Interactions between Brassinosteroids and Strigolactones in Alleviating Salt Stress in Maize
Abstract
:1. Introduction
2. Results
2.1. Changes of Maize Leaf Phenotype and Growth Parameters under Exogenous Hormone Treatment
2.2. Changes of Antioxidant Enzyme Activities in Maize under Exogenous Hormone Treatment
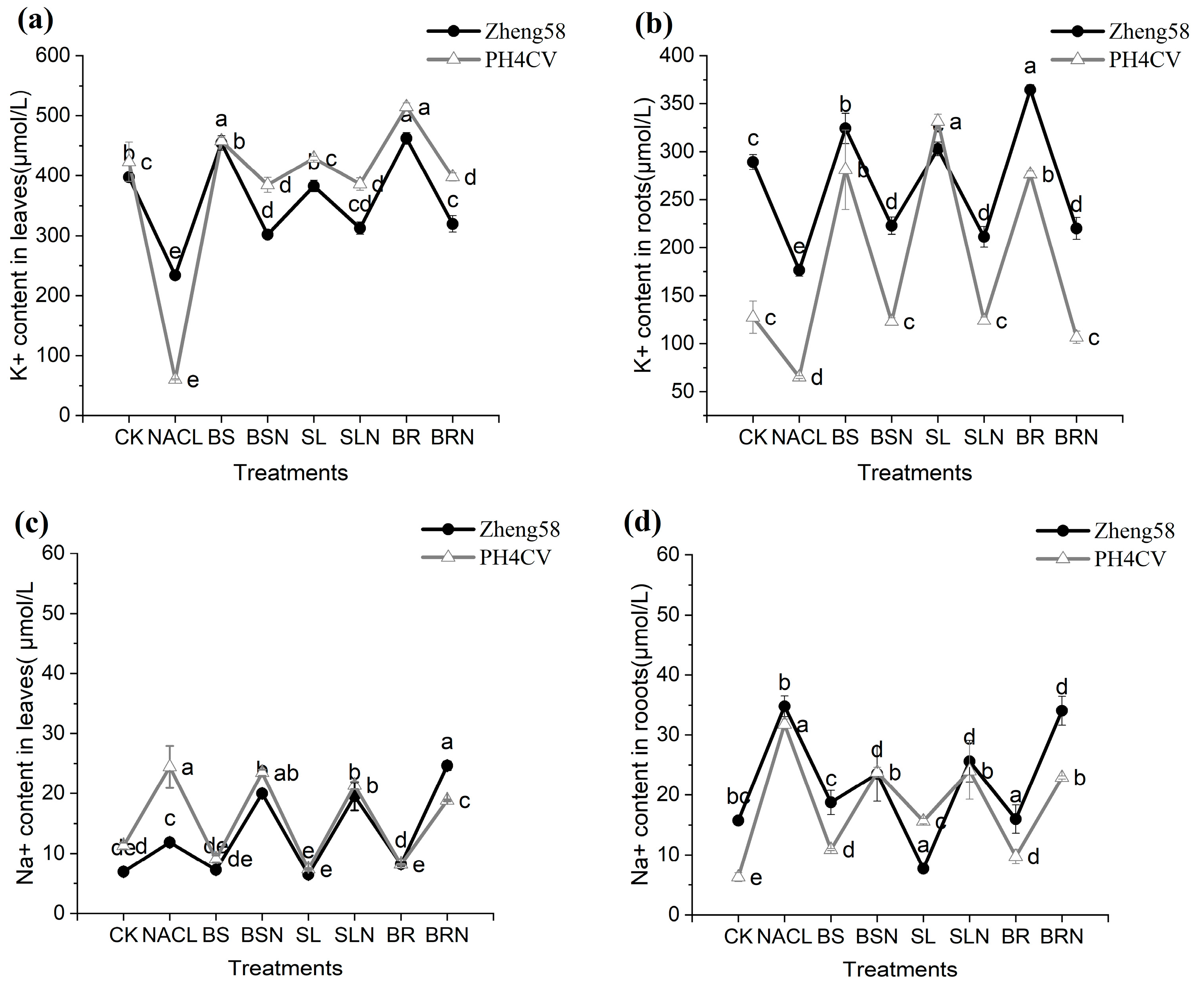
2.3. Changes of Ion Content in Corn Roots and Leaves under Exogenous Hormone Treatment
2.4. RNA Quality Inspection and Sequencing Results
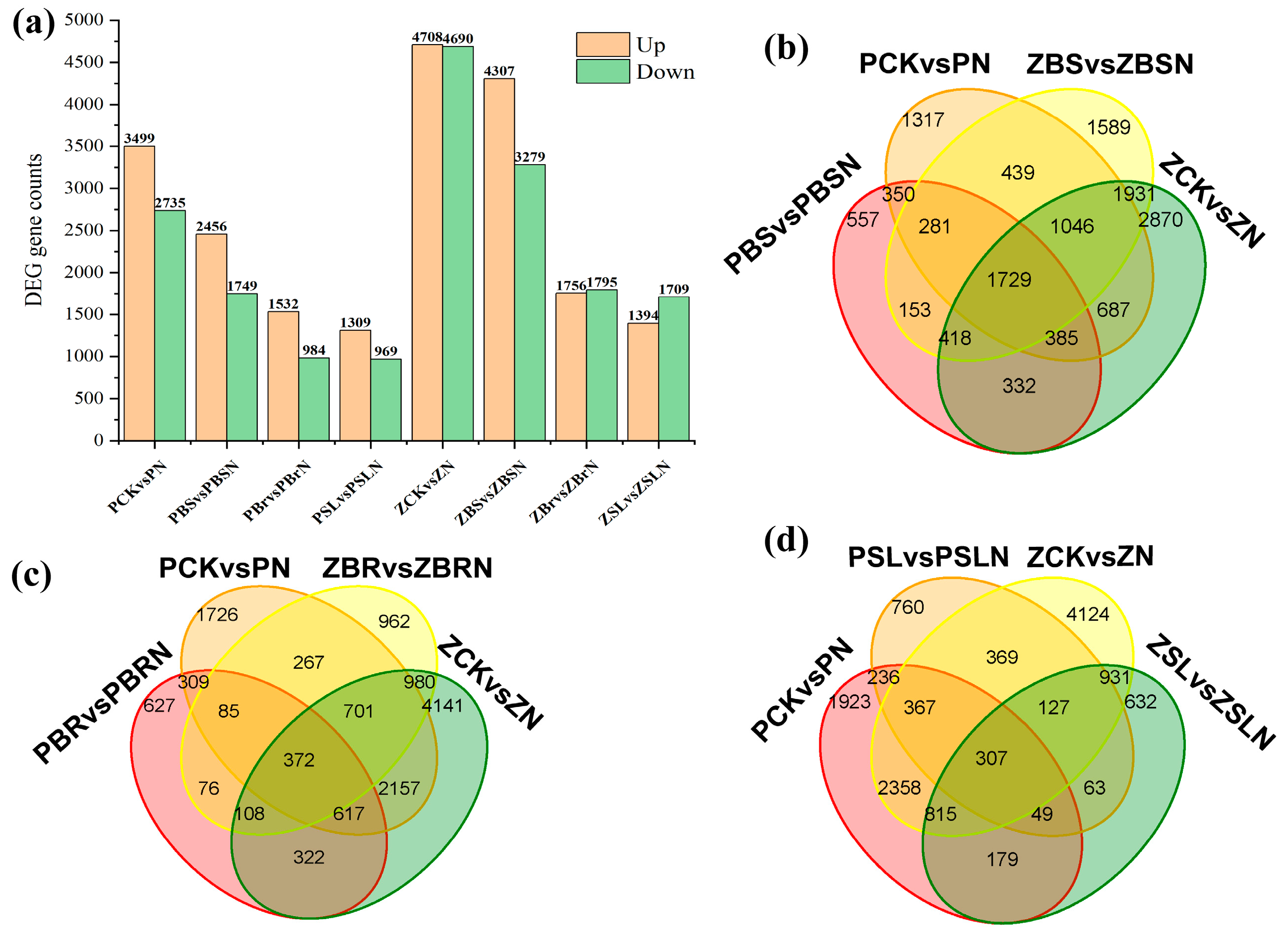
2.5. Differential Gene Analysis
2.6. GO Enrichment Analysis
2.7. KEGG Enrichment Analysis
2.8. Weighted Gene Co-Expression Network Analysis (WGCNA) Analysis
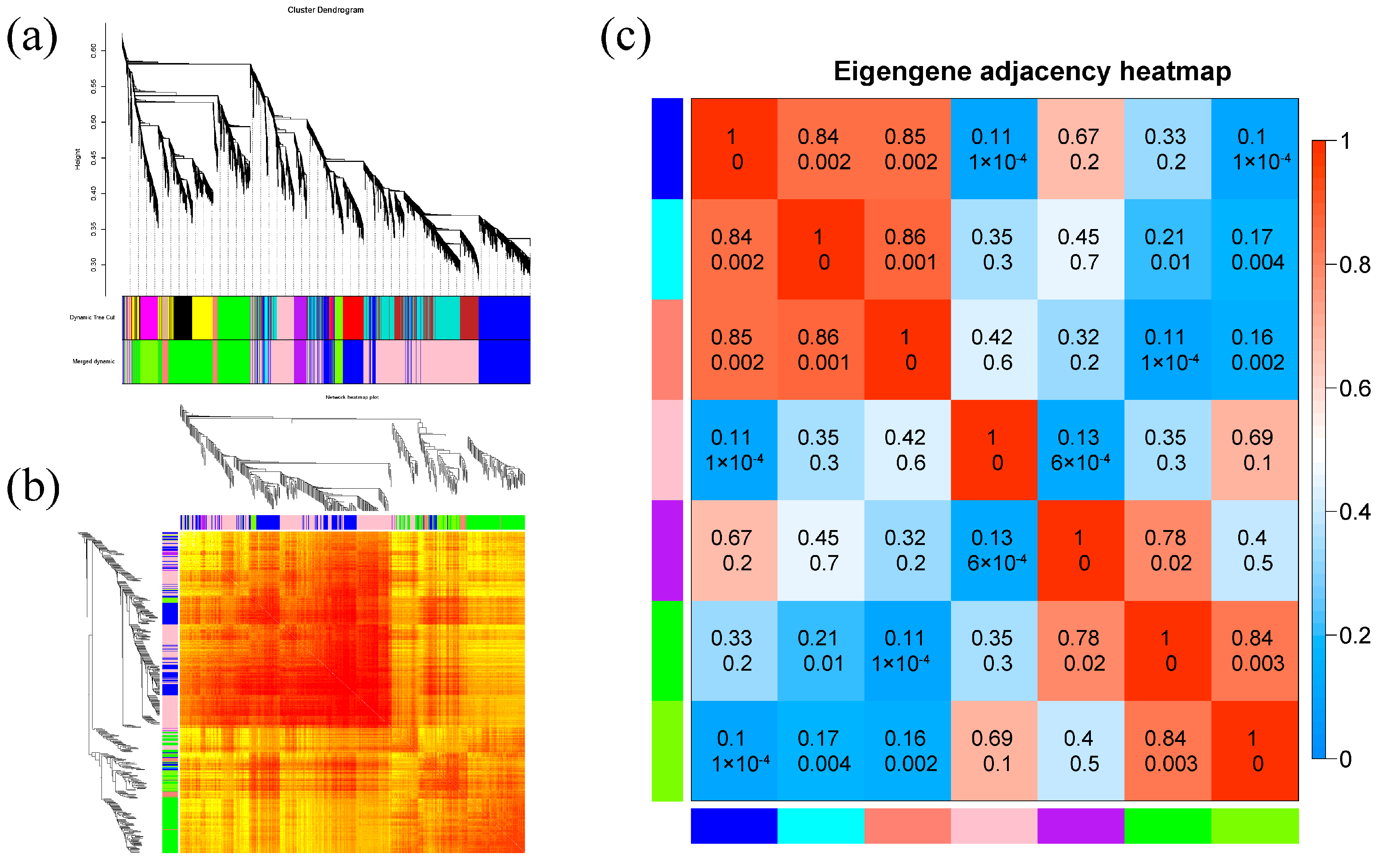
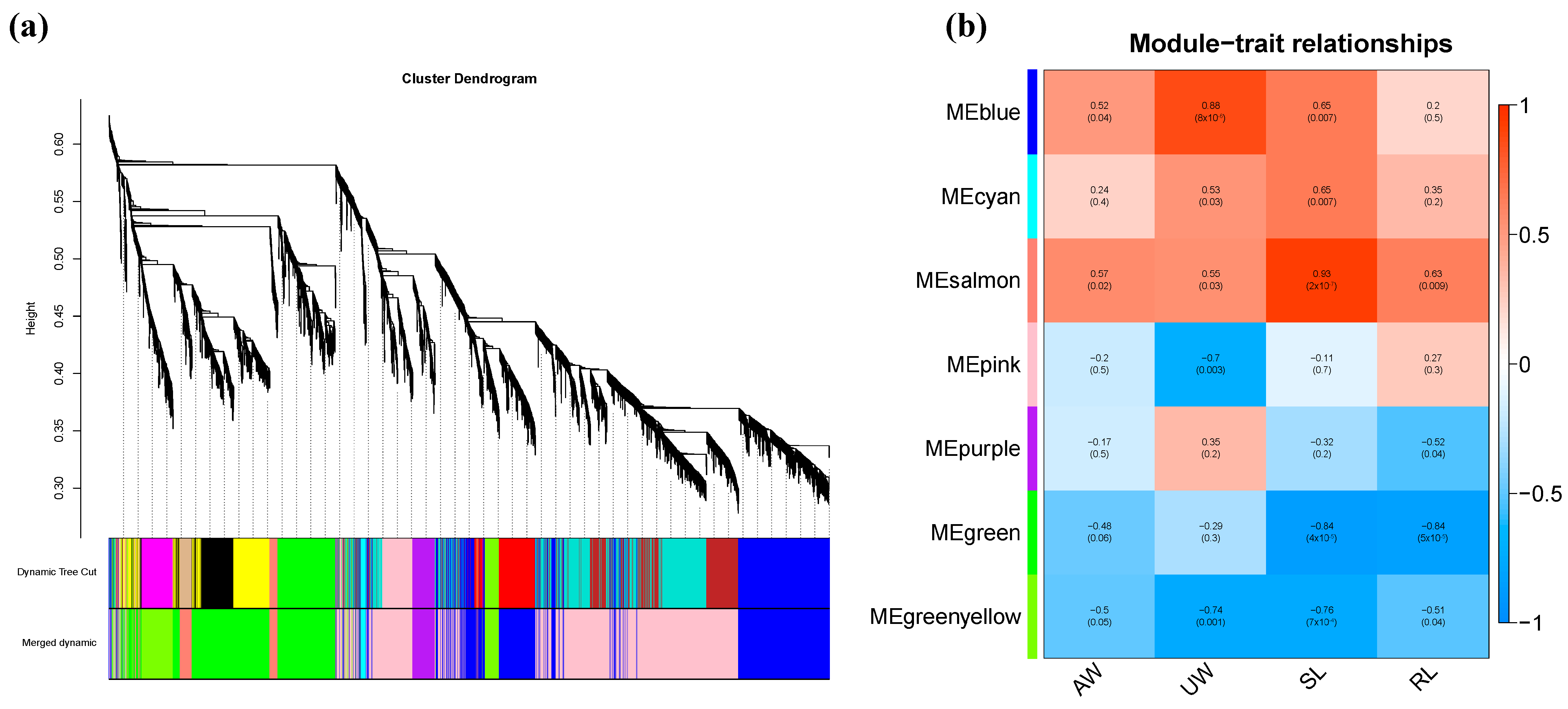
2.8.1. Construction of Gene Co-Expression Module
2.8.2. Analysis of the Correlation between Modules and Corresponding Phenotypic Characteristics
2.8.3. Functional Analysis of Gene Module
2.8.4. Visualization Analysis of Core Genes of the Gene Module
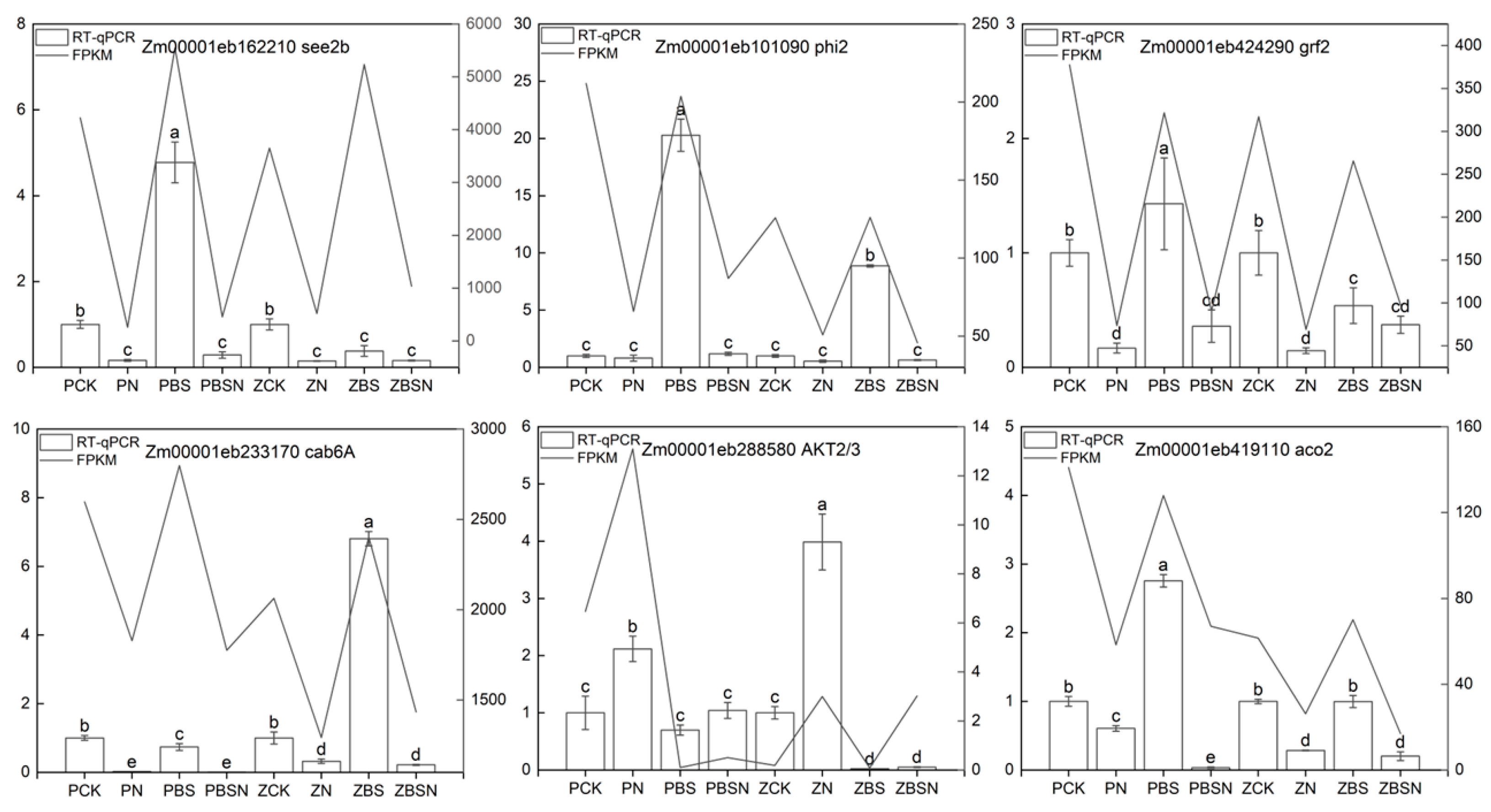
2.9. RT-qPCR Verification
3. Discussion
3.1. Physiological Response of Exogenous Hormones to Maize under Salt Stress
3.2. Analysis of Tolerance of Two Inbred Lines to Salt Stress
3.2.1. Effect of BR on Differential Gene Expression in Maize under Salt Stress
3.2.2. Effect of SLs on Differential Gene Expression in Maize under Salt Stress
3.2.3. Effect of BS on Differential Gene Expression in Maize under Salt Stress
4. Materials and Methods
4.1. Experimental Materials
4.2. Experimental Design and Treatment
4.3. Index Measurement
4.3.1. Growth Index Measurement
4.3.2. Measurement of Physiological Indexes
4.3.3. Determination of Ion Content
4.4. Transcriptomic Analysis
4.4.1. Total RNA Extraction and Illumina Deep Sequencing
4.4.2. Quality Assessment of Sequencing Results
4.4.3. Differential Expression Gene Analysis
4.4.4. Analysis of the Gene Co-Expression Network
4.4.5. Real-Time Fluorescence Quantitative PCR (RT-qPCR) Verification of DEGs
4.5. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mukhopadhyay, R.; Sarkar, B.; Jat, H.S.; Sharma, P.C.; Bolan, N.S. Soil salinity under climate change: Challenges for sustainable agriculture and food security. J. Environ. Manag. 2021, 280, 111736. [Google Scholar] [CrossRef] [PubMed]
- Del Buono, D. Can biostimulants be used to mitigate the effect of anthropogenic climate change on agriculture? It is time to respond. Sci. Total Environ. 2021, 751, 141763. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Zhou, X.; Tao, M.; Yuan, F.; Liu, L.; Wu, F.; Wu, X.; Xiang, Y.; Niu, Y.; Liu, F.; et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 2019, 572, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhu, Y.; Li, S.; Zhang, W.; Yin, C.; Lin, Y. Regulation of Phytohormones on the Growth and Development of Plant Root Hair. Front. Plant Sci. 2022, 13, 865302. [Google Scholar] [CrossRef]
- Ciccarelli, D.; Bottega, S.; Spano, C. Study of functional and physiological response of co-occurring shrub species to the Mediterranean climate. Saudi J. Biol. Sci. 2019, 26, 1668–1675. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef]
- Cortleven, A.; Leuendorf, J.E.; Frank, M.; Pezzetta, D.; Bolt, S.; Schmuelling, T. Cytokinin action in response to abiotic and biotic stresses in plants. Plant Cell Environ. 2019, 42, 998–1018. [Google Scholar] [CrossRef]
- Ku, Y.-S.; Sintaha, M.; Cheung, M.-Y.; Lam, H.-M. Plant Hormone Signaling Crosstalks between Biotic and Abiotic Stress Responses. Int. J. Mol. Sci. 2018, 19, 3206. [Google Scholar] [CrossRef]
- Tanveer, M.; Shahzad, B.; Sharma, A.; Biju, S.; Bhardwaj, R. 24-Epibrassinolide; an active brassinolide and its role in salt stress tolerance in plants: A review. Plant Physiol. Biochem. 2018, 130, 69–79. [Google Scholar] [CrossRef]
- Liu, L.; Sun, Y.; Di, P.; Cui, Y.; Meng, Q.; Wu, X.; Chen, Y.; Yuan, J. Overexpression of a Zea mays Brassinosteroid-Signaling Kinase Gene ZmBSK1 Confers Salt Stress Tolerance in Maize. Front. Plant Sci. 2022, 13, 894710. [Google Scholar] [CrossRef]
- Zeng, H.; Tang, Q.; Hua, X. Arabidopsis Brassinosteroid Mutants det2-1 and bin2-1 Display Altered Salt Tolerance. J. Plant Growth Regul. 2010, 29, 44–52. [Google Scholar] [CrossRef]
- Shang, Y.; Dai, C.; Lee, M.M.; Kwak, J.M.; Nam, K.H. BRI1-Associated Receptor Kinase 1 Regulates Guard Cell ABA Signaling Mediated by Open Stomata 1 in Arabidopsis. Mol. Plant 2016, 9, 447–460. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, X.; Wang, Q.; Chen, M.; Liu, X.; Gao, D.; Li, D.; Li, L. MdBZR1 and MdBZR1-2like Transcription Factors Improves Salt Tolerance by Regulating Gibberellin Biosynthesis in Apple. Front. Plant Sci. 2019, 10, 1473. [Google Scholar] [CrossRef]
- Que, F.; Khadr, A.; Wang, G.L.; Li, T.; Wang, Y.H.; Xu, Z.S.; Xiong, A.S. Exogenous brassinosteroids altered cell length, gibberellin content, and cellulose deposition in promoting carrot petiole elongation. Plant Sci. 2018, 277, 110–120. [Google Scholar] [CrossRef]
- Cook, C.E.; Whichard, L.P.; Turner, B.; Wall, M.E.; Egley, G.H. Germination of Witchweed (Striga lutea Lour.): Isolation and Properties of a Potent Stimulant. Science 1966, 154, 1189–1190. [Google Scholar] [CrossRef] [PubMed]
- Braun, N.; de Saint Germain, A.; Pillot, J.-P.; Boutet-Mercey, S.; Dalmais, M.; Antoniadi, I.; Li, X.; Maia-Grondard, A.; Le Signor, C.; Bouteiller, N.; et al. The Pea TCP Transcription Factor PsBRC1 Acts Downstream of Strigolactones to Control Shoot Branching. Plant Physiol. 2012, 158, 225–238. [Google Scholar] [CrossRef]
- Guan, J.C.; Koch, K.E.; Suzuki, M.; Wu, S.; Latshaw, S.; Petruff, T.; Goulet, C.; Klee, H.J.; McCarty, D.R. Diverse Roles of Strigolactone Signaling in Maize Architecture and the Uncoupling of a Branching-Specific Subnetwork. Plant Physiol. 2012, 160, 1303–1317. [Google Scholar] [CrossRef]
- Minakuchi, K.; Kameoka, H.; Yasuno, N.; Umehara, M.; Luo, L.; Kobayashi, K.; Hanada, A.; Ueno, K.; Asami, T.; Yamaguchi, S.; et al. FINE CULM1 (FC1) Works Downstream of Strigolactones to Inhibit the Outgrowth of Axillary Buds in Rice. Plant Cell Physiol. 2010, 51, 1127–1135. [Google Scholar] [CrossRef]
- Tian, M.-Q.; Jiang, K.; Takahashi, I.; Li, G.-D. Strigolactone-induced senescence of a bamboo leaf in the dark is alleviated by exogenous sugar. J. Pestic. Sci. 2018, 43, 173–179. [Google Scholar] [CrossRef]
- Xu, X.; Jibran, R.; Wang, Y.; Dong, L.; Flokova, K.; Esfandiari, A.; McLachlan, A.R.G.; Heiser, A.; Sutherland-Smith, A.J.; Brummell, D.A.; et al. Strigolactones regulate sepal senescence in Arabidopsis. J. Exp. Bot. 2021, 72, 5462–5477. [Google Scholar] [CrossRef]
- Seleiman, M.F.; Ahmad, A.; Tola, E.; Alhammad, B.A.; Almutairi, K.F.; Madugundu, R.; Al-Gaadi, K.A. Exogenous Application of 24-Epibrassinolide Confers Saline Stress and Improves Photosynthetic Capacity, Antioxidant Defense, Mineral Uptake, and Yield in Maize. Plants 2023, 12, 3559. [Google Scholar] [CrossRef]
- Iftikhar, I.; Shahbaz, M.; Wahid, M.A. Potential Role of Foliage Applied Strigolactone (GR24) on Photosynthetic Pigments, Gas Exchange Attributes, Mineral Nutrients and Yield Components of Zea mays (L.) Under Saline Regimes. Gesunde Pflanz. 2023, 75, 577–591. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Denaxa, N.-K.; Nomikou, A.; Malamos, N.; Liveri, E.; Roussos, P.A.; Papasotiropoulos, V. Salinity Effect on Plant Growth Parameters and Fruit Bioactive Compounds of Two Strawberry Cultivars, Coupled with Environmental Conditions Monitoring. Agronomy 2022, 12, 2279. [Google Scholar] [CrossRef]
- Weng, M.; Cui, L.; Liu, F.; Zhang, M.; Shan, L.; Yang, S.; Deng, X. Effects of Drought Stress on Antioxidant Enzymes in Seedlings of Different Wheat Genotypes. Pak. J. Bot. 2015, 47, 49–56. [Google Scholar]
- Xia, W.; Meng, W.; Peng, Y.; Qin, Y.; Zhang, L.; Zhu, N. Effects of Exogenous Isosteviol on the Physiological Characteristics of Brassica napus Seedlings under Salt Stress. Plants 2024, 13, 217. [Google Scholar] [CrossRef]
- Zulfiqar, H.; Shahbaz, M.; Ahsan, M.; Nafees, M.; Nadeem, H.; Akram, M.; Maqsood, A.; Ahmar, S.; Kamran, M.; Alamri, S.; et al. Strigolactone (GR24) Induced Salinity Tolerance in Sunflower (Helianthus annuus L.) by Ameliorating Morpho-Physiological and Biochemical Attributes Under In Vitro Conditions. J. Plant Growth Regul. 2020, 40, 2079–2091. [Google Scholar] [CrossRef]
- Kobayashi, N.I.; Yamaji, N.; Yamamoto, H.; Okubo, K.; Ueno, H.; Costa, A.; Tanoi, K.; Matsumura, H.; Fujii-Kashino, M.; Horiuchi, T.; et al. OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J. 2017, 91, 657–670. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.-H.; Gao, J.-P.; Li, L.-G.; Cai, X.-L.; Huang, W.; Chao, D.-Y.; Zhu, M.-Z.; Wang, Z.-Y.; Luan, S.; Lin, H.-X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 37, 1141–1146. [Google Scholar] [CrossRef]
- Chen, C.; Xu, L.; Zhang, X.; Wang, H.; Nisa, Z.U.; Jin, X.; Yu, L.; Jing, L.; Chen, C. Exogenous strigolactones enhance tolerance in soybean seedlings in response to alkaline stress. Physiol. Plant. 2022, 174, e13784. [Google Scholar] [CrossRef]
- Golldack, D.; Quigley, F.; Michalowski, C.B.; Kamasani, U.R.; Bohnert, H.J. Salinity stress-tolerant and -sensitive rice (Oryza sativa L.) regulate AKT1-type potassium channel transcripts differently. Plant Mol. Biol. 2003, 51, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Nanda, S.; Ashraf, M.; Siddiqui, A.; Masood, S.; Khaskheli, M.; Suleman, M.; Zhu, L.; Zhu, C.; Cao, X.; et al. Interplay Impact of Exogenous Application of Abscisic Acid (ABA) and Brassinosteroids (BRs) in Rice Growth, Physiology, and Resistance under Sodium Chloride Stress. Life 2023, 13, 498. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Q.; Ma, B.; Lu, X.; Huang, Y.-H.; He, S.-J.; Yang, C.; Yin, C.-C.; Zhao, H.; Zhou, Y.; Zhang, W.-K.; et al. Ethylene-Inhibited Jasmonic Acid Biosynthesis Promotes Mesocotyl/Coleoptile Elongation of Etiolated Rice Seedlings. Plant Cell 2017, 29, 1053–1072. [Google Scholar] [CrossRef] [PubMed]
- Fukazawa, J.; Ito, T.; Kamiya, Y.; Yamaguchi, S.; Takahashi, Y. Binding of GID1 to DELLAs promotes dissociation of GAF1 from DELLA in GA dependent manner. Plant Signal. Behav. 2015, 10, e1052923. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Chiang, Y.H.; Kieber, J.J.; Schaller, G.E. SCF(KMD) controls cytokinin signaling by regulating the degradation of type-B response regulators. Proc. Natl. Acad. Sci. USA 2013, 110, 10028–10033. [Google Scholar] [CrossRef]
- Xie, M.; Chen, H.; Huang, L.; O’Neil, R.C.; Shokhirev, M.N.; Ecker, J.R. A B-ARR-mediated cytokinin transcriptional network directs hormone cross-regulation and shoot development. Nat. Commun. 2018, 9, 1604. [Google Scholar] [CrossRef]
- Shakeel, S.N.; Gao, Z.; Amir, M.; Chen, Y.-F.; Rai, M.I.; Haq, N.U.; Schaller, G.E. Ethylene Regulates Levels of Ethylene Receptor/CTR1 Signaling Complexes in Arabidopsis thaliana. J. Biol. Chem. 2015, 290, 12415–12424. [Google Scholar] [CrossRef]
- Zhu, T.; Deng, X.; Zhou, X.; Zhu, L.; Zou, L.; Li, P.; Zhang, D.; Lin, H. Ethylene and hydrogen peroxide are involved in brassinosteroid-induced salt tolerance in tomato. Sci. Rep. 2016, 6, 35392. [Google Scholar] [CrossRef]
- Wilson, R.L.; Kim, H.; Bakshi, A.; Binder, B.M. The Ethylene Receptors ETHYLENE RESPONSE1 and ETHYLENE RESPONSE2 Have Contrasting Roles in Seed Germination of Arabidopsis during Salt Stress. Plant Physiol. 2014, 165, 1353–1366. [Google Scholar] [CrossRef]
- Zhang, C.; He, M.L.; Jiang, Z.X.; Liu, L.; Pu, J.B.; Zhang, W.J.; Wang, S.L.; Xu, F.S. The Xyloglucan Endotransglucosylase/Hydrolase Gene Regulates Plant Growth by Disrupting the Cell Wall Homeostasis in under Boron Deficiency. Int. J. Mol. Sci. 2022, 23, 1250. [Google Scholar] [CrossRef]
- Kasirajan, L.; Hoang, N.V.; Furtado, A.; Botha, F.C.; Henry, R.J. Transcriptome analysis highlights key differentially expressed genes involved in cellulose and lignin biosynthesis of sugarcane genotypes varying in fiber content. Sci. Rep. 2018, 8, 11612. [Google Scholar] [CrossRef] [PubMed]
- Weiste, C.; Pedrotti, L.; Selvanayagam, J.; Muralidhara, P.; Froeschel, C.; Novak, O.; Ljung, K.; Hanson, J.; Droege-Laser, W. The Arabidopsis bZIP11 transcription factor links low-energy signalling to auxin-mediated control of primary root growth. PLoS Genet. 2017, 13, e1006607. [Google Scholar] [CrossRef]
- Gutierrez, L.; Mongelard, G.; Floková, K.; Păcurar, D.I.; Novák, O.; Staswick, P.; Kowalczyk, M.; Păcurar, M.; Demailly, H.; Geiss, G.; et al. Auxin Controls Arabidopsis Adventitious Root Initiation by Regulating Jasmonic Acid Homeostasis. Plant Cell 2012, 24, 2515–2527. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Huang, D.; Lin, X. Interlinked regulator loops of ABA and JA respond to salt and drought stress in Caragana korshinskii. Environ. Exp. Bot. 2024, 225, 105829. [Google Scholar] [CrossRef]
- Jia, X.-M.; Wang, H.; Svetla, S.; Zhu, Y.-F.; Hu, Y.; Cheng, L.; Zhao, T.; Wang, Y.-X. Comparative physiological responses and adaptive strategies of apple Malus halliana to salt, alkali and saline-alkali stress. Sci. Hortic. 2019, 245, 154–162. [Google Scholar] [CrossRef]
- Yuan, Y.; Xu, X.; Gong, Z.; Tang, Y.; Wu, M.; Yan, F.; Zhang, X.; Zhang, Q.; Yang, F.; Hu, X.; et al. Auxin response factor 6A regulates photosynthesis, sugar accumulation, and fruit development in tomato. Hortic. Res. 2019, 6, 85. [Google Scholar] [CrossRef]
- Ye, J.-J.; Lin, X.-Y.; Yang, Z.-X.; Wang, Y.-Q.; Liang, Y.-R.; Wang, K.-R.; Lu, J.-L.; Lu, P.; Zheng, X.-Q. The light-harvesting chlorophyll a/b-binding proteins of photosystem II family members are responsible for temperature sensitivity and leaf color phenotype in albino tea plant. J. Adv. Res. 2023; online ahead of print. [Google Scholar] [CrossRef]
- Silva, J.; Kim, Y.J.; Sukweenadhi, J.; Rahimi, S.; Kwon, W.S.; Yang, D.C. Molecular characterization of 5-chlorophyll a/b-binding protein genes from Panax ginseng Meyer and their expression analysis during abiotic stresses. Photosynthetica 2016, 54, 446–458. [Google Scholar] [CrossRef]
- Meng, L.; Fan, Z.; Zhang, Q.; Wang, C.; Gao, Y.; Deng, Y.; Zhu, B.; Zhu, H.; Chen, J.; Shan, W.; et al. BEL1-LIKE HOMEODOMAIN 11 regulates chloroplast development and chlorophyll synthesis in tomato fruit. Plant J. 2018, 94, 1126–1140. [Google Scholar] [CrossRef]
- Liang, D.; Shen, Y.; Ni, Z.; Wang, Q.; Lei, Z.; Xu, N.; Deng, Q.; Lin, L.; Wang, J.; Lv, X.; et al. Exogenous Melatonin Application Delays Senescence of Kiwifruit Leaves by Regulating the Antioxidant Capacity and Biosynthesis of Flavonoids. Front. Plant Sci. 2018, 9, 426. [Google Scholar] [CrossRef] [PubMed]
- Tuan, P.A.; Shafai, T.; Kaur, G.; Grenier, G.; Ayele, B.T. Molecular and functional characterization of a jasmonate resistant gene of wheat (Triticum aestivum L.). J. Plant Physiol. 2022, 270, 153637. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, Z. PIF4 and PIF4-Interacting Proteins: At the Nexus of Plant Light, Temperature and Hormone Signal Integrations. Int. J. Mol. Sci. 2021, 22, 10304. [Google Scholar] [CrossRef]
- Yang, Z.; Yan, B.; Dong, H.; He, G.; Zhou, Y.; Sun, J. BIC1 acts as a transcriptional coactivator to promote brassinosteroid signaling and plant growth. EMBO J. 2021, 40, e104615. [Google Scholar] [CrossRef] [PubMed]
- Martinez, C.; Espinosa-Ruiz, A.; de Lucas, M.; Bernardo-Garcia, S.; Franco-Zorrilla, J.M.; Prat, S. PIF4-induced BR synthesis is critical to diurnal and thermomorphogenic growth. EMBO J. 2018, 37, e99552. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, W.-H. Potassium Transport and Signaling in Higher Plants. Annu. Rev. Plant Biol. 2013, 64, 451–476. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Murillo, F.; Marin-Guirao, L.; Sola, I.; Carbonell-Garzon, E.; Rodriguez-Rojas, F.; Sanchez-Lizaso, J.L.; Saez, C.A. Metabolic responses to desalination brine discharges in field-transplanted Posidonia oceanica: Advances for the development of specific early warning biomarkers. Desalination 2024, 576, 117395. [Google Scholar] [CrossRef]
- Chérel, I.; Michard, E.; Platet, N.; Mouline, K.; Alcon, C.; Sentenac, H.; Thibaud, J.-B. Physical and Functional Interaction of the Arabidopsis K+ Channel AKT2 and Phosphatase AtPP2CA. Plant Cell 2002, 14, 1133–1146. [Google Scholar] [CrossRef]
- Deeken, R.; Geiger, D.; Fromm, J.; Koroleva, O.; Ache, P.; Langenfeld-Heyser, R.; Sauer, N.; May, S.T.; Hedrich, R. Loss of the AKT2/3 potassium channel affects sugar loading into the phloem of Arabidopsis. Planta 2002, 216, 334–344. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Y.; Li, M.; Jia, H.; Wei, F.; Xia, Z.; Zhang, X.; Chang, J.; Wang, Z. Overexpression of the WRKY transcription factor gene NtWRKY65 enhances salt tolerance in tobacco (Nicotiana tabacum). BMC Plant Biol. 2024, 24, 326. [Google Scholar] [CrossRef]
- Wang, X.; Li, Z.; Shi, Y.; Liu, Z.; Zhang, X.; Gong, Z.; Yang, S. Strigolactones promote plant freezing tolerance by releasing the WRKY41-mediated inhibition of CBF/DREB1 expression. EMBO J. 2023, 42, e112999. [Google Scholar] [CrossRef]
- Li, G.D.; Pan, L.N.; Jiang, K.; Takahashi, I.; Nakamura, H.; Xu, Y.W.; Asami, T.; Shen, R.F. Strigolactones are involved in sugar signaling to modulate early seedling development in Arabidopsis. Plant Biotechnol. 2016, 33, 87–97. [Google Scholar] [CrossRef]
- Lu, X.; Liu, X.; Xu, J.; Liu, Y.; Chi, Y.; Yu, W.; Li, C. Strigolactone-Mediated Trehalose Enhances Salt Resistance in Tomato Seedlings. Horticulturae 2023, 9, 770. [Google Scholar] [CrossRef]
- Bertheloot, J.; Barbier, F.; Boudon, F.; Perez-Garcia, M.D.; Peron, T.; Citerne, S.; Dun, E.; Beveridge, C.; Godin, C.; Sakr, S. Sugar availability suppresses the auxin-induced strigolactone pathway to promote bud outgrowth. New Phytol. 2020, 225, 866–879. [Google Scholar] [CrossRef] [PubMed]
- Tal, L.; Palayam, M.; Ron, M.; Young, A.; Britt, A.; Shabek, N. A conformational switch in the SCF-D3/MAX2 ubiquitin ligase facilitates strigolactone signalling. Nat. Plants 2022, 8, 561–573. [Google Scholar] [CrossRef]
- Zhang, J.; Kirkham, M.B. Enzymatic responses of the ascorbate-glutathione cycle to drought in sorghum and sunflower plants. Plant Sci. 1996, 113, 139–147. [Google Scholar] [CrossRef]
- Hanson, D.; Horneck, D.A. Determination of Potassium and Sodium by Flame Emission Spectrophotometry. 1997. Available online: https://www.semanticscholar.org/paper/Determination-Of-Potassium-And-Sodium-By-Flame-Hanson-Horneck/af88bc4b730fc03dfee5517711e2ca2fbe8fd541 (accessed on 20 August 2024).
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]






| Treatments | SOD Activity (U/g) | POD Activity (U/g) | CAT Activity (U/g) |
|---|---|---|---|
| ZCK | 142.336 ± 18.597 e | 29.778 ± 3.031 e | 0.76 ± 0.062 e |
| ZN | 254.594 ± 6.1 c | 116 ± 14.967 cd | 2.033 ± 0.21 d |
| ZBS | 196.502 ± 10.565 d | 124.445 ± 22.732 d | 2.56 ± 0.333 d |
| ZBSN | 273.489 ± 12.886 bc | 233.334 ± 31.085 b | 4.66 ± 0.086 bc |
| ZSL | 192.889 ± 3.979 d | 182.667 ± 31.284 c | 4.94 ± 0.571 b |
| ZSLN | 297.119 ± 8.247 b | 206.223 ± 20.963 b | 5.96 ± 0.353 a |
| ZBR | 152.384 ± 12.616 e | 135.556 ± 22.985 cd | 2.18 ± 0.174 d |
| ZBRN | 337.839 ± 15.161 a | 276.889 ± 8.555 a | 3.94 ± 0.405 c |
| PCK | 131.707 ± 7.903 e | 85.528 ± 3.195 d | 1.227 ± 0.029 b |
| PN | 290.244 ± 5.174 b | 123.772 ± 18.504 bc | 3.013 ± 0.171 b |
| PBS | 185.366 ± 7.903 d | 163.355 ± 22.709 c | 1.693 ± 0.17 b |
| PBSN | 307.317 ± 10.77 b | 202.895 ± 4.881 b | 3.334 ± 0.177 b |
| PSL | 178.049 ± 7.903 d | 192.74 ± 25.232 bc | 1.687 ± 0.108 a |
| PSLN | 292.683 ± 13.021 b | 256.494 ± 11.281 a | 2.187 ± 0.064 ab |
| PBR | 241.463 ± 15.807 c | 182.655 ± 8.654 bc | 1.767 ± 0.153 b |
| PBRN | 358.537 ± 7.9033 a | 198.794 ± 3.289 b | 2.32 ± 0.11 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Qi, X.; Zhuang, Z.; Bian, J.; Li, J.; Chen, J.; Li, Z.; Peng, Y. Interactions between Brassinosteroids and Strigolactones in Alleviating Salt Stress in Maize. Int. J. Mol. Sci. 2024, 25, 10505. https://doi.org/10.3390/ijms251910505
Wang X, Qi X, Zhuang Z, Bian J, Li J, Chen J, Li Z, Peng Y. Interactions between Brassinosteroids and Strigolactones in Alleviating Salt Stress in Maize. International Journal of Molecular Sciences. 2024; 25(19):10505. https://doi.org/10.3390/ijms251910505
Chicago/Turabian StyleWang, Xinqi, Xue Qi, Zelong Zhuang, Jianwen Bian, Jiawei Li, Jiangtao Chen, Zhiming Li, and Yunling Peng. 2024. "Interactions between Brassinosteroids and Strigolactones in Alleviating Salt Stress in Maize" International Journal of Molecular Sciences 25, no. 19: 10505. https://doi.org/10.3390/ijms251910505
APA StyleWang, X., Qi, X., Zhuang, Z., Bian, J., Li, J., Chen, J., Li, Z., & Peng, Y. (2024). Interactions between Brassinosteroids and Strigolactones in Alleviating Salt Stress in Maize. International Journal of Molecular Sciences, 25(19), 10505. https://doi.org/10.3390/ijms251910505





