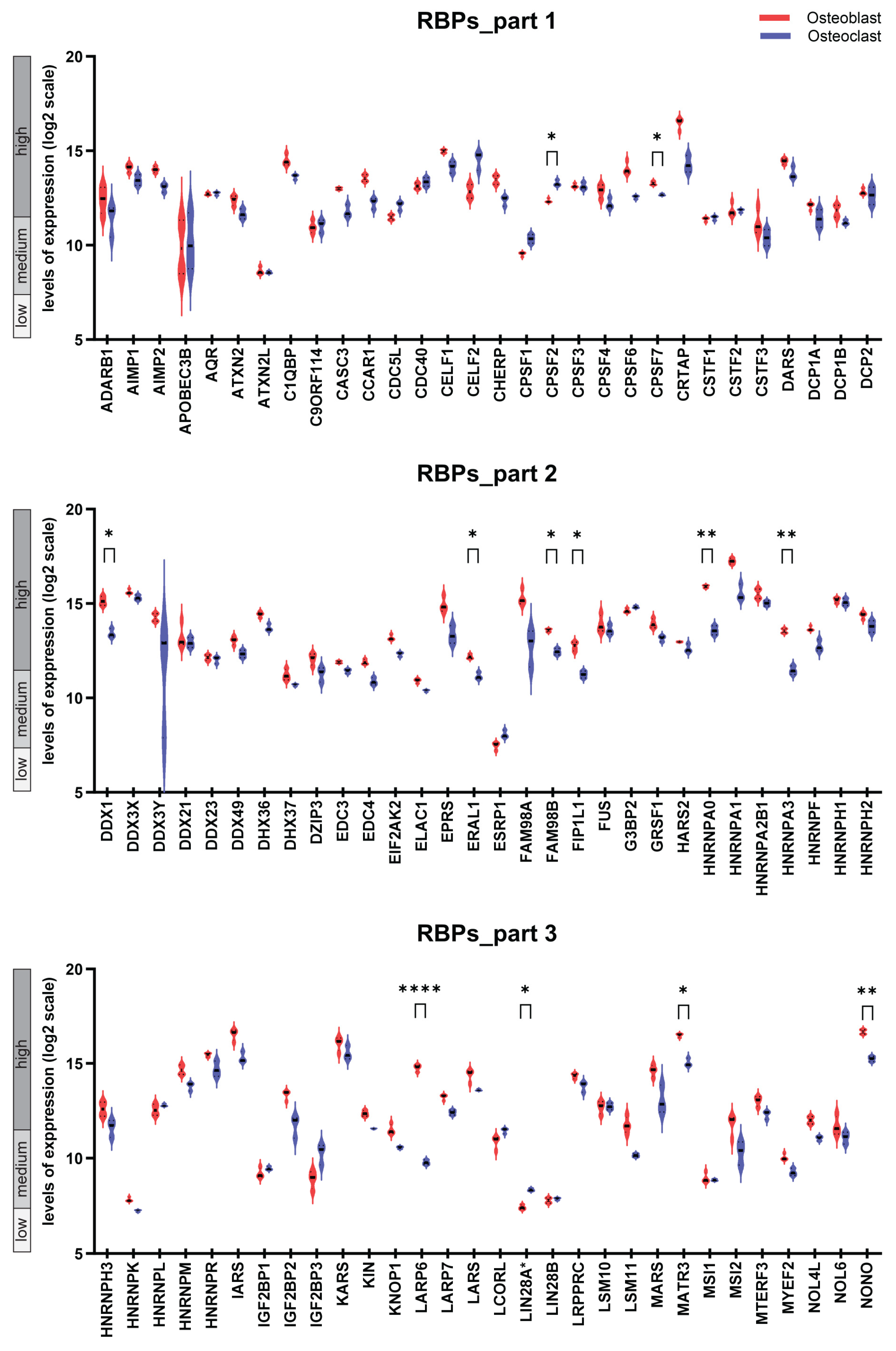
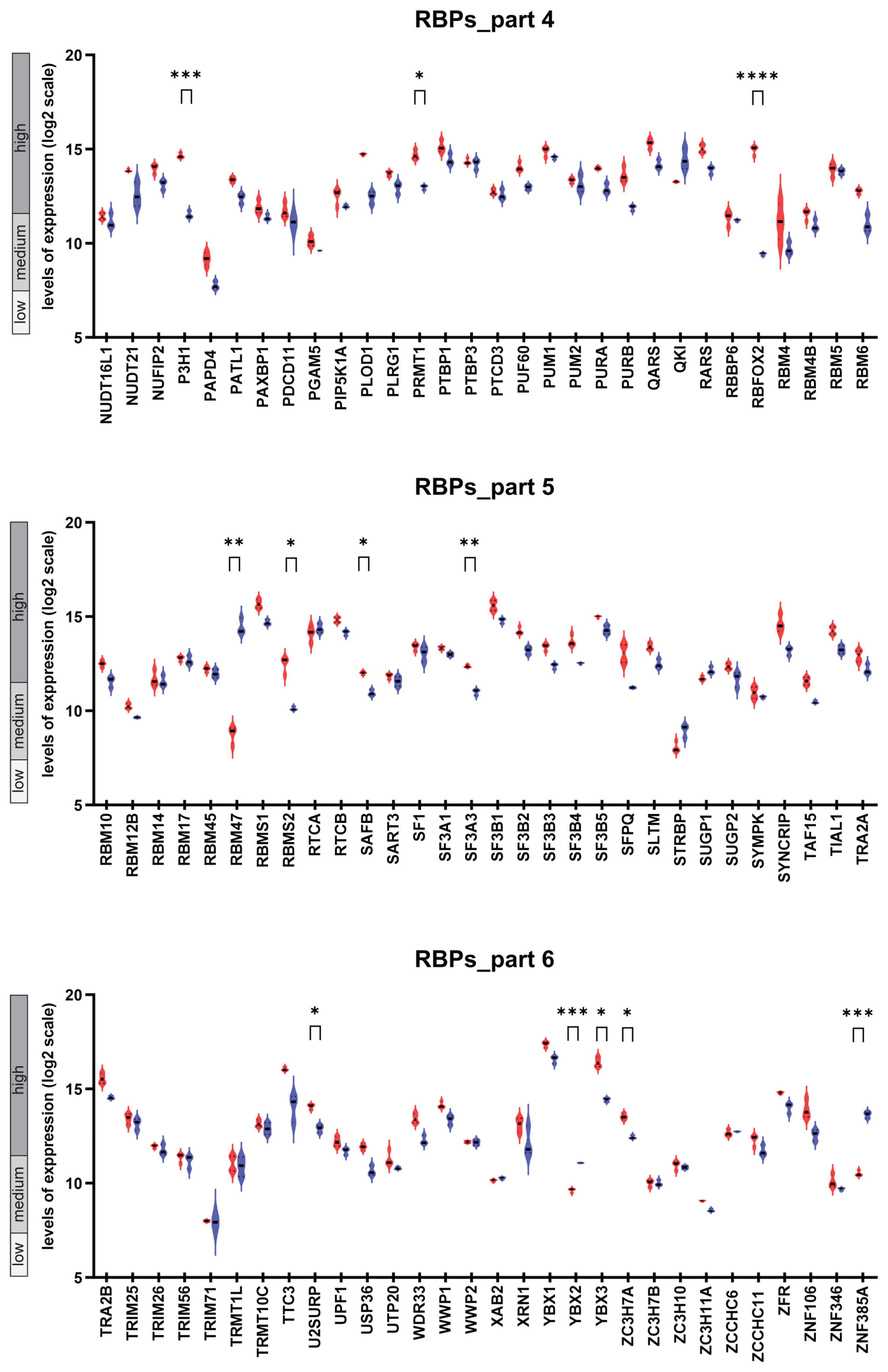





| Gene Name | Expression Level, OB | Mean Value, OB | Difference | Mean Value, OC | Expression Level, OC | p-Value |
|---|---|---|---|---|---|---|
| RBFOX2 | high | 14.97 | 5.54 | 9.43 | medium | <0.0001 |
| LARP6 | high | 14.77 | 5.00 | 9.77 | medium | <0.0001 |
| P3H1 | high | 14.64 | 3.12 | 11.52 | medium/high | <0.001 |
| RBMS2 | high | 12.51 | 2.40 | 10.11 | medium | 0.031 |
| HNRNPA0 | high | 15.9 | 2.33 | 13.57 | high | 0.001 |
| HNRNPA3 | high | 13.55 | 2.09 | 11.46 | medium/high | 0.004 |
| YBX3 | high | 16.41 | 1.99 | 14.42 | high | 0.011 |
| DDX1 | high | 15.13 | 1.72 | 13.41 | high | 0.042 |
| PRMT1 | high | 14.69 | 1.72 | 12.97 | high | 0.015 |
| FIP1L1 | high | 12.74 | 1.50 | 11.24 | medium/high | 0.049 |
| MATR3 | high | 16.49 | 1.46 | 15.03 | high | 0.011 |
| NONO | high | 16.67 | 1.43 | 15.24 | high | 0.007 |
| SF3A3 | high | 12.36 | 1.37 | 10.99 | medium | 0.003 |
| U2SURP | high | 14.07 | 1.17 | 12.9 | high | 0.037 |
| FAM98B | high | 13.57 | 1.12 | 12.45 | high | 0.014 |
| ZC3H7A | high | 13.52 | 1.08 | 12.44 | high | 0.023 |
| SAFB | high | 12.01 | 1.07 | 10.94 | medium | 0.014 |
| ERAL1 | high | 12.18 | 1.03 | 11.15 | medium | 0.047 |
| CPSF7 | high | 13.28 | 0.61 | 12.67 | high | 0.039 |
| LIN28A * | low | 7.41 | −0.93 | 8.34 | low | 0.013 |
| CPSF2 | high | 12.34 | −0.94 | 13.28 | high | 0.032 |
| YBX2 | medium | 9.62 | −1.45 | 11.07 | medium | < 0.001 |
| ZNF385A | medium | 10.5 | −3.13 | 13.63 | high | < 0.001 |
| RBM47 | medium | 8.77 | −5.68 | 14.45 | high | 0.002 |
| Clusters | Expression Level, OB | Mean Value, OB | Comparison | Mean Value, OC | Expression Level, OC |
|---|---|---|---|---|---|
| Cluster I | High | 14.97 | > | 9.43 | Medium |
| Cluster II | High | 15.54 | > | 13.43 | High |
| Cluster III | High | 13.66 | > | 13.04 | High |
| Cluster IV | High | 12.53 | > | 11.86 | High |
| Cluster V | Medium | 10.06 | > | 9.82 | Medium |
| Cluster VI | Medium | 9.63 | < | 14.04 | High |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meshcheryakova, A.; Bohdan, S.; Zimmermann, P.; Jaritz, M.; Pietschmann, P.; Mechtcheriakova, D. RNA-Binding Proteins as Novel Effectors in Osteoblasts and Osteoclasts: A Systems Biology Approach to Dissect the Transcriptional Landscape. Int. J. Mol. Sci. 2024, 25, 10417. https://doi.org/10.3390/ijms251910417
Meshcheryakova A, Bohdan S, Zimmermann P, Jaritz M, Pietschmann P, Mechtcheriakova D. RNA-Binding Proteins as Novel Effectors in Osteoblasts and Osteoclasts: A Systems Biology Approach to Dissect the Transcriptional Landscape. International Journal of Molecular Sciences. 2024; 25(19):10417. https://doi.org/10.3390/ijms251910417
Chicago/Turabian StyleMeshcheryakova, Anastasia, Serhii Bohdan, Philip Zimmermann, Markus Jaritz, Peter Pietschmann, and Diana Mechtcheriakova. 2024. "RNA-Binding Proteins as Novel Effectors in Osteoblasts and Osteoclasts: A Systems Biology Approach to Dissect the Transcriptional Landscape" International Journal of Molecular Sciences 25, no. 19: 10417. https://doi.org/10.3390/ijms251910417






