Transcriptomic Approach Reveals Contrasting Patterns of Differential Gene Expression during Tannin Biodegredation by Aspergillus tubingensis in Liquid and Solid Cultures
Abstract
:1. Introduction
2. Results
2.1. The Morphology of A. tubingensis in Different Cultures
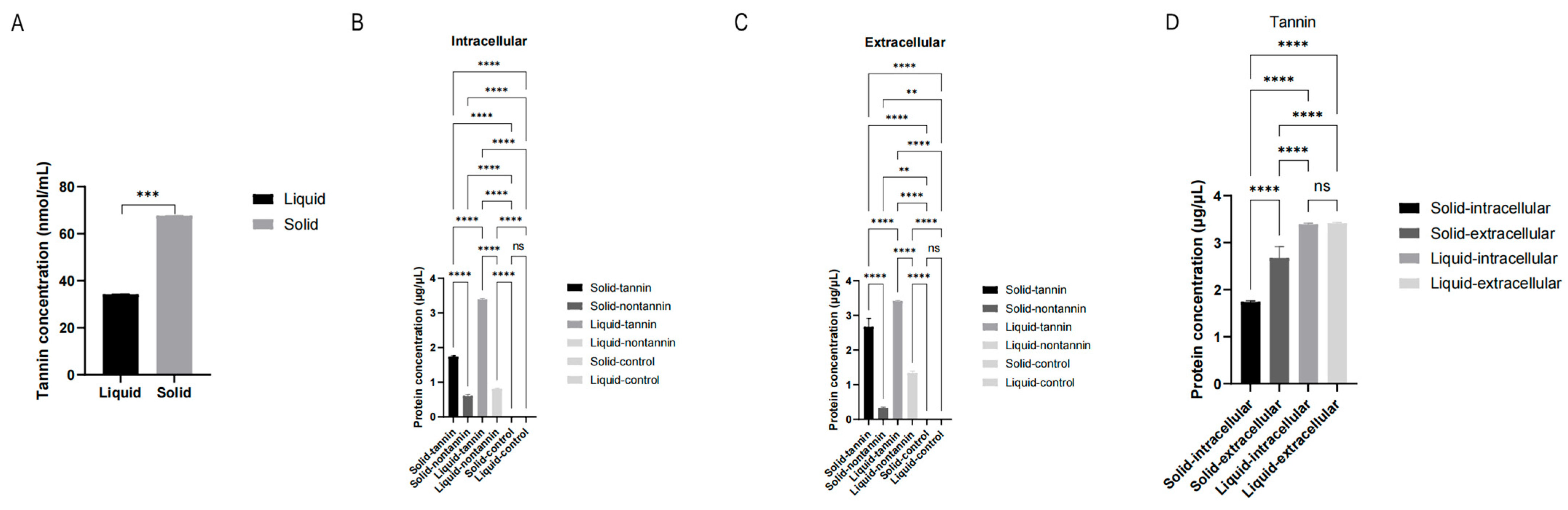
2.2. The Efficiency of Tannin Degradation by A. tubingensis in Different Cultures
2.3. The Protein Concentration Secreted by A. tubingensis in Different Cultures
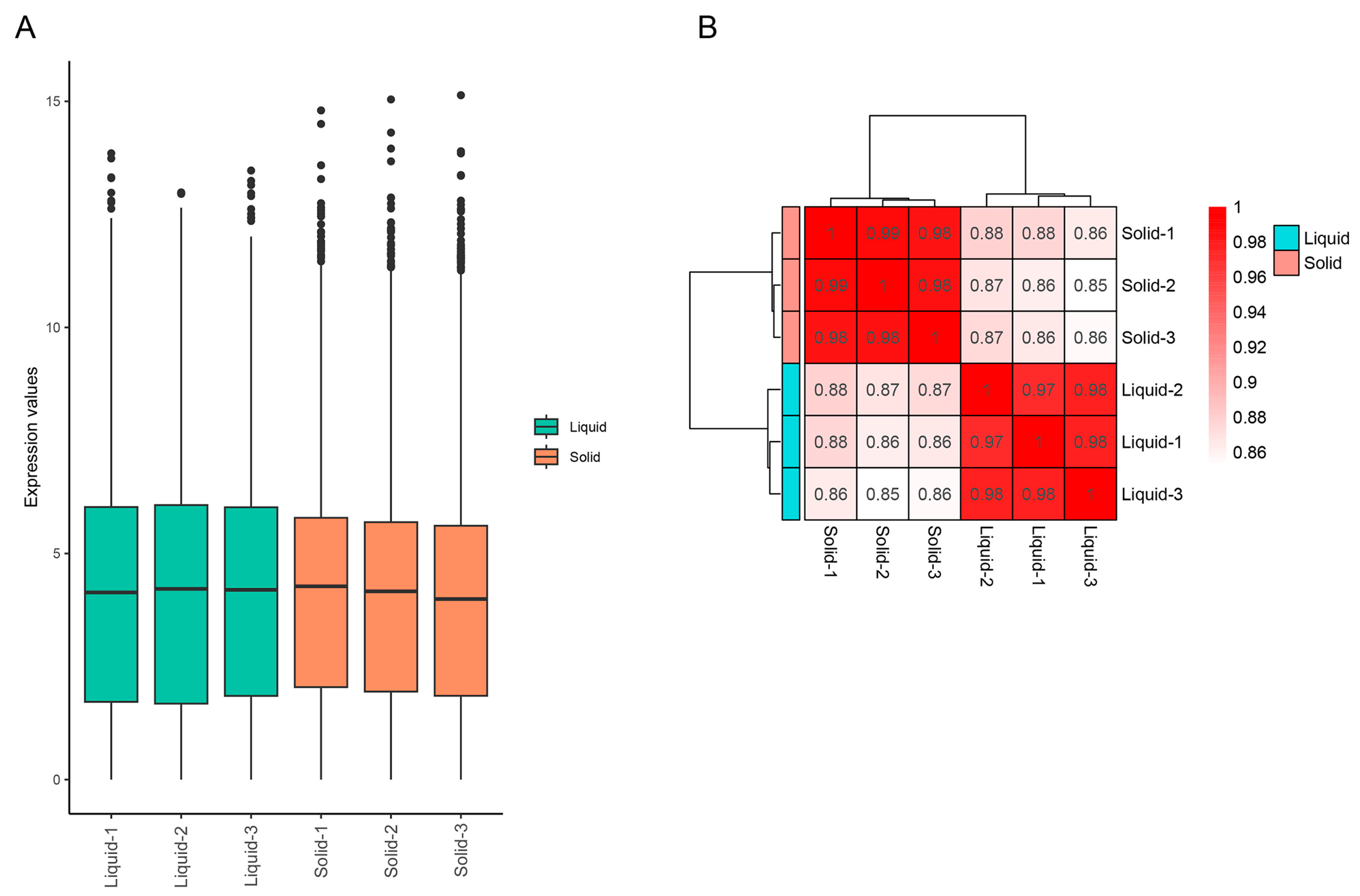
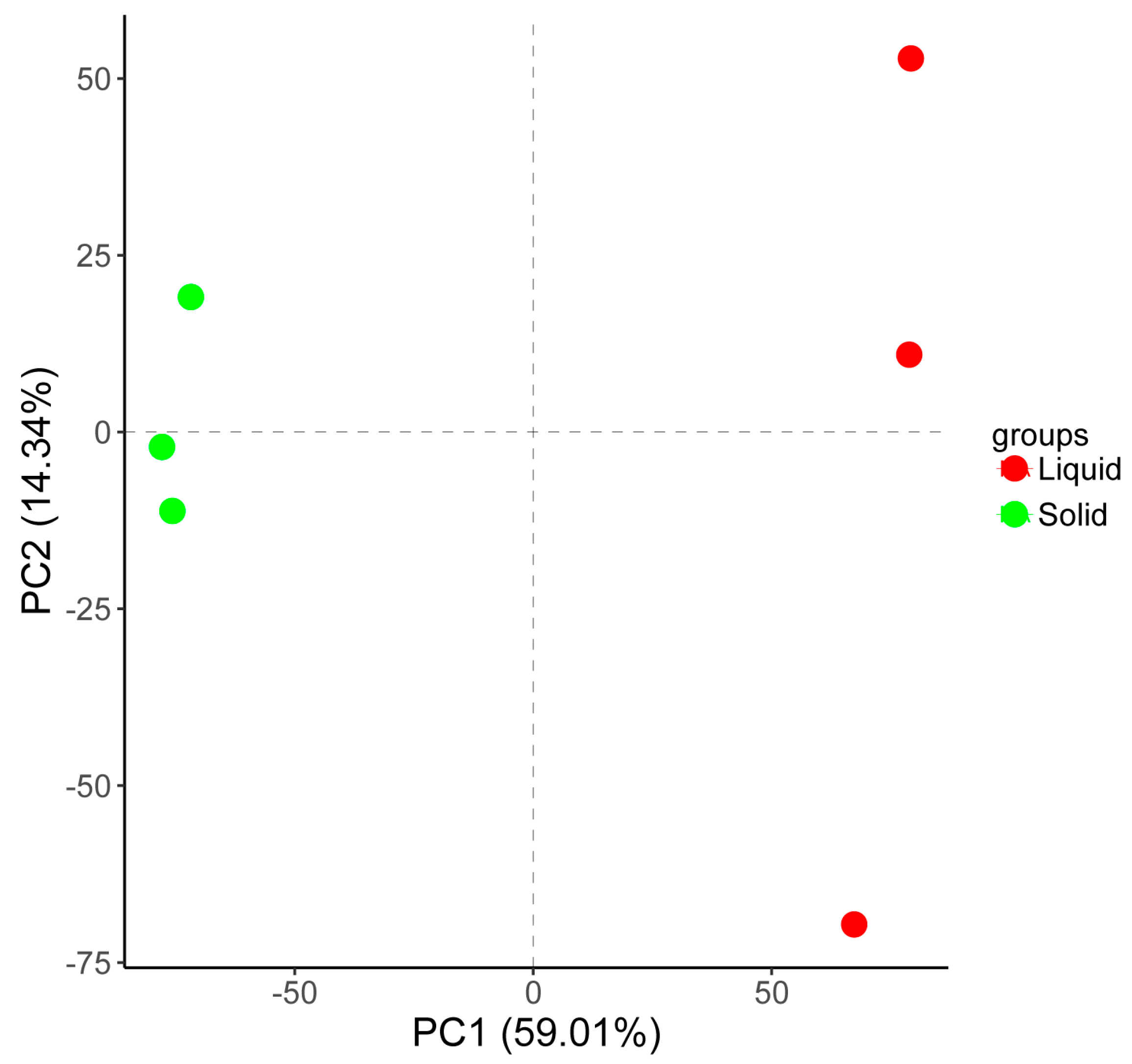
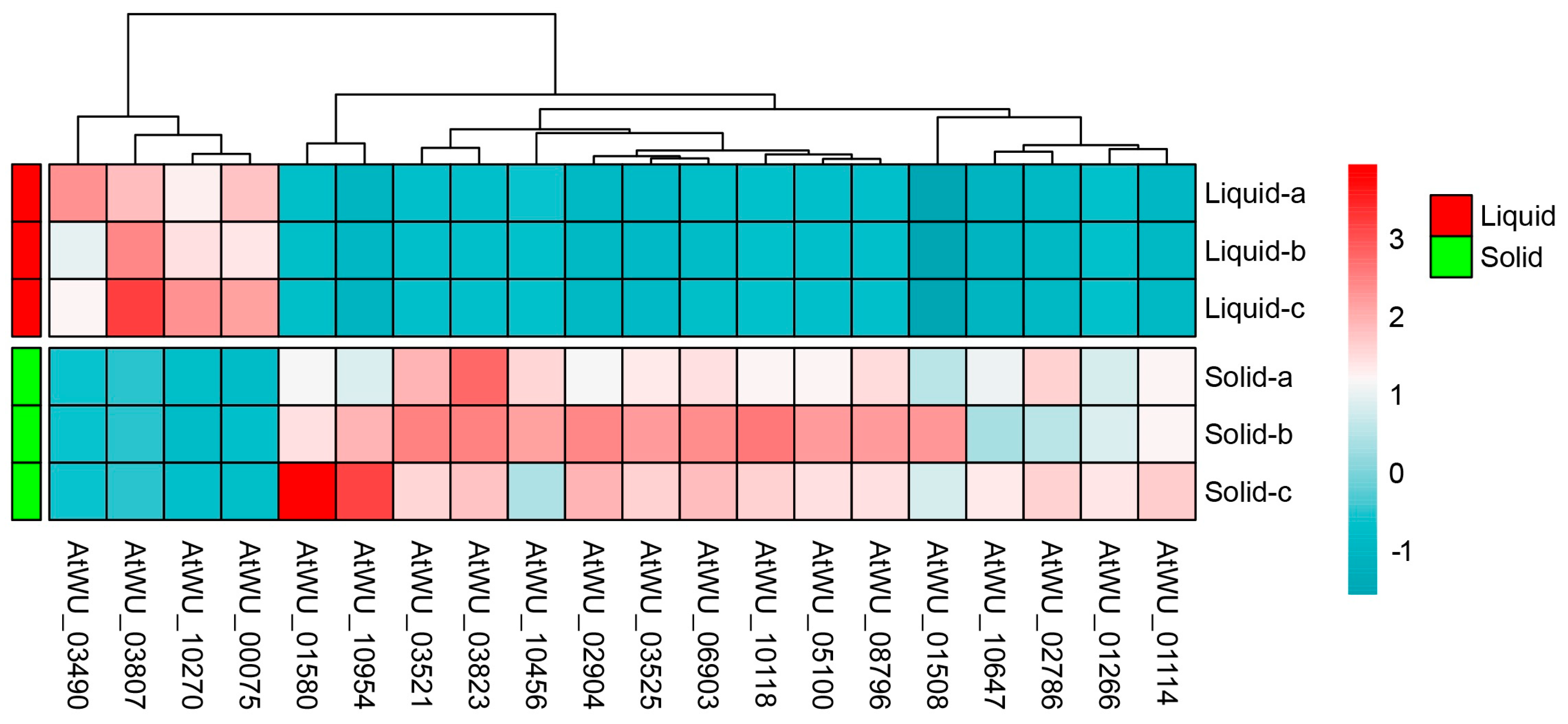
2.4. The Transcriptomic Responses to Tannin Biodegredation of A. tubingensis
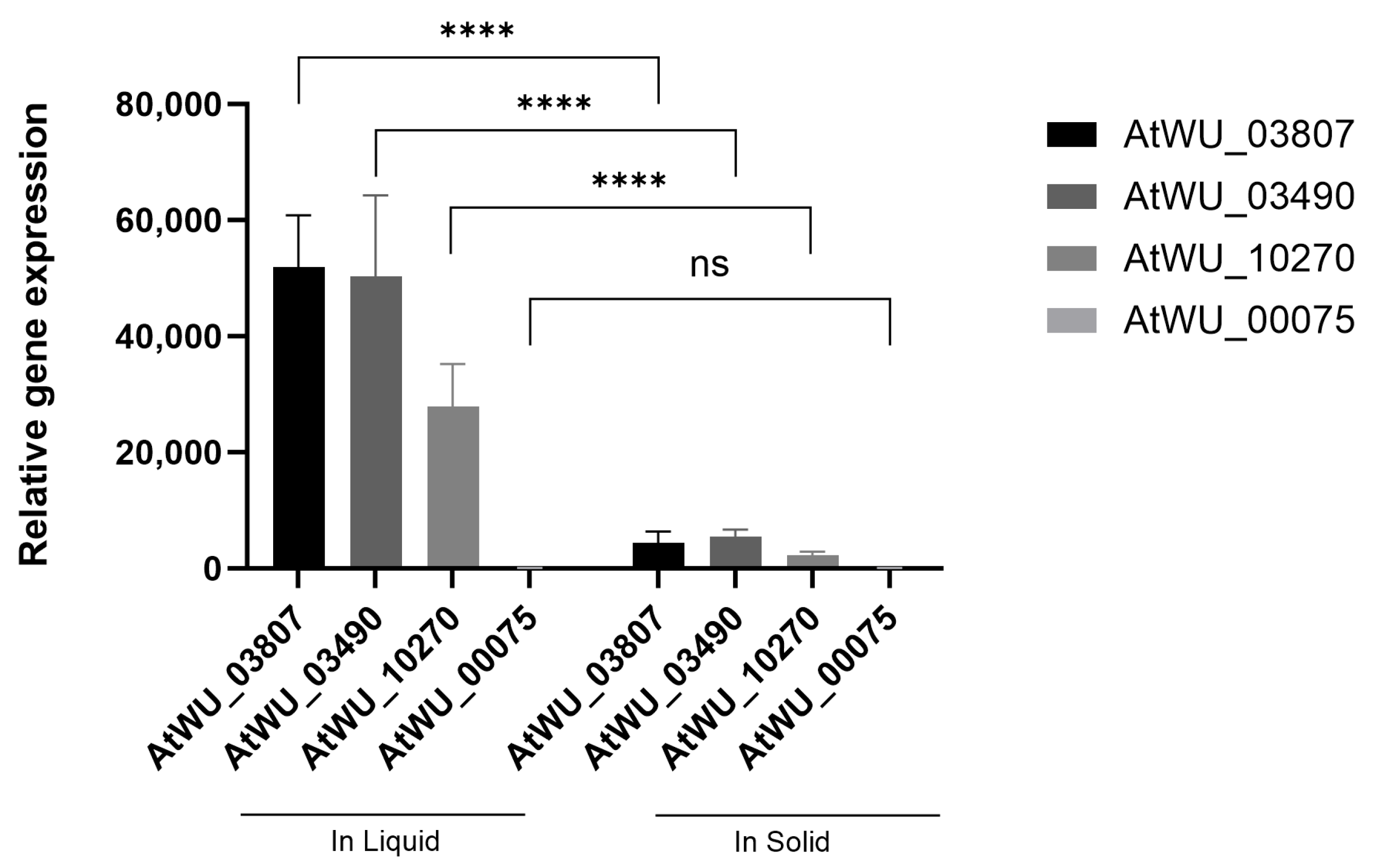
2.5. Validation of RNA-Seq Results by RT-qPCR
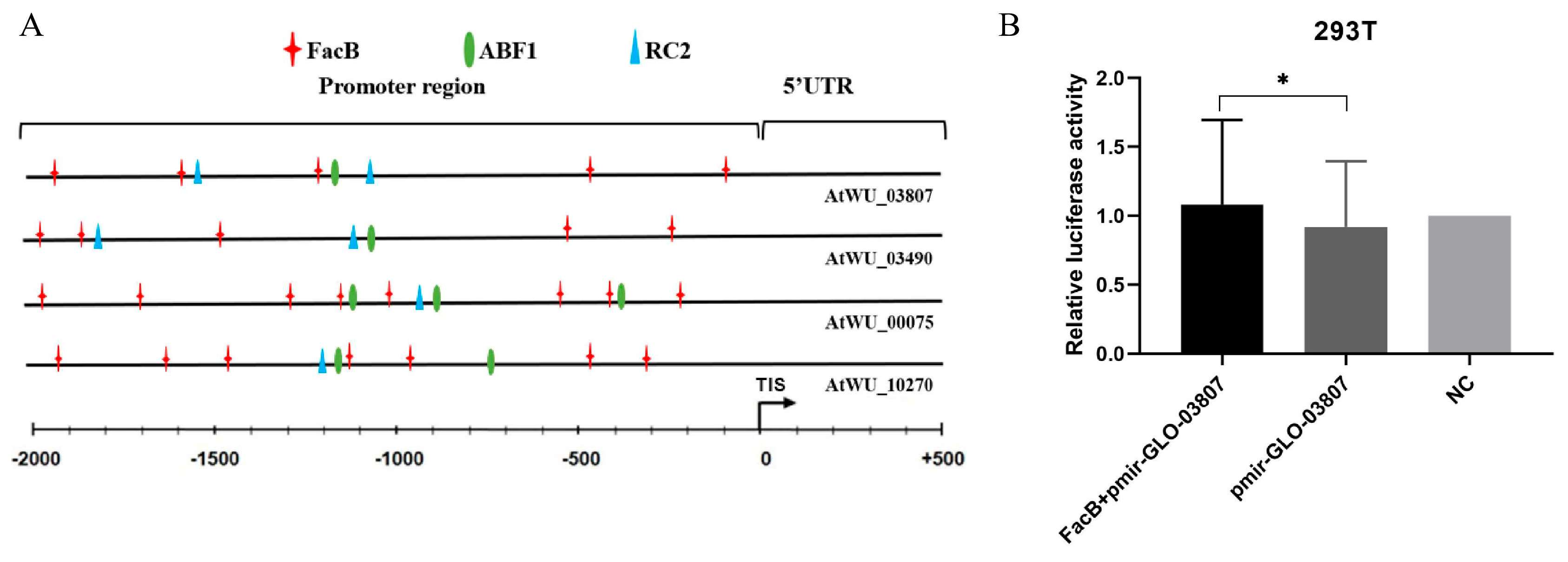
2.6. Confirmation of Transcription Factor by Dual-Luciferase Reporter Assay
2.7. Formatting of Mathematical Components
3. Discussion
4. Materials and Methods
4.1. Strain and Culture Conditions
4.2. Mycelia Harvest and Electron Microscopy Observation
4.3. Tannin and Protein Quantification Assays
4.4. RNA Extraction and Transcriptome Sequencing
4.5. Validation by RT- qPCR
4.6. Dual-Luciferase Reporter Gene Assay
4.7. Data Availability
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Besharati, M.; Maggiolino, A.; Palangi, V.; Kaya, A.; Jabbar, M.; Eseceli, H.; De Palo, P.; Lorenzo, J.M. Tannin in Ruminant Nutrition: Review. Molecules 2022, 27, 8273. [Google Scholar] [CrossRef]
- Duraiswamy, A.; Sneha, A.N.M.; Jebakani, K.S.; Selvaraj, S.; Pramitha, J.L.; Selvaraj, R.; Kumar, P.R.; Sheriff, S.K.; Thinakaran, J.; Rathinamoorthy, S.; et al. Genetic manipulation of anti-nutritional factors in major crops for a sustainable diet in future. Front. Plant Sci. 2023, 13, 1070398. [Google Scholar] [CrossRef]
- Cireșan, C.-A.; Cocan, I.; Alexa, E.; Cărpinișan, L.; Sîrbu, C.B.; Obiștioiu, D.; Jitea, B.A.-M.; Florea, T.; Dărăbuș, G. Research on the Control of Gastrointestinal Strongyles in Sheep by Using Lotus corniculatus or Cichorium intybus in Feed. Pathogens 2023, 12, 986. [Google Scholar] [CrossRef]
- Tian, K.E.; Aldian, D.; Luo, G.; Sossou, A.; Yayota, M. Condensed tannin-induced variations in the rumen metabolome and the correlation with fermentation characteristics in goats. Anim. Sci. J. 2024, 95, e13925. [Google Scholar] [CrossRef]
- Badjona, A.; Bradshaw, R.; Millman, C.; Howarth, M.; Dubey, B. Faba Bean Processing: Thermal and Non-Thermal Processing on Chemical, Antinutritional Factors, and Pharmacological Properties. Molecules 2023, 28, 5431. [Google Scholar] [CrossRef]
- Cui, R.; Zhang, C.; Pan, Z.H.; Hu, T.G.; Wu, H. Probiotic-fermented edible herbs as functional foods: A review of current status, challenges, and strategies. Compr. Rev. Food Sci. Food Saf. 2024, 23, e13305. [Google Scholar] [CrossRef]
- Tachie, C.Y.; Onuh, J.O.; Aryee, A.N.A. Nutritional and potential health benefits of fermented food proteins. J. Sci. Food Agric. 2023, 104, 1223–1233. [Google Scholar] [CrossRef]
- Aguilera-Carbo, A.; Augur, C.; Prado-Barragan, L.A.; Favela-Torres, E.; Aguilar, C.N. Microbial production of ellagic acid and biodegradation of ellagitannins. Appl. Microbiol. Biotechnol. 2008, 78, 189–199. [Google Scholar] [CrossRef]
- Rogowska-van, D.M.M.; Berasategui-Lopez, A.; Coolen, S.; Jansen, R.S.; Welte, C.U. Microbial degradation of plant toxins. Environ. Microbiol. 2023, 25, 2988–3010. [Google Scholar] [CrossRef]
- Jiménez, N.; Barcenilla, J.M.; de Felipe, F.L.; Rivas, B.d.L.; Muñoz, R. Characterization of a bacterial tannase from Streptococcus gallolyticus UCN34 suitable for tannin biodegradation. Appl. Microbiol. Biotechnol. 2014, 98, 6329–6337. [Google Scholar] [CrossRef]
- Dhiman, S.; Mukherjee, G.; Singh, A.K. Recent trends and advancements in microbial tannase-catalyzed biotransformation of tannins: A review. Int. Microbiol. 2018, 21, 175–195. [Google Scholar] [CrossRef]
- Prigione, V.; Spina, F.; Tigini, V.; Giovando, S.; Varese, G.C. Biotransformation of industrial tannins by filamentous fungi. Appl. Microbiol. Biotechnol. 2018, 102, 10361–10375. [Google Scholar] [CrossRef]
- Osete-Alcaraz, A.; Gómez-Plaza, E.; Martínez-Pérez, P.; Weiller, F.; Schückel, J.; Willats, W.G.; Moore, J.P.; Ros-García, J.M.; Bautista-Ortín, A.B. The Influence of Hydrolytic Enzymes on Tannin Adsorption-Desorption onto Grape Cell Walls in a Wine-Like Matrix. Molecules 2021, 26, 770. [Google Scholar] [CrossRef]
- Ristinmaa, A.S.; Coleman, T.; Cesar, L.; Weinmann, A.L.; Mazurkewich, S.; Brändén, G.; Hasani, M.; Larsbrink, J. Structural diversity and substrate preferences of three tannase enzymes encoded by the anaerobic bacterium Clostridium butyricum. J. Biol. Chem. 2022, 298, 101758. [Google Scholar] [CrossRef]
- Borin, G.P.; Sanchez, C.C.; De Souza, A.P.; De Santana, E.S.; De Souza, A.T.; Leme, A.F.P.; Squina, F.M.; Buckeridge, M.; Goldman, G.H.; De Castro Oliveira, J.V. Comparative Secretome Analysis of Trichoderma reesei and Aspergillus niger during Growth on Sugarcane Biomass. PLoS ONE 2015, 10, e0129275. [Google Scholar] [CrossRef]
- Zhang, N.; Jiang, J.-C.; Yang, J.; Wei, M.; Zhao, J.; Xu, H.; Xie, J.-C.; Tong, Y.-J.; Yu, L. Citric Acid Production from Acorn Starch by Tannin Tolerance Mutant Aspergillus niger AA120. Appl. Biochem. Biotechnol. 2019, 188, 1–11. [Google Scholar] [CrossRef]
- Sikandar, A.; Gao, F.; Mo, Y.; Chen, Q.; Ullah RM, K.; Wu, H. Efficacy of Aspergillus tubingensis GX3′ Fermentation against Meloidogyne enterolobii in Tomato (Solanum lycopersicum L.). Plants 2023, 12, 2724. [Google Scholar] [CrossRef]
- Xu, G.; Guo, H.; Yan, M.; Jia, Z.; Li, Z.; Chen, M.; Bao, X. An actin-like protein PoARP9 involves in the regulation of development and cellulase and amylase expression in Penicillium oxalicum. J. Appl. Microbiol. 2022, 132, 2894–2905. [Google Scholar] [CrossRef]
- Liu, J.; Yang, J.; Wang, R.; Liu, L.; Zhang, Y.; Bao, H.; Jang, M.J.; Wang, E.; Yuan, H. Comparative characterization of extracellular enzymes secreted by Phanerochaete chrysosporium during solid-state and submerged fermentation. Int. J. Biol. Macromol. 2020, 152, 288–294. [Google Scholar] [CrossRef]
- Salgado-Bautista, D.; Volke-Sepúlveda, T.; Figueroa-Martínez, F.; Carrasco-Navarro, U.; Chagolla-López, A.; Favela-Torres, E. Solid-state fermentation increases secretome complexity in Aspergillus brasiliensis. Fungal Biol. 2020, 124, 723–734. [Google Scholar] [CrossRef]
- Kim, H.Y.; Heo, D.Y.; Park, H.M.; Singh, D.; Lee, C.H. Metabolomic and Transcriptomic Comparison of Solid-State and Submerged Fermentation of Penicillium expansum KACC 40815. PLoS ONE 2016, 11, e0149012. [Google Scholar] [CrossRef]
- Garrigues, S.; Kun, R.S.; Peng, M.; Gruben, B.S.; Gelber, I.B.; Mäkelä, M.; de Vries, R.P. The Cultivation Method Affects the Transcriptomic Response of Aspergillus niger to Growth on Sugar Beet Pulp. Microbiol. Spectr. 2021, 9, e0106421. [Google Scholar] [CrossRef]
- Andersen, M.R.; Vongsangnak, W.; Panagiotou, G.; Salazar, M.P.; Lehmann, L.; Nielsen, J. A trispecies Aspergillus microarray: Comparative transcriptomics of three Aspergillus species. Proc. Natl. Acad. Sci. USA 2008, 105, 4387–4392. [Google Scholar] [CrossRef]
- Keller-Pearson, M.; Bortolazzo, A.; Willems, L.; Smith, B.; Peterson, A.; Ané, J.-M.; Silva, E.M. A Dual Transcriptomic Approach Reveals Contrasting Patterns of Differential Gene Expression during Drought in Arbuscular Mycorrhizal Fungus and Carrot. Mol. Plant-Microbe Interact. 2023, 36, 821–832. [Google Scholar] [CrossRef]
- Zhang, L.; Xiong, Z.; Xiao, M. A Review of the Application of Spatial Transcriptomics in Neuroscience. Interdiscip. Sci. Comput. Life Sci. 2024, 16, 243–260. [Google Scholar] [CrossRef]
- Mäkelä, M.R.; Aguilar-Pontes, M.V.; van Rossen-Uffink, D.; Peng, M.; de Vries, R.P. The fungus Aspergillus niger consumes sugars in a sequential manner that is not mediated by the carbon catabolite repressor CreA. Sci. Rep. 2018, 8, 6655. [Google Scholar] [CrossRef]
- de Vries, R.P.; Visser, J.A.A.P. Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol. Mol. Biol. Rev. 2001, 65, 497–522. [Google Scholar] [CrossRef]
- Iwanicki, N.S.; Júnior, I.D.; de Carvalho, L.L.; Eilenberg, J.; Henrik, H. Comparative transcriptomics of growth metabolism and virulence reveal distinct morphogenic profiles of yeast-like cells and hyphae of the fungus Metarhizium rileyi. Fungal Genet. Biol. 2023, 164, 103766. [Google Scholar]
- Benoit, I.; Zhou, M.; Duarte, A.V.; Downes, D.J.; Todd, R.B.; Kloezen, W.; Post, H.; Heck, A.J.R.; Altelaar, A.F.M.; de Vries, R.P. Spatial differentiation of gene expression in Aspergillus niger colony grown for sugar beet pulp utilization. Sci. Rep. 2015, 5, 13592. [Google Scholar] [CrossRef]
- Tanaka, T.; Terauchi, Y.; Yoshimi, A.; Abe, K. Aspergillus Hydrophobins: Physicochemical Properties, Biochemical Properties, and Functions in Solid Polymer Degradation. Microorganisms 2022, 10, 1498. [Google Scholar] [CrossRef]
- Ball, S.R.; Kwan, A.H.; Sunde, M. Sunde, Hydrophobin Rodlets on the Fungal Cell Wall. Fungal Cell Wall Armour A Weapon Hum. Fungal Pathog. 2020, 425, 29–51. [Google Scholar]
- Valsecchi, I.; Dupres, V.; Stephen-Victor, E.; Guijarro, J.I.; Gibbons, J.; Beau, R.; Bayry, J.; Coppee, J.-Y.; Lafont, F.; Latgé, J.-P.; et al. Role of Hydrophobins in Aspergillus fumigatus. J. Fungi 2017, 4, 2. [Google Scholar] [CrossRef]
- Cai, F.; Gao, R.; Zhao, Z.; Ding, M.; Jiang, S.; Yagtu, C.; Zhu, H.; Zhang, J.; Ebner, T.; Mayrhofer-Reinhartshuber, M.; et al. Evolutionary compromises in fungal fitness: Hydrophobins can hinder the adverse dispersal of conidiospores and challenge their survival. ISME J. 2020, 14, 2610–2624. [Google Scholar] [CrossRef]
- Zutz, A.; Hoffmann, J.; Hellmich, U.A.; Glaubitz, C.; Ludwig, B.; Brutschy, B.; Tampé, R. Asymmetric ATP hydrolysis cycle of the heterodimeric multidrug ABC transport complex TmrAB from Thermus thermophilus. J. Biol. Chem. 2011, 286, 7104–7115. [Google Scholar] [CrossRef]
- Orelle, C.; Mathieu, K.; Jault, J.-M. Multidrug ABC transporters in bacteria. Res. Microbiol. 2019, 170, 381–391. [Google Scholar] [CrossRef]
- Eustermann, S.; Patel, A.B.; Hopfner, K.-P.; He, Y.; Korber, P. Energy-driven genome regulation by ATP-dependent chromatin remodellers. Nat. Rev. Mol. Cell Biol. 2024, 25, 309–332. [Google Scholar] [CrossRef]
- MacRae, I.J.; A Doudna, J. Ribonuclease revisited: Structural insights into ribonuclease III family enzymes. Curr. Opin. Struct. Biol. 2007, 17, 138–145. [Google Scholar] [CrossRef]
- Das, G.; Varshney, U. Peptidyl-tRNA hydrolase and its critical role in protein biosynthesis. Microbiology 2006, 152 Pt 8, 2191–2195. [Google Scholar] [CrossRef]
- Jamaldheen, S.B.; Thakur, A.; Moholkar, V.S.; Goyal, A. Enzymatic hydrolysis of hemicellulose from pretreated Finger millet (Eleusine coracana) straw by recombinant endo-1,4-beta-xylanase and exo-1,4-beta-xylosidase. Int. J. Biol. Macromol. 2019, 135, 1098–1106. [Google Scholar] [CrossRef]
- Hynes, M.J.; Murray, S.L. ATP-citrate lyase is required for production of cytosolic acetyl coenzyme A and development in Aspergillus nidulans. Eukaryot. Cell 2010, 9, 1039–1048. [Google Scholar] [CrossRef]
- Bland, J. Jeffrey Bland, Phd, FACN, FACB: Heart disease, inflammation, and the revolution in health care. Interview by Craig Gustafson. Altern. Ther. Health Med. 2013, 19 (Suppl. 1), 50–55. [Google Scholar]
- Ries, L.N.A.; de Castro, P.A.; Silva, L.P.; Valero, C.; dos Reis, T.F.; Saborano, R.; Duarte, I.F.; Persinoti, G.F.; Steenwyk, J.L.; Rokas, A.; et al. Aspergillus fumigatus Acetate Utilization Impacts Virulence Traits and Pathogenicity. mBio 2021, 12, e0168221. [Google Scholar] [CrossRef]
- Cairns, T.C.; Zheng, X.; Zheng, P.; Sun, J.; Meyer, V. Moulding the mould: Understanding and reprogramming filamentous fungal growth and morphogenesis for next generation cell factories. Biotechnol. Biofuels 2019, 12, 77. [Google Scholar] [CrossRef]
- Śliżewska, K.; Chlebicz-Wójcik, A. Growth Kinetics of Probiotic Lactobacillus Strains in the Alternative, Cost-Efficient Semi-Solid Fermentation Medium. Biology 2020, 9, 423. [Google Scholar] [CrossRef]
- Hanzalova, K.; Rossner, P.; Sram, R.J. Oxidative damage induced by carcinogenic polycyclic aromatic hydrocarbons and organic extracts from urban air particulate matter. Mutat. Res. 2010, 696, 114–121. [Google Scholar] [CrossRef]
- Sampaio, G.R.; Guizellini, G.M.; da Silva, S.A.; de Almeida, A.P.; Pinaffi-Langley, A.C.C.; Rogero, M.M.; de Camargo, A.C.; Torres, E.A.F.S. Polycyclic Aromatic Hydrocarbons in Foods: Biological Effects, Legislation, Occurrence, Analytical Methods, and Strategies to Reduce Their Formation. Int. J. Mol. Sci. 2021, 22, 6010. [Google Scholar] [CrossRef]
- Zhang, W.; Kou, Y.; Xu, J.; Cao, Y.; Zhao, G.; Shao, J.; Wang, H.; Wang, Z.; Bao, X.; Chen, G.; et al. Two major facilitator superfamily sugar transporters from Trichoderma reesei and their roles in induction of cellulase biosynthesis. J. Biol. Chem. 2013, 288, 32861–32872. [Google Scholar] [CrossRef]
- Huang, Z.B.; Chen, X.Z.; Qin, L.N.; Wu, H.Q.; Su, X.Y.; Dong, Z.Y. A novel major facilitator transporter TrSTR1 is essential for pentose utilization and involved in xylanase induction in Trichoderma reesei. Biochem. Biophys. Res. Commun. 2015, 460, 663–669. [Google Scholar] [CrossRef]
- Zeng, X.; Tang, S.; Dong, X.; Dong, M.; Shao, R.; Liu, R.; Li, T.; Zhang, X.; Wong, Y.H.; Xie, Q. Analysis of metagenome and metabolome disclosed the mechanisms of Dendrobium officinale polysaccharide on DSS-induced ulcerative colitis-affected mice. Int. J. Biol. Macromol. 2024, 277, 134229. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]


| ID | Description * | Set Size | Category | |
|---|---|---|---|---|
| GSEA | GO:0043933 | Protein-containing complex subunit organization | 488 | Process |
| GO:1903506 | Regulation of nucleic acid-templated transcription | 475 | Process | |
| GO:2001141 | Regulation of RNA biosynthetic process | 475 | Process | |
| GO:0006355 | Regulation of transcription, DNA-templated | 470 | Process | |
| GO:0006396 | RNA processing | 466 | Process | |
| GO:0042886 | Amide transport | 446 | Process | |
| GO:0009892 | Negative regulation of metabolic process | 435 | Process | |
| GO:0015833 | Peptide transport | 434 | Process | |
| GO:0045184 | Establishment of protein localization | 432 | Process | |
| GO:0005694 | Chromosome | 424 | Component | |
| ORA | GO:0006396 | RNA processing | 101 | Process |
| GO:0005783 | Endoplasmic reticulum | 100 | Component | |
| GO:0006259 | DNA metabolic process | 97 | Process | |
| GO:0005886 | Plasma membrane | 93 | Component | |
| GO:0019438 | Aromatic compound biosynthetic process | 90 | Process | |
| GO:0006355 | Regulation of transcription, DNA-templated | 87 | Process | |
| GO:1903506 | Regulation of nucleic acid-templated transcription | 87 | Process | |
| GO:2001141 | Regulation of RNA biosynthetic process | 87 | Process | |
| GO:0051276 | Chromosome organization | 83 | Process | |
| GO:0005694 | Chromosome | 82 | Component |
| ID | Description * | Set Size | |
|---|---|---|---|
| GSEA | ko01110 | Biosynthesis of secondary metabolites | 326 |
| ko01120 | Microbial metabolism in diverse environments | 169 | |
| ko05014 | Amyotrophic lateral sclerosis | 145 | |
| ko05022 | Pathways of neurodegeneration—multiple diseases | 139 | |
| ko05016 | Huntington disease | 127 | |
| ko05010 | Alzheimer disease | 120 | |
| ko01240 | Biosynthesis of cofactors | 119 | |
| ko05012 | Parkinson disease | 110 | |
| ko05020 | Prion disease | 105 | |
| ko03010 | Ribosome | 104 | |
| ORA | ko01240 | Biosynthesis of cofactors | 37 |
| ko01230 | Biosynthesis of amino acids | 25 | |
| ko04111 | Cell cycle—yeast | 21 | |
| ko01200 | Carbon metabolism | 21 | |
| ko03040 | Spliceosome | 18 | |
| ko04113 | Meiosis—yeast | 17 | |
| ko04141 | Protein processing in endoplasmic reticulum | 17 | |
| ko04146 | Peroxisome | 16 | |
| ko00230 | Purine metabolism | 16 | |
| ko00500 | Starch and sucrose metabolism | 15 |
| Primer Name | NCBI Gene ID | Description | Product Size (bp) | Primer Sequence (5′-3′) |
|---|---|---|---|---|
| AtWU_03490 | 56002975 | ABC multidrug transporter | 128 | F: GCGTTCACATTCGGCTGGAAG |
| R: GAGTAGACCTTGGCGTTCATTGC | ||||
| AtWU_03807 | 56003291 | Ribonuclease III | 101 | F: GACACTTCAAGGCAACGAGCAATC |
| R: ACTCTTTGACGGCATCCTGTTTTCC | ||||
| AtWU_00075 | 55999564 | Arabinogalactan en-do-1,4-beta-galactosidase A | 129 | F: GGCAAGACCAGCAACTATGACAAC |
| R: CTCGTCCCAGTCCCATCCATTG | ||||
| AtWU_10270 | 56009747 | PeptidyI-tRNA hydrolase | 110 | F: GGATTTGTCGGTCTTGAGGATTGG |
| R: CCTTCAACTCCTGTTCACTCATCTC | ||||
| β-tubulin | 37238 | Beta-Tubulin | 148 | F: AGCAGATGTTCGACCCCAA |
| R: TAGGTCTGGTTCTTGCTCTGGATG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, X.; Song, J.; Tang, S.; Dong, X.; Chen, S.; Kong, J.; Chen, L.; Li, Y.; Shao, G.; Wong, Y.-H.; et al. Transcriptomic Approach Reveals Contrasting Patterns of Differential Gene Expression during Tannin Biodegredation by Aspergillus tubingensis in Liquid and Solid Cultures. Int. J. Mol. Sci. 2024, 25, 10547. https://doi.org/10.3390/ijms251910547
Zeng X, Song J, Tang S, Dong X, Chen S, Kong J, Chen L, Li Y, Shao G, Wong Y-H, et al. Transcriptomic Approach Reveals Contrasting Patterns of Differential Gene Expression during Tannin Biodegredation by Aspergillus tubingensis in Liquid and Solid Cultures. International Journal of Molecular Sciences. 2024; 25(19):10547. https://doi.org/10.3390/ijms251910547
Chicago/Turabian StyleZeng, Xiaona, Jiabei Song, Shengqiu Tang, Xiaoying Dong, Sheng Chen, Jie Kong, Liyi Chen, Yajuan Li, Guanming Shao, Yung-Hou Wong, and et al. 2024. "Transcriptomic Approach Reveals Contrasting Patterns of Differential Gene Expression during Tannin Biodegredation by Aspergillus tubingensis in Liquid and Solid Cultures" International Journal of Molecular Sciences 25, no. 19: 10547. https://doi.org/10.3390/ijms251910547
APA StyleZeng, X., Song, J., Tang, S., Dong, X., Chen, S., Kong, J., Chen, L., Li, Y., Shao, G., Wong, Y.-H., & Xie, Q. (2024). Transcriptomic Approach Reveals Contrasting Patterns of Differential Gene Expression during Tannin Biodegredation by Aspergillus tubingensis in Liquid and Solid Cultures. International Journal of Molecular Sciences, 25(19), 10547. https://doi.org/10.3390/ijms251910547







