Role of Genetic Polymorphisms -238 G>A and -308 G>A, and Serum TNF-α Levels in a Cohort of Mexican Pediatric Neuroblastoma Patients: Preliminary Study
Abstract
:1. Introduction
2. Results
2.1. Distribution of TNF-α Genetic Polymorphisms between NB Cases and Control Group
2.2. Analysis of NB Patient Genotypes and Prognostic Factors
2.3. Relationship of TNF-α Serum Levels Between Cases of NB and Control Group
2.4. Association of Genotypes and Serum Levels
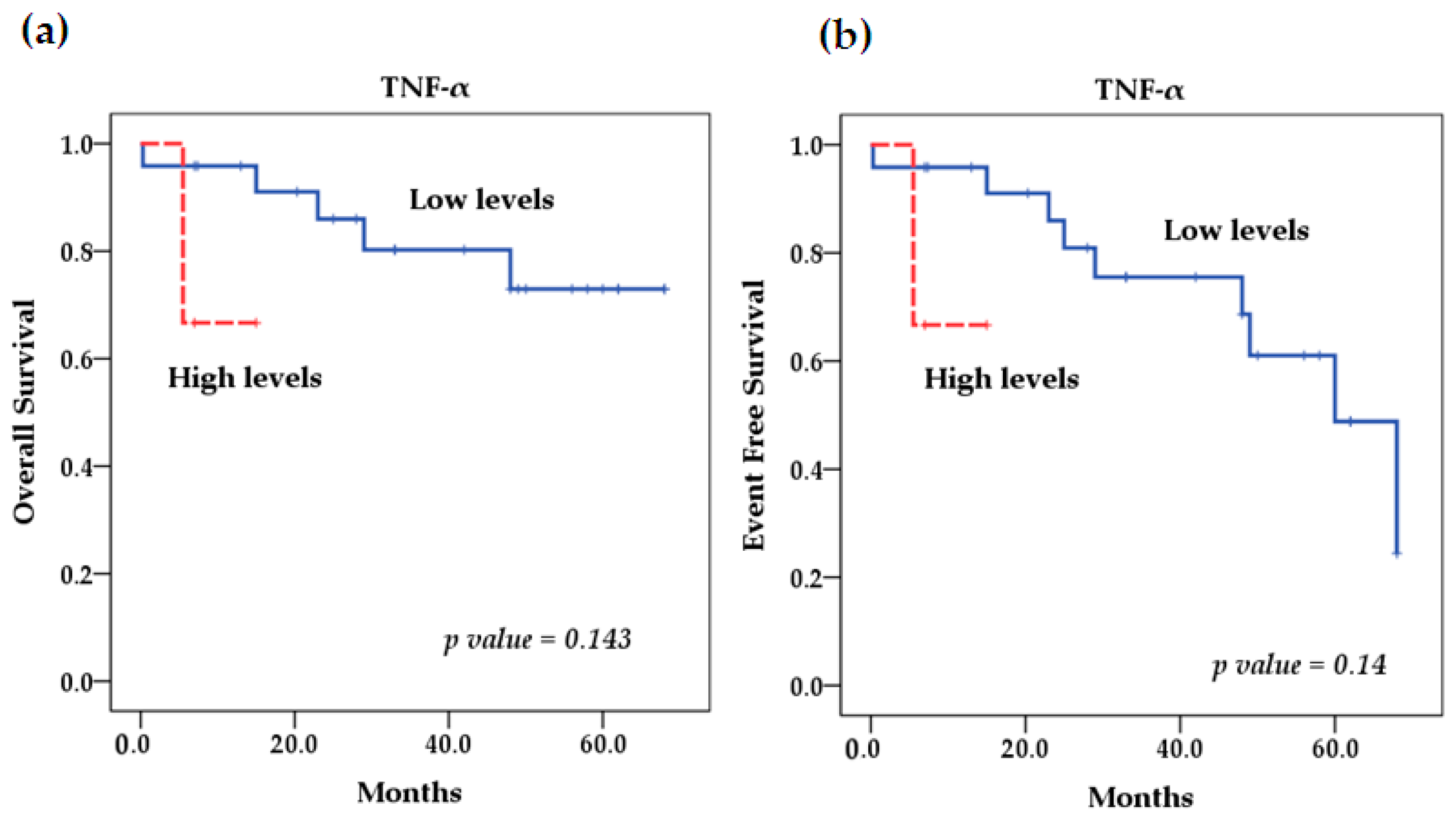
2.5. Survival Analysis
3. Discussion
4. Materials and Methods
4.1. Ethical Considerations
4.2. Determination of MYCN Gene Amplification
4.3. Analysis of TNF-α Genetic Polymorphisms
4.4. Measurement of TNF-α Serum Levels
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| SNP -238 G/A rs361525 n = 27 (%) | SNP -308 G/A rs1800629 n = 27 (%) | |||
|---|---|---|---|---|
| Variables | GG + GA | AA | GG | GA + AA |
| INSS stage | ||||
| 1 | 3 (11.1) | 2 (7.4) | 5 (18.5) | ---- |
| 2b | 2 (7.4) | ---- | 1 (3.7) | 1 (3.7) |
| 3 | 7 (25.9) | ---- | 3 (11.1) | 4 (14.8) |
| 4 | 12 (44.4) | 1 (3.7) | 4 (14.8) | 9 (33.3) |
| p value | 0.218 | 0.05 | ||
| INRG | ||||
| L1 | 3 (11.1) | 2 (7.4) | 3 (11.1) | 2 (7.4) |
| L2 | 8 (29.6) | ---- | 5 (18.5) | 3 (11.1) |
| M | 13 (48.1) | 1 (3.7) | 5 (18.5) | 9 (33.3) |
| MS | ---- | --- | --- | |
| p value | 0.146 | 0.51 | ||
| Risk | ||||
| Low | 3 (11.1) | 2 (7.4) | 4 (14.8) | 1 (3.7) |
| Intermediate | 7 (25.9) | ---- | 5 (18.5) | 2 (7.4) |
| High | 14 (51.9) | 1 (3.7) | 4 (14.8) | 11 (40.7) |
| p value | 0.126 | 0.05 | ||
| Differentiation | ||||
| Undifferentiated | 2 (7.4) | ---- | ---- | 2 (7.4) |
| Partially differentiated | 14 (51.9) | 2 (7.4) | 9 (33.3) | 7 (25.9) |
| Differentiated | 4 (14.8) | 1 (3.7) | 3 (11.1) | 2 (7.4) |
| Not specified | 4 (14.8) | ---- | 1 (3.7) | 3 (11.1) |
| p value | 1 | 0.414 | ||
| Histology (Shimada) | ||||
| Favorable | 13 (48.1) | 2 (7.4) | 10 (37) | 5 (18.5) |
| Unfavorable | 11 (40.7) | 1 (3.7) | 3 (11.1) | 9 (33.3) |
| p value | 0.586 | 0.038 | ||
| MYCN | ||||
| Amplified | 1 (3.7) | ----- | ---- | 1 (3.7) |
| Not amplified | 23 (85.2) | 3 (11.1) | 13 (48.1) | 13 (48.1) |
| p value | 0.296 | 0.3 | ||
| Low Serum TNF-α Levels | High Serum TNF-α Levels | Value p | OR (95% CI) | |
|---|---|---|---|---|
| TNF-α -308 G/A rs1800629 | ||||
| GG | 12 (44.44) | 1 (3.7) | Ref | ---- |
| GA | 4 (14.8) | 0 | --- | --- |
| AA | 8 (29.6) | 2 (7.4) | 0.145 | 2.63 (0.71–9.70) |
| GG + GA vs. AA | ----- | ----- | 0.119 | 7.27 (0.60–87.84) |
| GA + AA vs. GG | ----- | ----- | 0.214 | 0.20 (0.017–2.47) |
| TNF-α -238 G/A rs361525 | ||||
| GG | 13 (48.14) | 0 | Ref | ---- |
| GA | 8 (29.6) | 3 (11.1) | 0.118 | 3.87 (0.71–21.12) |
| AA | 3 (11.1) | 0 | ----- | ------ |
| GG + GA vs. AA | ----- | ---- | --- | NC |
| GA + AA vs. GG | ----- | ---- | ---- | NC |
| Model | HR (95% CI) | p Value |
|---|---|---|
| A | ||
| TNF-α rs361525 (GG + GA vs. AA) | NC | NC |
| Age (≥18 months vs. <18 months) | 1.16 (0.16–8.39) | 0.87 |
| INSS stage (4 vs. 1, 2, 3) | 5.62 (0.51–61.6) | 0.15 |
| MYCN (amplified vs. not amplified) | NC | NC |
| B | ||
| TNF-α rs1800629 (GG + GA vs. AA) | 2.08 (0.35–12.30) | 0.41 |
| Age (≥18 months vs. <18 months) | 1.04 (0.14–7.64) | 0.96 |
| INSS stage (4 vs. 1, 2, 3) | 5.16 (0.41–64.3) | 0.20 |
| MYCN (amplified vs. not amplified) | NC | NC |
| C | ||
| Serum TNF-α levels (High vs. Low) | 4.95 (0.30–79.5) | 0.25 |
| Age (≥18 months vs. <18 months) | 1.74 (0.20–15.12) | 0.61 |
| INSS stage (4 vs. 1, 2, 3) | 4.91 (0.48–49.35) | 0.17 |
| MYCN (amplified vs. not amplified) | NC | NC |
Appendix B
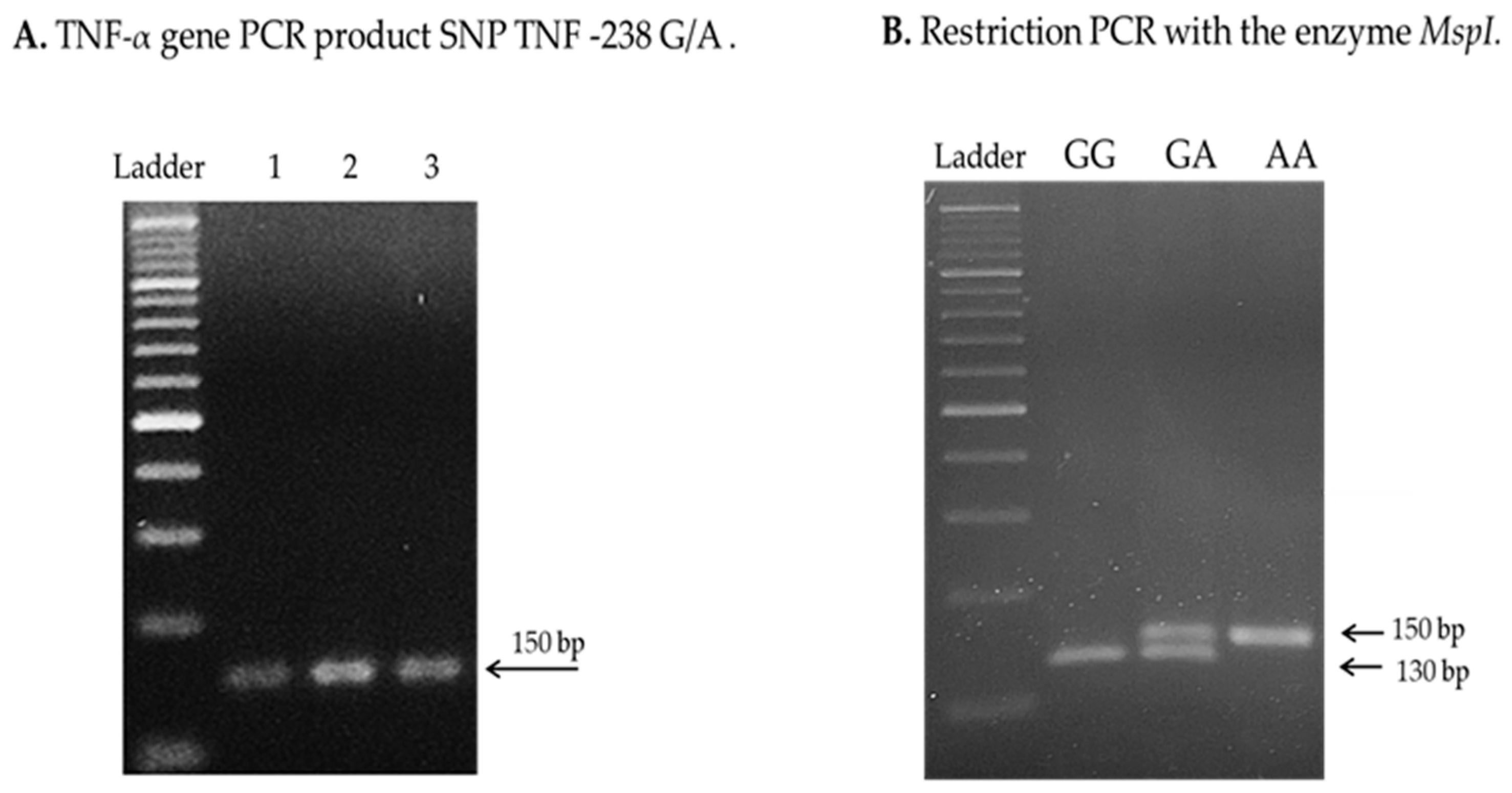
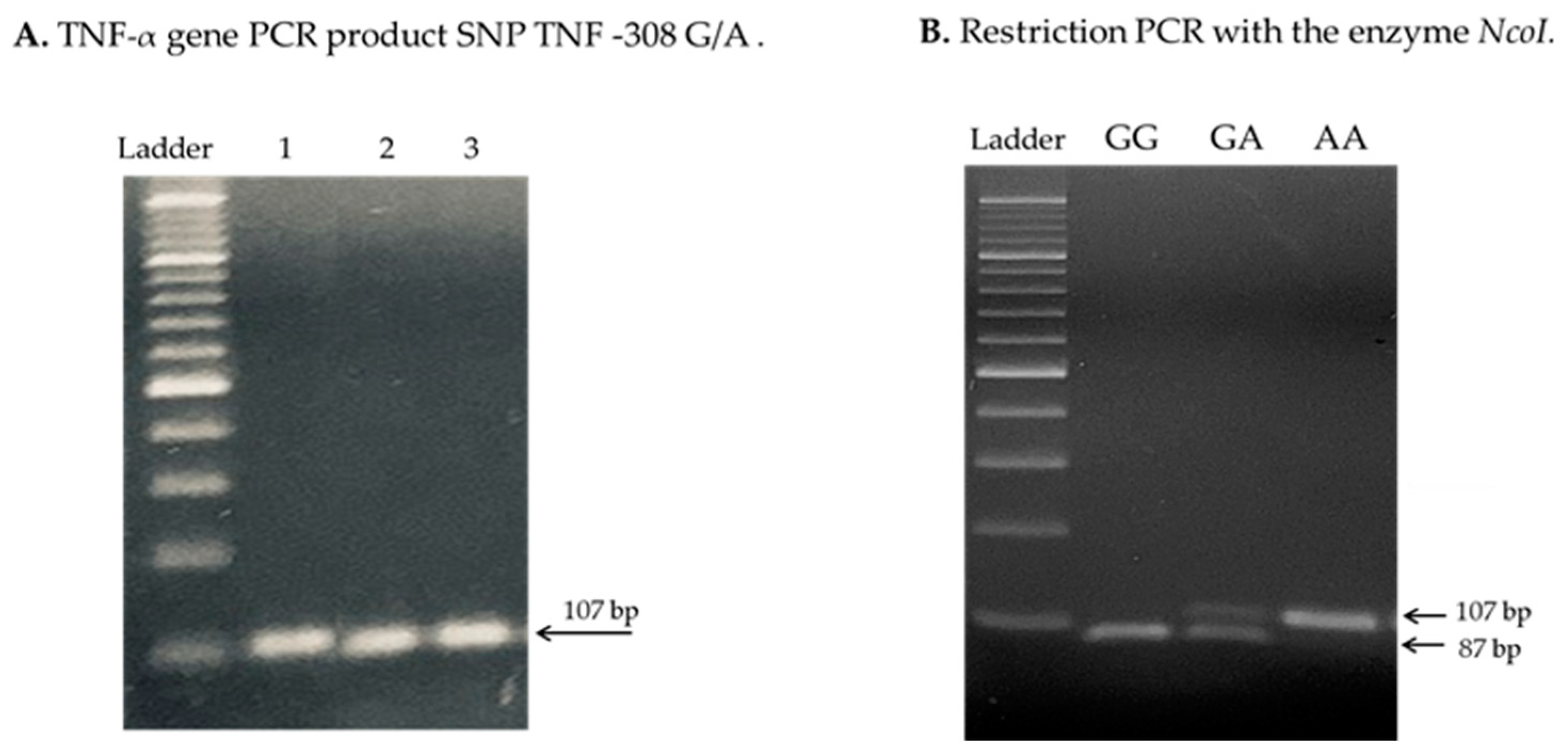
References
- Swift, C.C.; Eklund, M.J.; Kraveka, J.M.; Alazraki, A.L. Updates in Diagnosis, Management, and Treatment of Neuroblastoma, Radiographics: A review publication of the Radiological Society of North America. RadioGraphics 2018, 38, 566–580. [Google Scholar] [CrossRef] [PubMed]
- Newman, E.A.; Abdessalam, S.; Aldrink, J.H.; Austin, M.; Heaton, T.E.; Bruny, J.; Ehrlich, P.; Dasgupta, R.; Baertschiger, R.M.; Lautz, T.B.; et al. APSA Cancer committee. Update on neuroblastoma. J. Pediatr. Surg. 2019, 54, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Juárez-Ocaña, S.; Palma-Padilla, V.; González-Miranda, G.; Siordia-Reyes, A.G.; López-Aguilar, E.; Aguilar-Martínez, M.; Mejía-Aranguré, J.M.; Carreón-Cruz, R.; Rendón-Macías, M.E.; Fajardo-Gutiérrez, A. Epidemiological and some clinical characteristics of neuroblastoma in Mexican children (1996–2005). BMC Cancer 2009, 3, 266. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Palma-Padilla, V.; Juárez-Ocaña, S.; González-Miranda, G.; Siordia-Reyes, A.G.; Mejia-Aranguré, J.M.; Carreón-Cruz, R.; Fajardo-Gutiérrez, A. Incidencia y tendencia del Neuroblastoma en niños derechohabientes del IMSS. Rev. Med. Inst. Mex. Seguro Soc. 2010, 48, 151–158. [Google Scholar] [PubMed]
- Pinheiro, P.S.; Callahan, K.E.; Stern, M.C.; de Vries, E. Migration from Mexico to the United States: A high-speed cancer transition. Int. J. Cancer 2018, 142, 477–488. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tolbert, V.P.; Matthay, K.K. Neuroblastoma: Clinical and biological approach to risk stratification and treatment. Cell Tissue Res. 2018, 372, 195–209. [Google Scholar] [CrossRef]
- Juárez-Villegas, L.E.; Zapata-Tarrés, M.; Lezama del Valle, P. Resultados del tratamiento de niños con neuroblastoma en el Hospital Infantil de México “Federico Gómez”. Gac. Mex. Oncol. 2014, 13, 26–30. [Google Scholar]
- Takita, J. Molecular Basis and Clinical Features of Neuroblastoma. JMA J. 2021, 4, 321–331. [Google Scholar] [CrossRef]
- Liang, W.H.; Federico, S.M.; London, W.B.; Naranjo, A.; Irwin, M.S.; Volchenboum, S.L.; Cohn, S.L. Tailoring Therapy for Children With Neuroblastoma on the Basis of Risk Group Classification: Past, Present, and Future. JCO Clin. Cancer Inform. 2020, 4, 895–905. [Google Scholar] [CrossRef]
- Zeineldin, M.; Patel, A.G.; Dyer, M.A. Neuroblastoma: When differentiation goes awry. Neuron 2022, 110, 2916–2928. [Google Scholar] [CrossRef]
- Li, L.; Yu, R.; Cai, T.; Chen, Z.; Lan, M.; Zou, T.; Wang, B.; Wang, Q.; Zhao, Y.; Cai, Y. Effects of immune cells and cytokines on inflammation and immunosuppression in the tumor microenvironment. Int. Immunopharmacol. 2020, 88, 106939. [Google Scholar] [CrossRef] [PubMed]
- Laha, D.; Grant, R.; Mishra, P.; Nilubol, N. The Role of Tumor Necrosis Factor in Manipulating the Immunological Response of Tumor Microenvironment. Front. Immunol. 2021, 12, 656908. [Google Scholar] [CrossRef] [PubMed]
- Kartikasari, A.E.R.; Huertas, C.S.; Mitchell, A.; Plebanski, M. Tumor-Induced Inflammatory Cytokines and the Emerging Diagnostic Devices for Cancer Detection and Prognosis. Front. Oncol. 2021, 11, 692142. [Google Scholar] [CrossRef] [PubMed]
- Elahi, M.M.; Asotra, K.; Matata, B.M.; Mastana, S.S. Tumor necrosis factor alpha -308 gene locus promoter polymorphism: An analysis of association with health and disease. Biochim. Biophys. Acta 2009, 1792, 163–172. [Google Scholar] [CrossRef]
- Waters, J.P.; Pober, J.S.; Bradley, J.R. Tumour necrosis factor and cancer. J. Pathol. 2013, 230, 241–248. Available online: https://onlinelibrary.wiley.com/doi/pdf/10.1002/path.4188 (accessed on 22 April 2021). [CrossRef]
- Cereda, C.; Gagliardi, S.; Cova, E.; Diamanti, L.; Ceroni, M. The Role of TNF-Alpha in ALS: New Hypotheses for Future Therapeutic Approaches. In Amyotrophic Lateral Sclerosis; IntechOpen: London, UK, 2012. [Google Scholar] [CrossRef]
- de Oliveira, M.M.; da Silva, J.C.S.; Costa, J.F.; Amim, L.H.; Loredo, C.C.S.; Melo, H.; Queiroz, L.F.; Mello, F.C.Q.; e Silva, J.R.L.; Kritski, A.L.; et al. Single Nucleotide Polymorphisms (SNPs) of the TNF-alpha (-238/-308) gene among TB and nom TB patients: Susceptibility markers of TB occurrence? J. Bras. Pneumol. 2004, 30, 371–377. [Google Scholar] [CrossRef]
- Shih, C.M.; Lee, Y.L.; Chiou, H.L.; Chen, W.; Chang, G.C.; Chou, M.C.; Lin, L.Y. Association of TNF-alpha polymorphism with susceptibility to and severity of non-small cell lung cancer. Lung Cancer 2006, 52, 15–20. [Google Scholar] [CrossRef]
- Tomolonis, J.A.; Xu, X.; Dholakia, K.H.; Zhang, C.; Guo, L.; Courtney, A.N.; Wang, S.; Balzeau, J.; Barragán, G.A.; Tian, G.; et al. Interaction between tumor cell TNFR2 and monocyte membrane-bound TNF-α triggers tumorigenic inflammation in neuroblastoma. J. Immunother. Cancer 2023, 11, e005478. [Google Scholar] [CrossRef]
- Li, X.Y.; Liang, C.H.; Parkman, V.; Lv, Z.T. The association between TNF-α 238A/G and 308A/G polymorphisms and juvenile idiopathic arthritis: An updated PRISMA-compliant meta-analysis. Medicine 2018, 97, e12883. [Google Scholar] [CrossRef]
- Wilson, A.G.; Symons, J.A.; McDowell, T.L.; McDevitt, H.O.; Duff, G.W. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc. Natl. Acad. Sci. USA 1997, 94, 3195–3199. [Google Scholar] [CrossRef]
- Glossop, J.R.; Dawes, P.T.; Nixon, N.B.; Mattey, D.L. Polymorphism in the tumour necrosis factor receptor II gene is associated with circulating levels of soluble tumour necrosis factor receptors in rheumatoid arthritis. Arthritis Res. Ther. 2005, 7, R1227–R1234. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Bello, J.; Vargas-Alarcón, G.; Tovilla-Zárate, C.; Fragoso, J.M. Polimorfismos de un solo nucleótido (SNP): Implicaciones funcionales de los SNP reguladores (rSNP) y de los SNP-ARN estructurales (srSNP) en enfermedades complejas [Single nucleotide polymorphisms (SNPs): Functional implications of regulatory-SNP (rSNP) and structural RNA (srSNPs) in complex diseases]. Gac. Medica Mex. 2013, 149, 220–228. [Google Scholar]
- Jeong, P.; Kim, E.J.; Kim, E.G.; Byun, S.S.; Kim, C.S.; Kim, W.J. Association of bladder tumors and GA genotype of -308 nucleotide in tumor necrosis factor-alpha promoter with greater tumor necrosis factor-alpha expression. Urology 2004, 64, 1052–1056. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Sharma, A.; Kumar, S.; Sharma, S.K.; Ghosh, B. Association of TNF haplotypes with asthma, serum IgE levels, and correlation with serum TNF-alpha levels. Am. J. Respir. Cell Mol. Biol. 2006, 35, 488–495. [Google Scholar] [CrossRef]
- Zhou, P.; Lv, G.Q.; Wang, J.Z.; Li, C.W.; Du, L.F.; Zhang, C.; Li, J.P. The TNF-alpha-238 polymorphism and cancer risk: A meta-analysis. PLoS ONE 2011, 6, e22092. [Google Scholar] [CrossRef]
- Zhao, H.; Liu, L.; Liu, B.; Wang, Y.; Li, F.; Yu, H. An updated association between TNF-α -238G/A polymorphism and gastric cancer susceptibility in East Asians. Biosci. Rep. 2018, 38, BSR20181231. [Google Scholar] [CrossRef]
- Bounder, G.; Jouimyi, M.R.; Boura, H.; Touati, E.; Michel, V.; Badre, W.; Jouhadi, H.; Kadi, M.; Eljihad, M.; Benomar, H.; et al. Associations of the -238(G/A) and -308(G/A) TNF-α Promoter Polymorphisms and TNF-α Serum Levels with the Susceptibility to Gastric Precancerous Lesions and Gastric Cancer Related to Helicobacter pylori Infection in a Moroccan Population. Asian Pac. J. Cancer Prev. APJCP 2020, 21, 1623–1629. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Xu, X.H.; Xu, L.; Liu, Y.; Sun, M.; Ni, L.H.; Wang, X.L.; Chen, Z.; Zhang, K.; Zeng, G. No association of TNF-α-308G/A polymorphisms, with head and neck cancer risk: A PRISMA-compliant meta-analysis. Medicine 2017, 96, e7298. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Qin, S.; Liu, Y.; Tao, L.; Jiang, H. Associations of tumor necrosis factor-α polymorphisms with the risk of colorectal cancer: A meta-analysis. Biosci. Rep. 2019, 39, BSR20181750. [Google Scholar] [CrossRef]
- Hollegaard, M.V.; Bidwell, J.L. Cytokine gene polymorphism in human disease: On-line databases, Supplement 3. Genes Immun. 2006, 7, 269–276. [Google Scholar] [CrossRef]
- Vargas-Alarcon, G.; Ramírez-Bello, J.; Juárez-Cedillo, T.; Ramírez-Fuentes, S.; Carrillo-Sánchez, S.; Fragoso, J.M. Distribution of the IL-1RN, IL-6, IL-10, INF-γ, and TNF-α Gene Polymorphisms in the Mexican Population. Genet. Test. Mol. Biomark. 2012, 16, 1246–1253. [Google Scholar] [CrossRef] [PubMed]
- Parra-Rojas, I.; Ruíz-Madrigal, B.; Martínez-López, E.; Panduro, A. Influence of the -308 TNF-alpha and -174 IL-6 polymorphisms on lipid profile in Mexican subjects. Hereditas 2006, 143, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Gómez Flores-Ramos, L.; Escoto-De Dios, A.; Puebla-Pérez, A.M.; Figuera-Villanueva, L.E.; Ramos-Silva, A.; Ramírez-Patiño, R.; Delgado-Saucedo, J.I.; Salas-González, E.; Zúñiga-González, G.M.; Alonzo-Rojo, A.; et al. Association of the tumor necrosis factor-alpha -308G>A polymorphism with breast cancer in Mexican women. Genet. Mol. Res. GMR 2013, 12, 5680–5693. [Google Scholar] [CrossRef]
- Mohamed, A.A.; Rashed, L.A.; Shaker, S.M.; Ammar, R.I. Association of tumor necrosis factor-alpha polymorphisms with susceptibility and clinical outcomes of rheumatic heart disease. Saudi Med. J. 2010, 31, 644–649. [Google Scholar]
- Vázquez-Huerta, D.I.; Alvarez-Rodríguez, B.A.; Topete-Reyes, J.F.; Muñoz-Valle, J.F.; Parra-Michel, R.; Fuentes-Ramírez, F.; Salazar-López, M.A.; Valle, Y.; Reyes-Castillo, Z.; Cruz-González, A.; et al. Tumor necrosis factor alpha -238 G/A and -308 G/A polymorphisms and soluble TNF-α levels in chronic kidney disease: Correlation with clinical variables. Int. J. Clin. Exp. Med. 2014, 7, 2111–2119. [Google Scholar]
- Zheng, W.; Zhang, S.; Zhang, S.; Min, L.; Wang, Y.; Xie, J.; Hou, Y.; Tian, X.; Cheng, J.; Liu, K.; et al. The relationship between tumor necrosis factor-α polymorphisms and gastric cancer risk: An updated meta-analysis. Biomed. Rep. 2017, 7, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Montfort, A.; Colacios, C.; Levade, T.; Andrieu-Abadie, N.; Meyer, N.; Ségui, B. The TNF Paradox in Cancer Progression and Immunotherapy. Front. Immunol. 2019, 10, 1818. [Google Scholar] [CrossRef]
- Wungu, C.; Ariyanto, F.C.; Prabowo, G.I.; Soetjipto; Handajani, R. Association between five types of Tumor Necrosis Factor-α gene polymorphism and hepatocellular carcinoma risk: A meta-analysis. BMC Cancer 2020, 20, 1134. [Google Scholar] [CrossRef]
- Zmorzyński, S.; Popek-Marciniec, S.; Szudy-Szczyrek, A.; Wojcierowska-Litwin, M.; Korszeń-Pilecka, I.; Chocholska, S.; Styk, W.; Hus, M.; Filip, A.A. The Association of GSTT1, GSTM1, and TNF-α Polymorphisms With the Risk and Outcome in Multiple Myeloma. Front. Oncol. 2019, 9, 1056. [Google Scholar] [CrossRef]
- Basmaci, C.; Pehlivan, M.; Tomatir, A.; Sever, T.; Okan, V.; Yilmaz, M.; Oguzkan-Balci, S.; Pehlivan, S. Effects of TNFα, NOS3, MDR1 Gene Polymorphisms on Clinical Parameters, Prognosis and Survival of Multiple Myeloma Cases. Asian Pac. J. Cancer Prev. APJCP 2016, 17, 1009–1014. [Google Scholar] [CrossRef]
- Zhao, Z.; Tang, X.; Song, K.; Li, X.; Zhang, Y. Association of -308G/A and -238G/A polymorphisms of TNF-α and osteosarcoma risk. Int. J. Clin. Exp. Pathol. 2015, 8, 4177–4181. [Google Scholar] [PubMed]
- Zhang, P.; Wu, X.; Li, G.; He, Q.; Dai, H.; Ai, C.; Shi, J. Tumor necrosis factor-alpha gene polymorphisms and susceptibility to ischemic heart disease: A systematic review and meta-analysis. Medicine 2017, 96, e6569. [Google Scholar] [CrossRef] [PubMed]
- Ozhan, G.; Yanar, H.T.; Ertekin, C.; Alpertunga, B. Polymorphisms in tumour necrosis factor alpha (TNFalpha) gene in patients with acute pancreatitis. Mediat. Inflamm. 2010, 2010, 482950. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Hurtado, I.A.; Puebla-Pérez, A.M.; Delgado-Saucedo, J.I.; Figuera, L.E.; Zúñiga-González, G.M.; Gomez-Mariscal, K.; Ronquillo-Carreón, C.A.; Gallegos-Arreola, M.P. Association between TNF-α-308G>A and -238G>A gene polymorphisms and TNF-α serum levels in Mexican colorectal cancer patients. Genet. Mol. Res. GMR 2016, 15, gmr.15028199. [Google Scholar] [CrossRef]
- Mandal, R.K.; Khan, M.A.; Hussain, A.; Akhter, N.; Jawed, A.; Dar, S.A.; Wahid, M.; Panda, A.K.; Lohani, M.; Mishra, B.N.; et al. A trial sequential meta-analysis of TNF-α -308G>A (rs800629) gene polymorphism and susceptibility to colorectal cancer. Biosci. Rep. 2019, 39, BSR20181052. [Google Scholar] [CrossRef]
- Kosałka-Węgiel, J.; Lichołai, S.; Dziedzina, S.; Milewski, M.; Kuszmiersz, P.; Rams, A.; Gąsior, J.; Matyja-Bednarczyk, A.; Kwiatkowska, H.; Korkosz, M.; et al. Genetic Association between TNFA Polymorphisms (rs1799964 and rs361525) and Susceptibility to Cancer in Systemic Sclerosis. Life 2022, 12, 698. [Google Scholar] [CrossRef]
- Miao, Z.; Wang, K.; Wang, X.; Zhang, C.; Xu, Y. TNF-α-308G/A polymorphism and the risk of colorectal cancer: A systematic review and an updated meta-analysis. J. BUON 2018, 23, 1616–1624. [Google Scholar]
- Korobeinikova, E.; Myrzaliyeva, D.; Ugenskiene, R.; Raulinaityte, D.; Gedminaite, J.; Smigelskas, K.; Juozaityte, E. The prognostic value of IL10 and TNF alpha functional polymorphisms in premenopausal early-stage breast cancer patients. BMC Genet. 2015, 16, 70. [Google Scholar] [CrossRef]
- Balkwill, F. Tumor necrosis factor or tumor promoting factor? Cytokine Growth Factor Rev. 2002, 13, 135–141. [Google Scholar] [CrossRef]
- Grigorova, A.A.; Trenova, A.G.; Stanilova, S.A. Association of polymorphism -308G/A in tumor necrosis factor-alpha gene (TNF-α) and TNF-α serum levels in patients with relapsing-remitting multiple sclerosis. Neurol. Res. 2021, 43, 291–298. [Google Scholar] [CrossRef]
- Pistoia, V.; Bianchi, G.; Borgonovo, G.; Raffaghello, L. Cytokines in neuroblastoma: From pathogenesis to treatment. Immunotherapy 2011, 3, 895–907. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Gao, Y.; Lin, T. Expression of interleukin-1 (IL-1), IL-6, and tumor necrosis factor-α (TNF-α) in non-small cell lung cancer and its relationship with the occurrence and prognosis of cancer pain. Ann. Palliat. Med. 2021, 10, 12759–12766. [Google Scholar] [CrossRef] [PubMed]
- Coşkun, Ö.; Öztopuz, Ö.; Özkan, Ö.F. Determination of IL-6, TNF-α and VEGF levels in the serums of patients with colorectal cancer. Cell. Mol. Biol. 2017, 63, 97–101. [Google Scholar] [CrossRef]
- Xiao, H.; Chen, L.; Luo, G.; Son, H.; Prectoni, J.H.; Zheng, W. Effect of the cytokine levels in serum on osteosarcoma. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2014, 35, 1023–1028. [Google Scholar] [CrossRef]
- Berdat, P.A.; Wehrle, T.J.; Küng, A.; Achermann, F.; Sutter, M.; Carrel, T.P.; Nydegger, U.E. Age-specific analysis of normal cytokine levels in healthy infants. Clin. Chem. Lab. Med. 2003, 41, 1335–1339. [Google Scholar] [CrossRef]
- Decker, M.L.; Grobusch, M.P.; Ritz, N. Influence of Age and Other Factors on Cytokine Expression Profiles in Healthy Children-A Systematic Review. Front. Pediatr. 2017, 5, 255. [Google Scholar] [CrossRef]
- Abraham, L.J.; Kroeger, K.M. Impact of the -308 TNF promoter polymorphism on the transcriptional regulation of the TNF gene: Relevance to disease. J. Leukoc. Biol. 1999, 66, 562–566. [Google Scholar] [CrossRef] [PubMed]
- Prasad, K.N.; Nyati, K.K.; Verma, A.; Rizwan, A.; Paliwal, V.K. Tumor necrosis factor-alpha polymorphisms and expression in Guillain-Barré syndrome. Hum. Immunol. 2010, 71, 905–910. [Google Scholar] [CrossRef]
- Kaluza, W.; Reuss, E.; Grossmann, S.; Hug, R.; Schopf, R.E.; Galle, P.R.; Maerker-Hermann, E.; Hoehler, T. Different transcriptional activity and in vitro TNF-alpha production in psoriasis patients carrying the TNF-alpha 238A promoter polymorphism. J. Investig. Dermatol. 2000, 114, 1180–1183. [Google Scholar] [CrossRef]
- Skoog, T.; van’t Hooft, F.M.; Kallin, B.; Jovinge, S.; Boquist, S.; Nilsson, J.; Eriksson, P.; Hamsten, A. A common functional polymorphism (C → A substitution at position -863) in the promoter region of the tumour necrosis factor-alpha (TNF-alpha) gene associated with reduced circulating levels of TNF-alpha. Hum. Mol. Genet. 1999, 8, 1443–1449. [Google Scholar] [CrossRef]
- Wang, H.; Liu, J.; Hu, X.; Liu, S.; He, B. Prognostic and Therapeutic Values of Tumor Necrosis Factor-Alpha in Hepatocellular Carcinoma. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2016, 22, 3694–3704. [Google Scholar] [CrossRef] [PubMed]
- Stoll, J.R.; Vaidya, T.S.; Mori, S.; Dusza, S.W.; Lacouture, M.E.; Markova, A. Association of interleukin-6 and tumor necrosis factor-α with mortality in hospitalized patients with cancer. J. Am. Acad. Dermatol. 2021, 84, 273–282. [Google Scholar] [CrossRef] [PubMed]
- American Cancer Society. Available online: www.cancer.org/cancer/types/neuroblastoma/detection-diagnosis-staging/staging.html (accessed on 28 April 2021).
- Shohet, J.M.; Nuchtern, J.G. Clinical Presentation, Diagnosis, and Staging Evaluation of Neuroblastoma, UpToDate. 2021. Available online: https://www.medilib.ir/uptodate/show/5187 (accessed on 7 April 2021).
- Sokol, E.; Desai, A.V. The Evolution of Risk Classification for Neuroblastoma. Children 2019, 6, 27. [Google Scholar] [CrossRef] [PubMed]
- Navarro, S.; Llombart-Bosch, A.; Pellin, A.; Burgués, O.; Ruiz, A.; Piqueras, M.; Noguera, R. Patología de los tumores neuroblásticos: Evaluación pronóstica. Experiencia del centro español de referencia de la SEOP para estudios biopatológicos del neuroblastoma (1992–2005). Rev. Esp. Patol. 2007, 40, 70–90. [Google Scholar] [CrossRef]
- Noori, N.M.; Shahramian, I.; Teimouri, A.; Keyvani, B.; Mahjoubifard, M. Serum Levels of Tumor Necrosis Factor-α and Interleukins in Children with Congenital Heart Disease. J. Tehran Heart Cent. 2017, 12, 15–22. [Google Scholar]



| Characteristics | Patients n = 27 (%) | Healthy Controls n = 38 (%) | |
|---|---|---|---|
| Age | <18 months | 16 (59.3) | 22 (57.9) |
| 18 months–5 years | 6 (22.2) | 9 (23.7) | |
| >5 years | 5 (18.5) | 7 (18.4) | |
| Sex | Male | 10 (37.03) | 14 (36.85) |
| Female | 17 (62.97) | 24 (63.15) | |
| INSS stage | 1 | 5 (18.5) | ----- |
| 2a | 0 (0) | ----- | |
| 2b | 2 (7.5) | ----- | |
| 3 | 7 (25.9) | ----- | |
| 4 | 13 (48.1) | ----- | |
| 4S | 0 (0) | ----- | |
| INRG | L1 | 5 (18.5) | ----- |
| L2 | 8 (29.6) | ----- | |
| M | 14 (51.9) | ----- | |
| MS | 0 (0) | ----- | |
| Risk | Low | 5 (18.5) | ----- |
| Intermediate | 7 (25.9) | ----- | |
| High | 15 (55.6) | ----- | |
| Primary Tumor Site | Adrenal | 8 (29.6) | ----- |
| Retroperitoneal | 11 (40.7) | ----- | |
| Paraspinal | 3 (11.1) | ----- | |
| Abdomen/Pelvic | 1 (3.7) | ----- | |
| Mediastinal | 4 (14.9) | ||
| Differentiation | Undifferentiated | 2 (7.4) | ----- |
| Partially differentiated | 16 (59.3) | ----- | |
| Differentiated | 5 (18.5) | ----- | |
| Not specified | 4 (14.8) | ----- | |
| Histology (Shimada) | Favorable | 15 (55.6) | ----- |
| Unfavorable | 12 (44.4) | ----- | |
| MYCN | Amplified | 1 (3.7) | ----- |
| Not amplified | 26 (96.3) | 38 (100) | |
| Relapse | Yes | 7 (25.9) | |
| Death | Yes | 6 (22.2) |
| SNP | Controls (%) n = 38 | NB Patients (%) n = 27 | OR (95% CI) | p Value |
|---|---|---|---|---|
| TNF-α -238 G/A rs361525 | ||||
| GG | 31 (81.6) | 13 (48.1) | Ref | |
| GA | 7 (18.4) | 11 (40.7) | 4.52 (1.61–12.67) | 0.004 |
| AA | 0 (0) | 3 (11.1) | ----- | --- |
| GA + AA vs. GG | --- | --- | 0.20 (0.068–0.63) | 0.006 |
| Allele G | 69 (90.79) | 37 (68.5) | ---- | ---- |
| Allele A | 7 (9.21) | 17 (31.5) | ---- | ---- |
| TNF-α -308 G/A rs1800629 | ||||
| GG | 33 (86.8) | 13(48.2) | Ref | |
| GA | 1 (2.6) | 4 (14.8) | ----- | |
| AA | 4 (10.5) | 10 (37) | 2.89 (1.452–5.769) | 0.003 |
| GG + GA vs. AA | ---- | ---- | 0.10 (0.030–0.393) | 0.001 |
| GA + AA vs. GG | ---- | ---- | 5 (1.36–18.3) | 0.015 |
| Allele G | 67 (88.15) | 30 (55.6) | --- | -- |
| Allele A | 9 (11.84) | 24 (44.4) | ---- | ---- |
| Variables | GG + GA | AA | OR * (95% CI) | p Value |
|---|---|---|---|---|
| Risk | ||||
| Low | 3 (11.1) | 2 (7.4) | 1.5 (0.20–10.8) | 0.05 |
| Intermediate/High | 21 (77.7) | 1 (3.7) | ||
| INSS stage | ||||
| stage 1, 2 | 5 (18.5) | 3 (11.1) | 0.043 (0.003–0.57) | 0.017 |
| stage 3, 4 | 19 (70.3) | 1 (3.7) | ||
| Histology (Shimada) | ||||
| Favorable | 13 (48.1) | 2 (7.4) | 1.69 (0.134–21.26) | 0.68 |
| Unfavorable | 11 (40.7) | 1 (3.7) | ||
| MYCN | ||||
| Amplified | 1 (3.7) | ---- | NC | NC |
| Not amplified | 23 (85.2) | 3 (11.1) | ||
| Death | ||||
| Yes | 6 (22.2) | -- | NC | NC |
| No | 18 (66.7) | 3 (11.1) |
| Variables | GG | GA + AA | OR * (95% CI) | p Value |
|---|---|---|---|---|
| Risk | ||||
| Low/Intermediate | 9 (33.3) | 3 (11.1) | 2.07 (0.39–10.8) | 0.04 |
| High | 4 (14.8) | 11 (40.7) | ||
| INSS stage | ||||
| stage 1, 2 | 6 (22.2) | 1 (3.7) | 0.71 (0.07–6.53) | 0.76 |
| stage 3,4 | 7 (25.9) | 13 (48.1) | ||
| Histology (Shimada) | ||||
| Favorable | 10 (37) | 5 (18.5) | 6 (1.10–32.5) | 0.03 |
| Unfavorable | 3 (11.1) | 9 (33.3) | ||
| MYCN | ||||
| Amplified | --- | 1 (3.7) | NC | NC |
| Not amplified | 13 (48.1) | 13 (48.1) | ||
| Death | ||||
| Yes | 1 (3.7) | 5 (18.5) | 0.15 (0.01–1.51) | 0.10 |
| No | 12 (44.4) | 9 (33.3) |
| TNF-α (pg/mL) | Controls n = 27 | NB Cases n = 27 |
|---|---|---|
| Range | 1.8–3 | 7.8–80.1 |
| Mean | 2.474 | 11.851 |
| Standard deviation (SD) | 0.358 | 14.86 |
| p value | <0.001 | |
| Low levels of TNF-α (≤20 pg/mL) | 27 (100) | 24 (88.9) |
| High levels of TNF-α (≥21 pg/mL) | ------ | 3 (11.1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramírez-Pacheco, A.; Moreno-Guerrero, S.S.; Rocha-Ramírez, L.M.; Hernández-Pliego, G.; Escobar-Sánchez, M.A.; Reyes-López, A.; Sienra-Monge, J.J.L.; Juárez-Villegas, L.E. Role of Genetic Polymorphisms -238 G>A and -308 G>A, and Serum TNF-α Levels in a Cohort of Mexican Pediatric Neuroblastoma Patients: Preliminary Study. Int. J. Mol. Sci. 2024, 25, 10590. https://doi.org/10.3390/ijms251910590
Ramírez-Pacheco A, Moreno-Guerrero SS, Rocha-Ramírez LM, Hernández-Pliego G, Escobar-Sánchez MA, Reyes-López A, Sienra-Monge JJL, Juárez-Villegas LE. Role of Genetic Polymorphisms -238 G>A and -308 G>A, and Serum TNF-α Levels in a Cohort of Mexican Pediatric Neuroblastoma Patients: Preliminary Study. International Journal of Molecular Sciences. 2024; 25(19):10590. https://doi.org/10.3390/ijms251910590
Chicago/Turabian StyleRamírez-Pacheco, Arturo, Silvia Selene Moreno-Guerrero, Luz María Rocha-Ramírez, Gabriela Hernández-Pliego, María Argelia Escobar-Sánchez, Alfonso Reyes-López, Juan José Luis Sienra-Monge, and Luis Enrique Juárez-Villegas. 2024. "Role of Genetic Polymorphisms -238 G>A and -308 G>A, and Serum TNF-α Levels in a Cohort of Mexican Pediatric Neuroblastoma Patients: Preliminary Study" International Journal of Molecular Sciences 25, no. 19: 10590. https://doi.org/10.3390/ijms251910590





