Identification of Potential Growth-Related Proteins in Chick Vitreous during Emmetropization Using SWATH-MS and Targeted-Based Proteomics (MRMHR)
Abstract
1. Introduction
2. Results
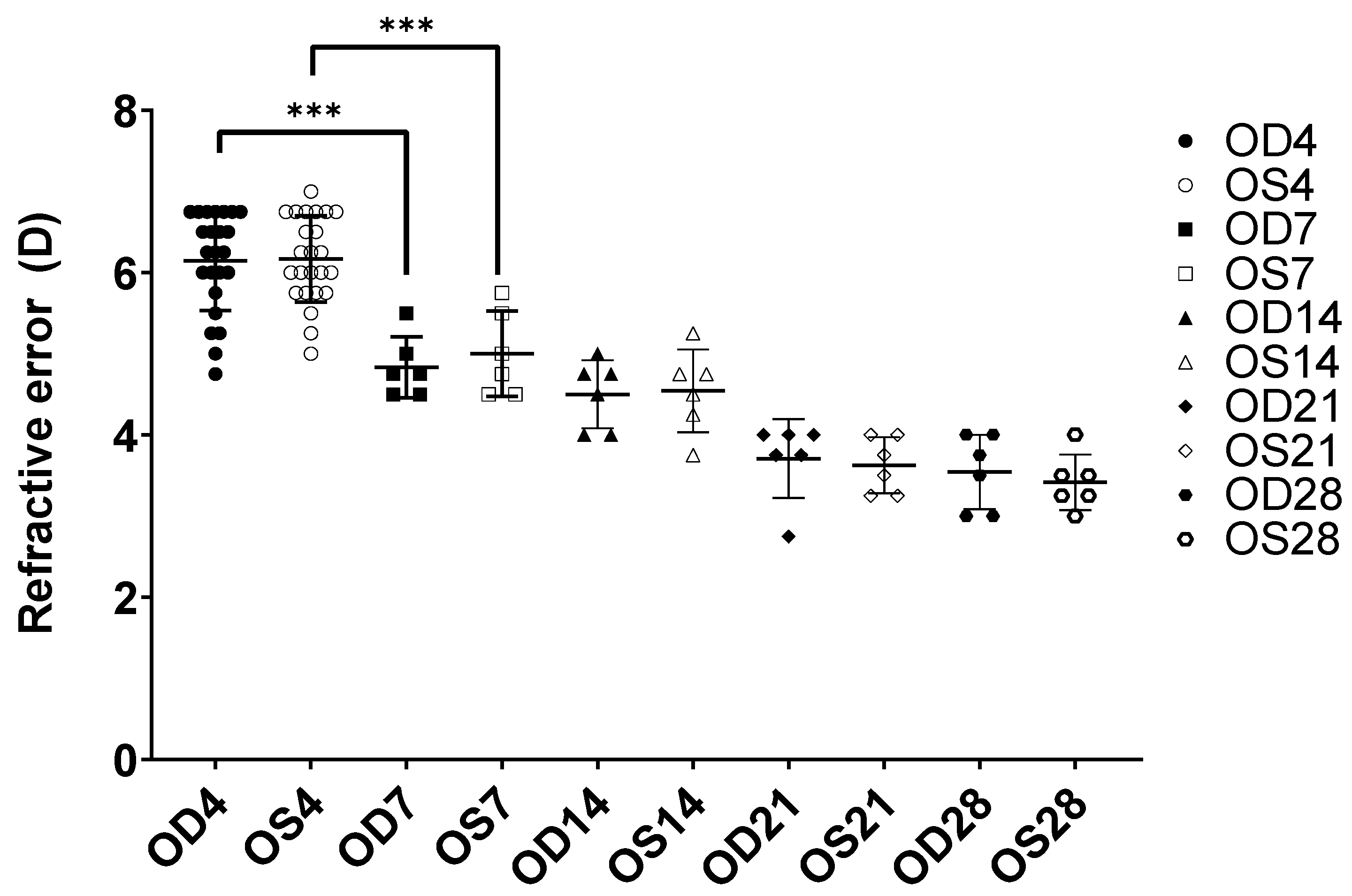
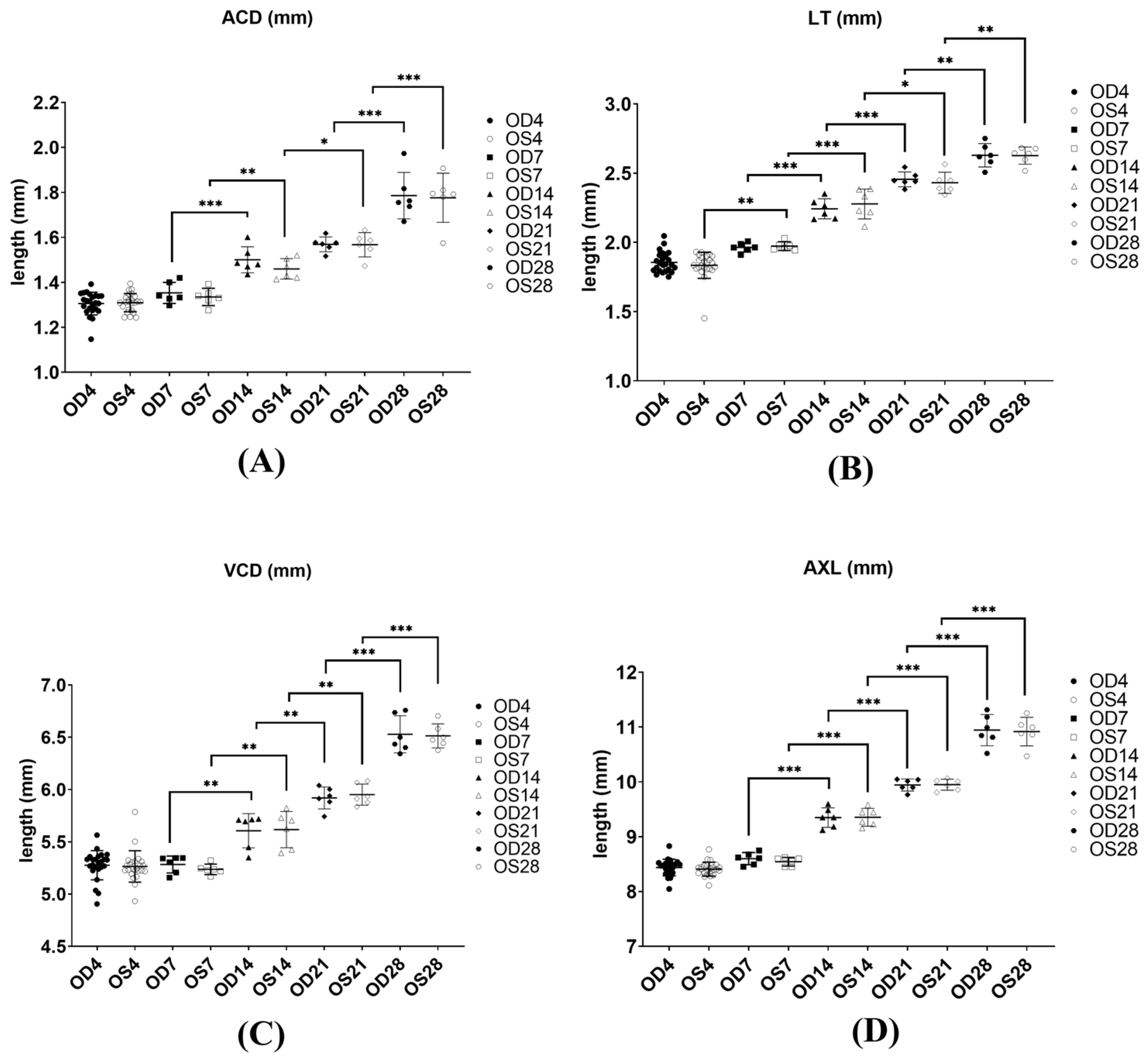
2.1. Biometric Measurements of Chicks during Normal Growth
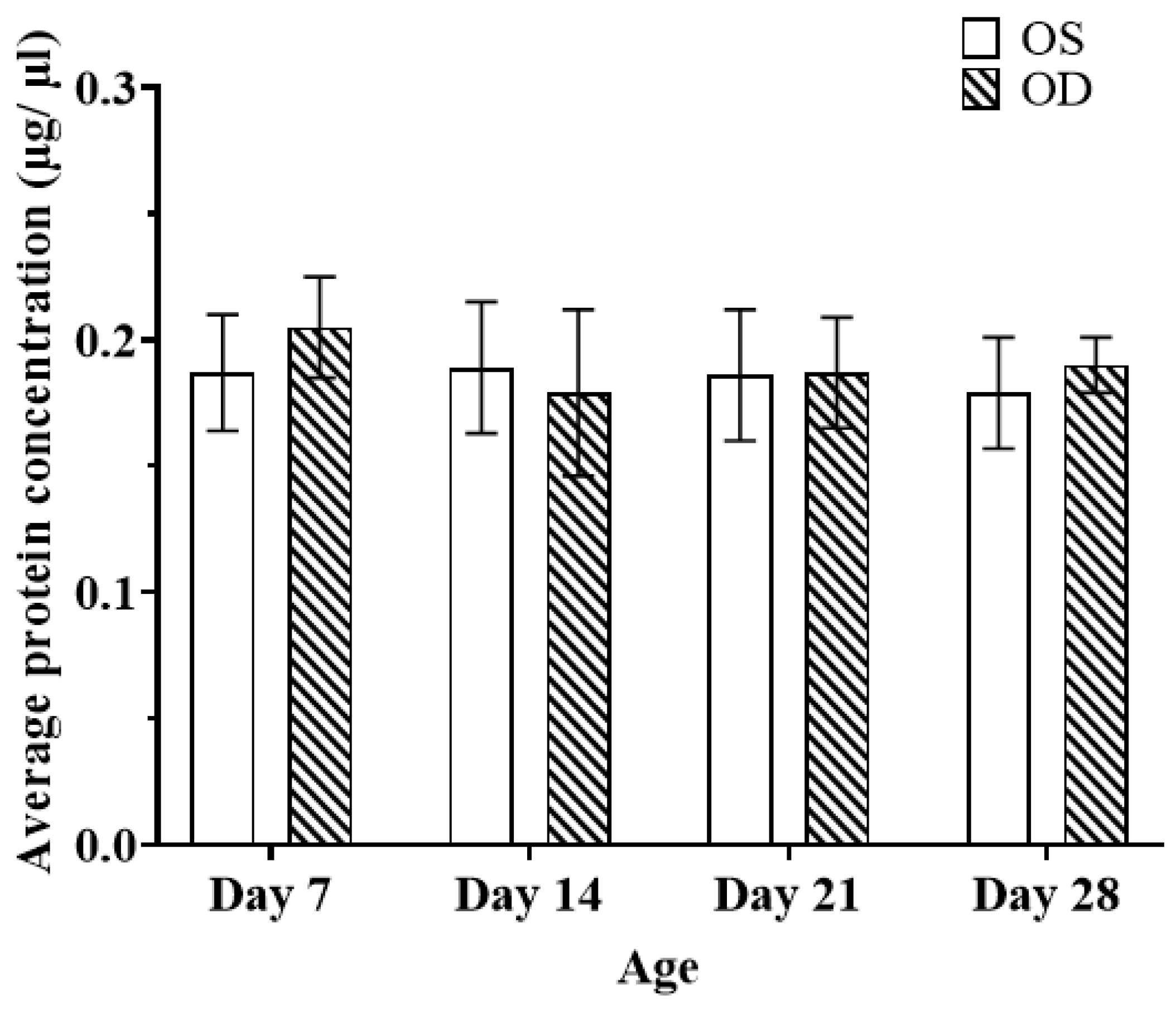
2.2. Generation of the Normal Growth Chick Vitreous Proteome
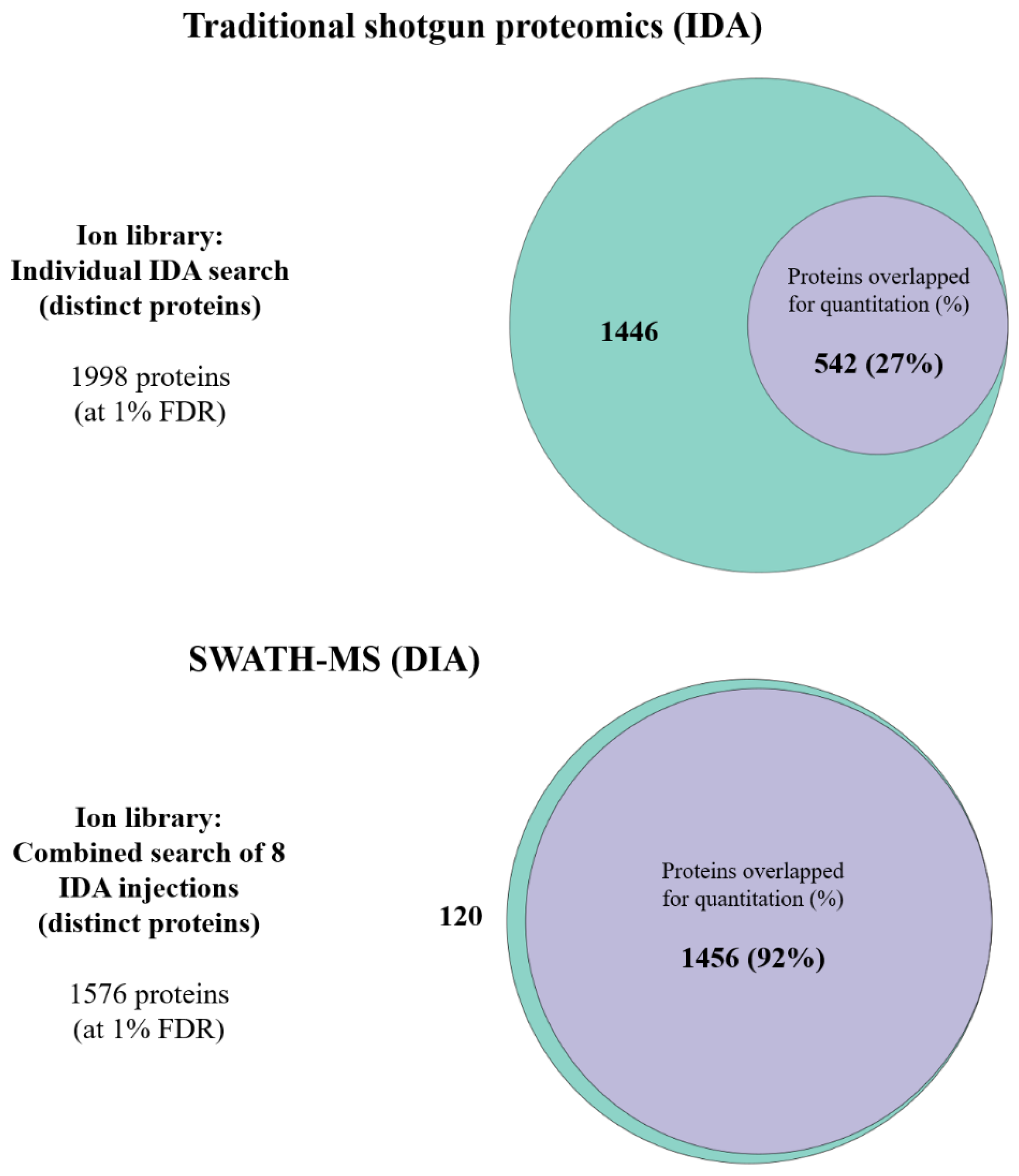
2.3. Ion Library Generation and Quantitation for SWATH-MS
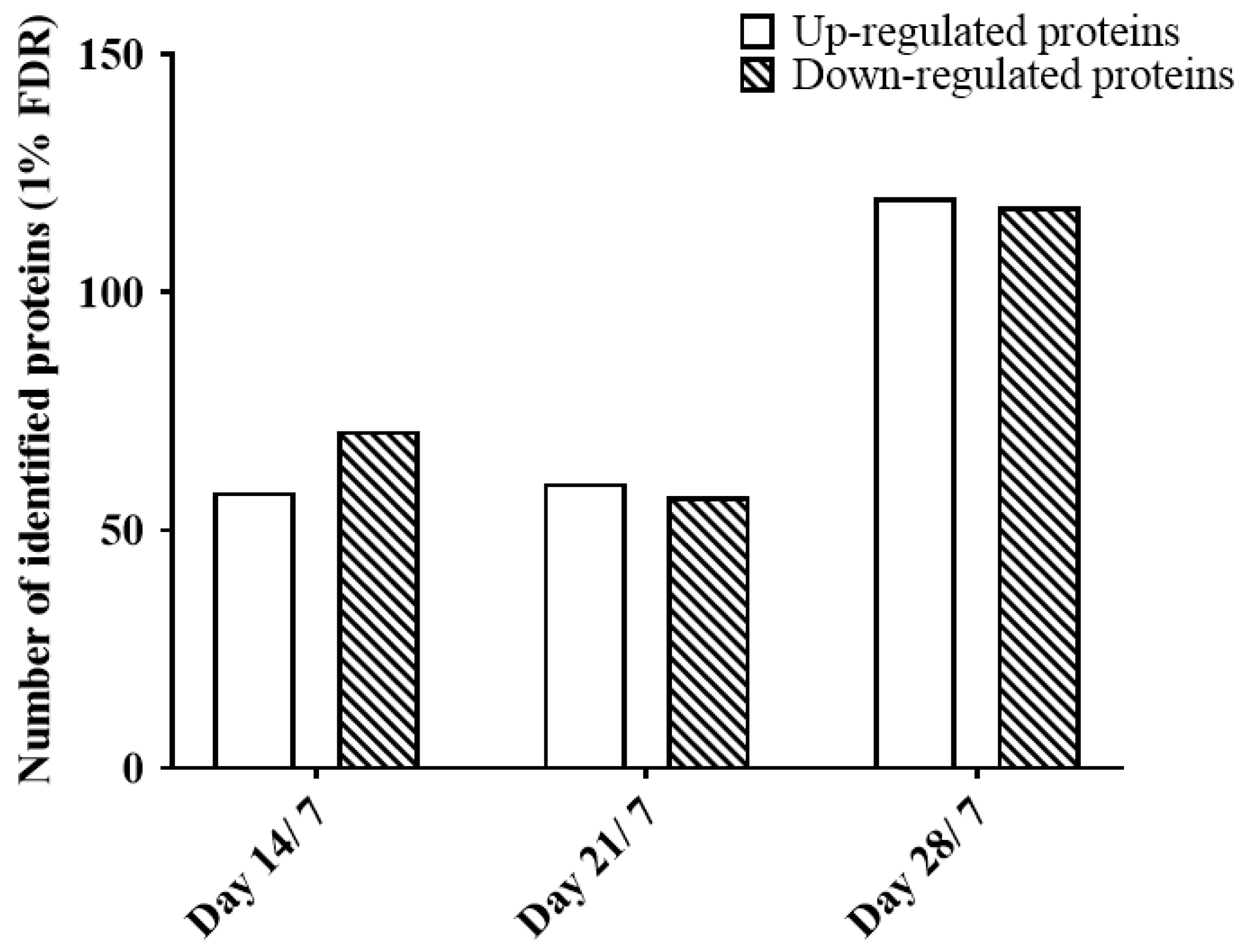
2.4. Protein Quantitation of Chick Vitreous Proteins during Normal Growth Using SWATH-MS
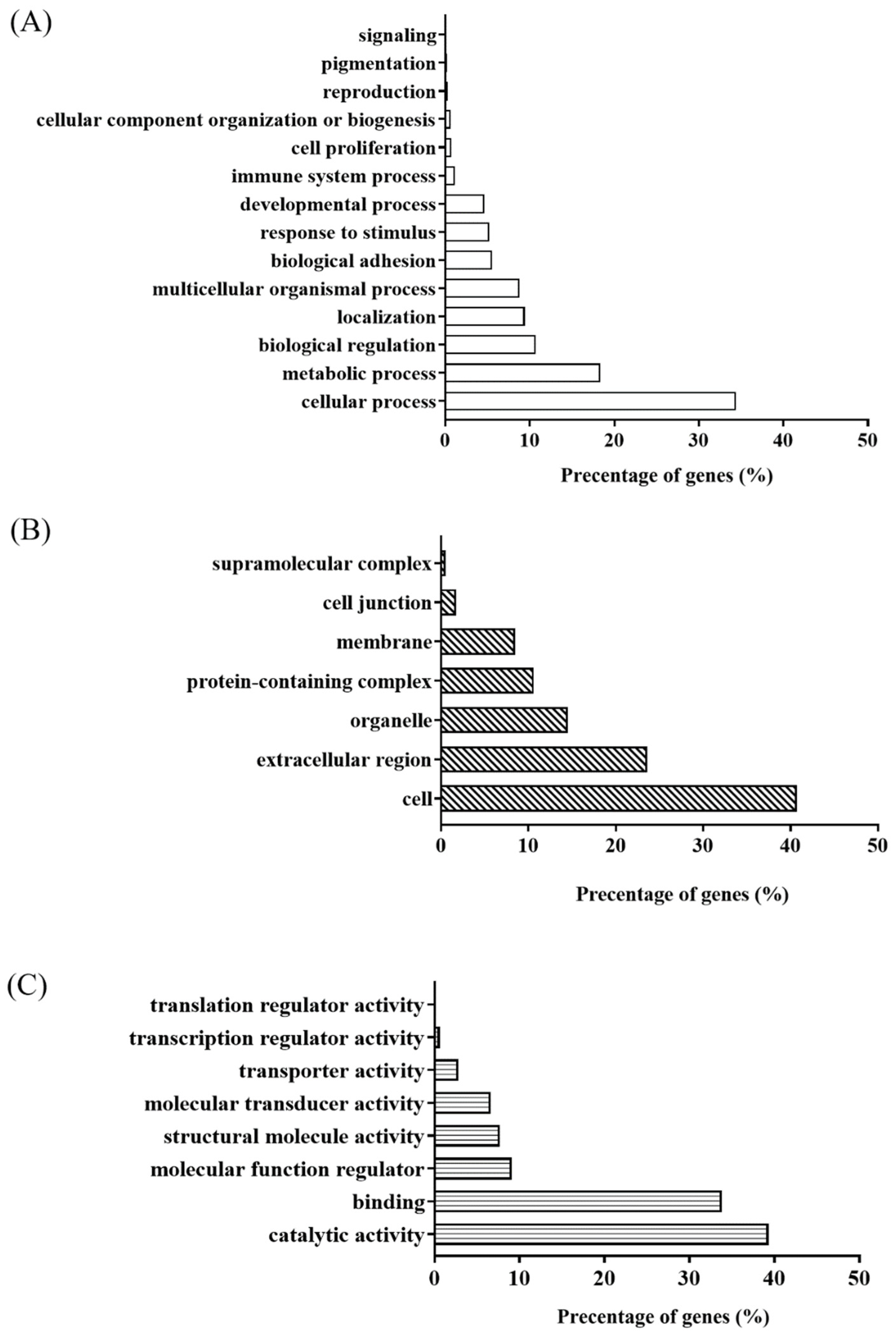
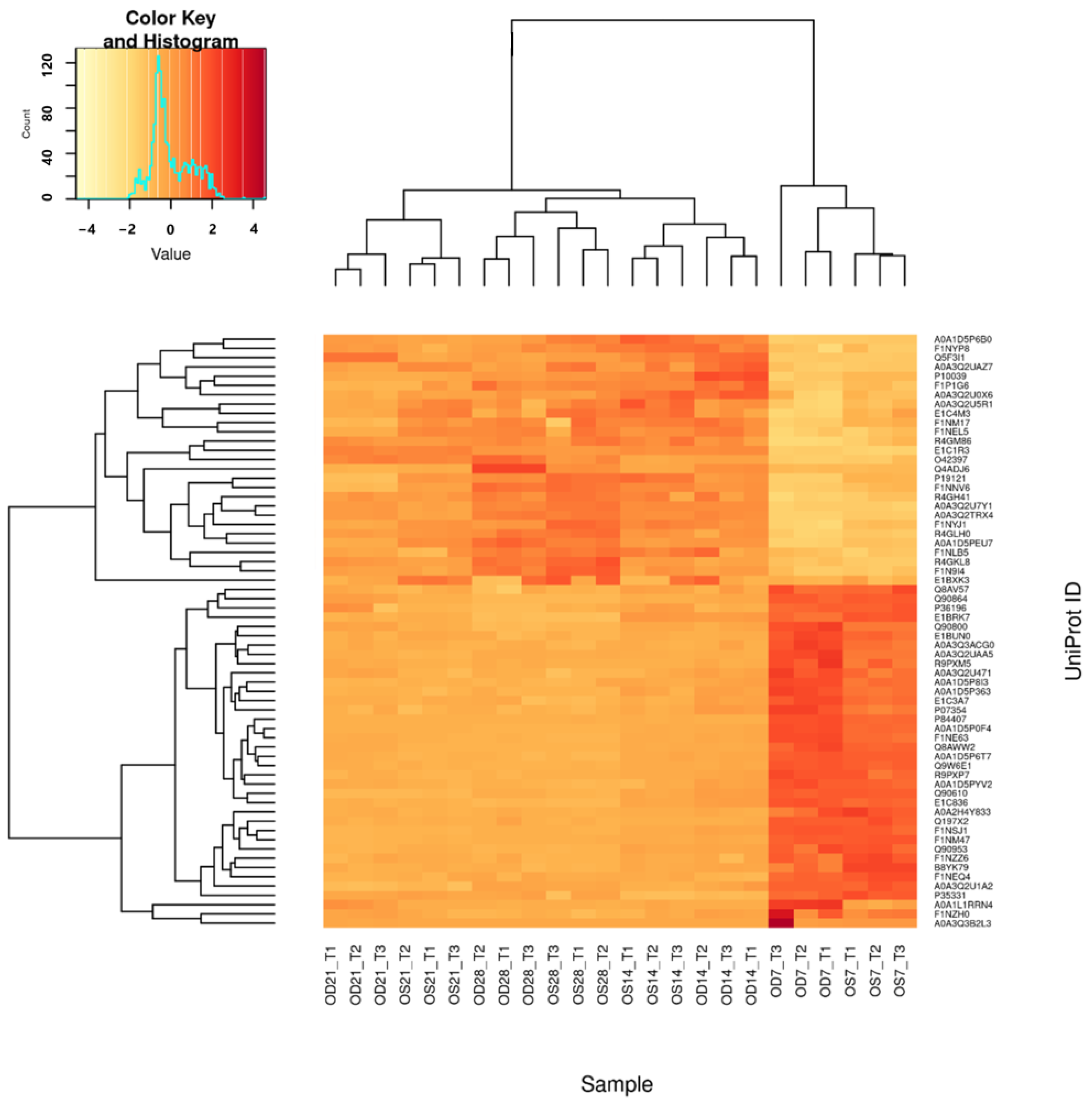
2.5. Data Analysis Using Bioinformatics Software and Confirmation Using Targeted Proteomics
2.6. Protein Validation Using High-Resolution MRM (MRMHR)
3. Discussion
3.1. Alpha-Fetoprotein (AFP)
3.2. Cadherin Superfamily Group
3.3. Contactin-2 (TAG-1)
3.4. Neurocan Core Protein (NCAM)
3.5. Reelin (RELN)
3.6. Ovotransferrin (TF)
4. Materials and Methods
4.1. Data Measurements and Vitreous Sample Collection
4.2. Ocular Growth and Weight Measurements
4.3. Vitreous Sample Preparation
4.4. In-Solution Digestion and Sample Loading
4.5. LC-MS Acquisition
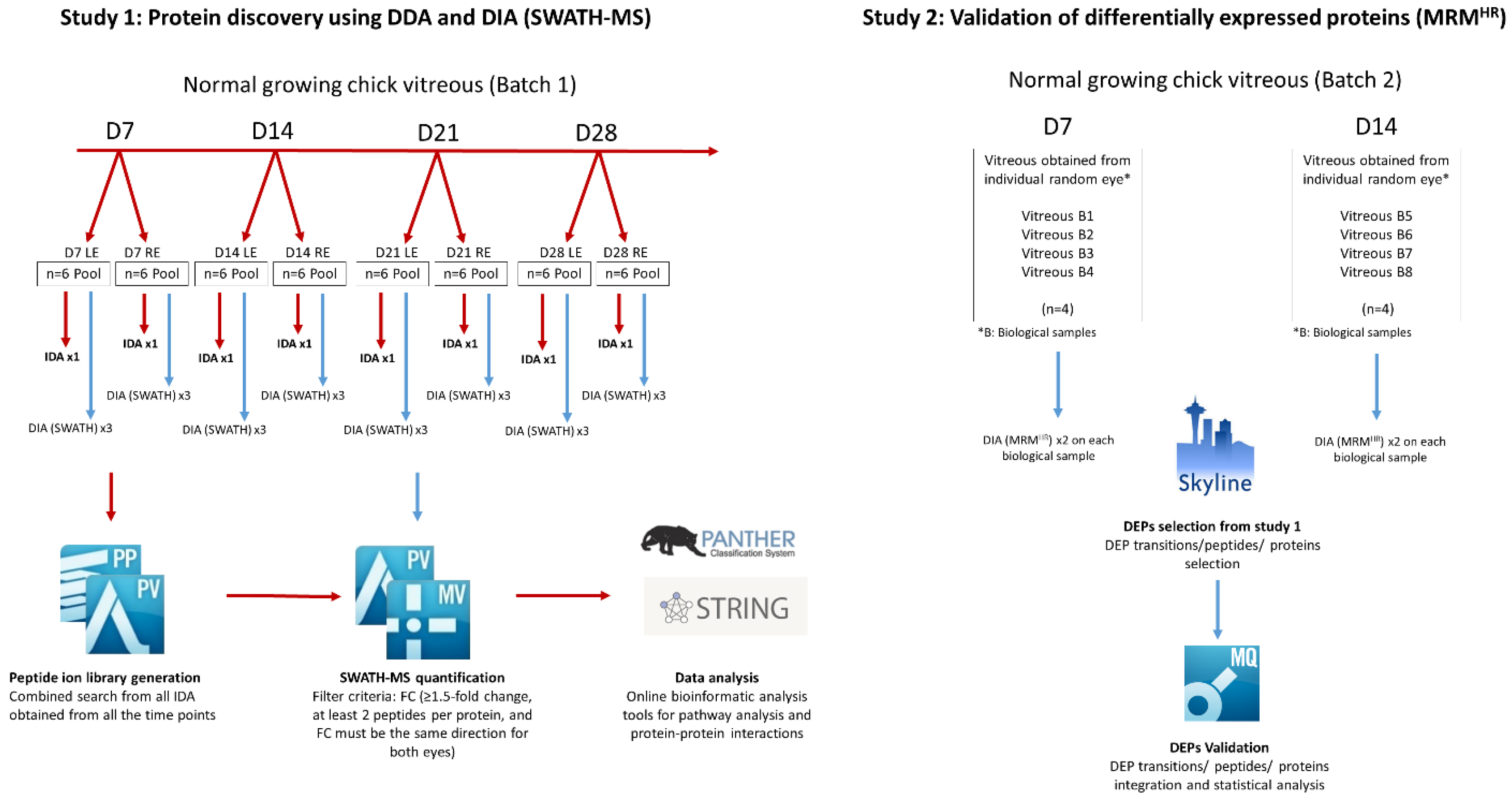
4.6. Study 1: Protein Identification and Quantification Using IDA and DIA (SWATH-MS)
4.7. Bioinformatic Analysis and Statistical Analysis
4.8. Study 2: Validation by High-Resolution Multiple Reaction Monitoring (MRMHR)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Holden, B.A.; Fricke, T.R.; Wilson, D.A.; Jong, M.; Naidoo, K.S.; Sankaridurg, P.; Wong, T.Y.; Naduvilath, T.J.; Resnikoff, S. Global Prevalence of Myopia and High Myopia and Temporal Trends from 2000 through 2050. Ophthalmology 2016, 123, 1036–1042. [Google Scholar] [CrossRef] [PubMed]
- Flitcroft, D.I. The complex interactions of retinal, optical and environmental factors in myopia aetiology. Prog. Retin. Eye Res. 2012, 31, 622–660. [Google Scholar] [CrossRef]
- Marcus, M.W.; de Vries, M.M.; Junoy Montolio, F.G.; Jansonius, N.M. Myopia as a risk factor for open-angle glaucoma: A systematic review and meta-analysis. Ophthalmology 2011, 118, 1989–1994.e1982. [Google Scholar] [CrossRef]
- Younan, C.; Mitchell, P.; Cumming, R.G.; Rochtchina, E.; Wang, J.J. Myopia and incident cataract and cataract surgery: The blue mountains eye study. Investig. Ophthalmol. Vis. Sci. 2002, 43, 3625–3632. [Google Scholar]
- Morgan, I.; Rose, K. How genetic is school myopia? Prog. Retin. Eye Res. 2005, 24, 1–38. [Google Scholar] [CrossRef] [PubMed]
- Foster, P.J.; Jiang, Y. Epidemiology of myopia. Eye 2014, 28, 202–208. [Google Scholar] [CrossRef]
- Rahi, J.S.; Cumberland, P.M.; Peckham, C.S. Myopia over the lifecourse: Prevalence and early life influences in the 1958 British birth cohort. Ophthalmology 2011, 118, 797–804. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, D.L.; Atkinson, J.; Braddick, O.; Bobier, W.; Durden, K. Reduction of infant myopia: A longitudinal cycloplegic study. Vis. Res. 1995, 35, 1313–1324. [Google Scholar] [CrossRef]
- Bradley, D.V.; Fernandes, A.; Lynn, M.; Tigges, M.; Boothe, R.G. Emmetropization in the rhesus monkey (Macaca mulatta): Birth to young adulthood. Investig. Ophthalmol. Vis. Sci. 1999, 40, 214–229. [Google Scholar]
- Wallman, J.; Adams, J.I.; Trachtman, J.N. The eyes of young chickens grow toward emmetropia. Investig. Ophthalmol. Vis. Sci. 1981, 20, 557–561. [Google Scholar]
- Howlett, M.H.; McFadden, S.A. Emmetropization and schematic eye models in developing pigmented guinea pigs. Vis. Res. 2007, 47, 1178–1190. [Google Scholar] [CrossRef] [PubMed]
- Kisilak, M.L.; Campbell, M.C.; Hunter, J.J.; Irving, E.L.; Huang, L. Aberrations of chick eyes during normal growth and lens induction of myopia. J. Comp. Physiol. A 2006, 192, 845–855. [Google Scholar] [CrossRef] [PubMed]
- Wallman, J.; Adams, J.I. Developmental aspects of experimental myopia in chicks: Susceptibility, recovery and relation to emmetropization. Vis. Res. 1987, 27, 1139–1163. [Google Scholar] [CrossRef]
- Wallman, J.; Turkel, J.; Trachtman, J. Extreme myopia produced by modest change in early visual experience. Science 1978, 201, 1249–1251. [Google Scholar] [CrossRef] [PubMed]
- Theocharis, A.D.; Papageorgakopoulou, N.; Feretis, E.; Theocharis, D.A. Occurrence and structural characterization of versican-like proteoglycan in human vitreous. Biochimie 2002, 84, 1237–1243. [Google Scholar] [CrossRef]
- Mahajan, V.B.; Skeie, J.M. Translational vitreous proteomics. PROTEOM.–Clin. Appl. 2014, 8, 204–208. [Google Scholar] [CrossRef]
- Halfter, W.; Dong, S.; Dong, A.; Eller, A.W.; Nischt, R. Origin and turnover of ECM proteins from the inner limiting membrane and vitreous body. Eye 2008, 22, 1207–1213. [Google Scholar] [CrossRef]
- Murthy, K.R.; Goel, R.; Subbannayya, Y.; Jacob, H.K.; Murthy, P.R.; Manda, S.S.; Patil, A.H.; Sharma, R.; Sahasrabuddhe, N.A.; Parashar, A.; et al. Proteomic analysis of human vitreous humor. Clin. Proteom. 2014, 11, 29. [Google Scholar] [CrossRef]
- Bertrand, E.; Fritsch, C.; Diether, S.; Lambrou, G.; Muller, D.; Schaeffel, F.; Schindler, P.; Schmid, K.L.; van Oostrum, J.; Voshol, H. Identification of apolipoprotein A-I as a “STOP” signal for myopia. Mol. Cell. Proteom. 2006, 5, 2158–2166. [Google Scholar] [CrossRef]
- Yu, F.J.; Lam, T.C.; Liu, L.Q.; Chun, R.K.; Cheung, J.K.; Li, K.K.; To, C.H. Isotope-coded protein label based quantitative proteomic analysis reveals significant up-regulation of apolipoprotein A1 and ovotransferrin in the myopic chick vitreous. Sci. Rep. 2017, 7, 12649. [Google Scholar] [CrossRef]
- Gao, B.B.; Chen, X.; Timothy, N.; Aiello, L.P.; Feener, E.P. Characterization of the vitreous proteome in diabetes without diabetic retinopathy and diabetes with proliferative diabetic retinopathy. J. Proteome Res. 2008, 7, 2516–2525. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Kim, S.J.; Kim, K.; Kang, U.B.; Lee, C.; Park, K.S.; Yu, H.G.; Kim, Y. Profiling of vitreous proteomes from proliferative diabetic retinopathy and nondiabetic patients. Proteomics 2007, 7, 4203–4215. [Google Scholar] [CrossRef] [PubMed]
- Loukovaara, S.; Nurkkala, H.; Tamene, F.; Gucciardo, E.; Liu, X.; Repo, P.; Lehti, K.; Varjosalo, M. Quantitative Proteomics Analysis of Vitreous Humor from Diabetic Retinopathy Patients. J. Proteome Res. 2015, 14, 5131–5143. [Google Scholar] [CrossRef] [PubMed]
- Tabb, D.L.; Vega-Montoto, L.; Rudnick, P.A.; Variyath, A.M.; Ham, A.J.; Bunk, D.M.; Kilpatrick, L.E.; Billheimer, D.D.; Blackman, R.K.; Cardasis, H.L.; et al. Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry. J. Proteome Res. 2010, 9, 761–776. [Google Scholar] [CrossRef]
- Gillet, L.C.; Navarro, P.; Tate, S.; Rost, H.; Selevsek, N.; Reiter, L.; Bonner, R.; Aebersold, R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: A new concept for consistent and accurate proteome analysis. Mol. Cell. Proteom. 2012, 11, O111 016717. [Google Scholar] [CrossRef]
- Elliott, M.H.; Smith, D.S.; Parker, C.E.; Borchers, C. Current trends in quantitative proteomics. J. Mass. Spectrom. 2009, 44, 1637–1660. [Google Scholar] [CrossRef]
- Anderson, L.; Hunter, C.L. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell. Proteom. 2006, 5, 573–588. [Google Scholar] [CrossRef] [PubMed]
- Gerber, S.A.; Rush, J.; Stemman, O.; Kirschner, M.W.; Gygi, S.P. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. USA 2003, 100, 6940–6945. [Google Scholar] [CrossRef]
- Peti, A.P.F.; Locachevic, G.A.; Prado, M.K.B.; de Moraes, L.A.B.; Faccioli, L.H. High-resolution multiple reaction monitoring method for quantification of steroidal hormones in plasma. J. Mass. Spectrom. 2018, 53, 423–431. [Google Scholar] [CrossRef]
- Schilling, B.; MacLean, B.; Held, J.M.; Sahu, A.K.; Rardin, M.J.; Sorensen, D.J.; Peters, T.; Wolfe, A.J.; Hunter, C.L.; MacCoss, M.J.; et al. Multiplexed, Scheduled, High-Resolution Parallel Reaction Monitoring on a Full Scan QqTOF Instrument with Integrated Data-Dependent and Targeted Mass Spectrometric Workflows. Anal. Chem. 2015, 87, 10222–10229. [Google Scholar] [CrossRef]
- Cheung, J.K.; Li, K.K.; Zhou, L.; To, C.H.; Lam, T.C. Data on protein changes of chick vitreous during normal eye growth using data-independent acquisition (SWATH-MS). Data Brief. 2020, 30, 105576. [Google Scholar] [CrossRef] [PubMed]
- Koss, M.J.; Hoffmann, J.; Nguyen, N.; Pfister, M.; Mischak, H.; Mullen, W.; Husi, H.; Rejdak, R.; Koch, F.; Jankowski, J.; et al. Proteomics of vitreous humor of patients with exudative age-related macular degeneration. PLoS ONE 2014, 9, e96895. [Google Scholar] [CrossRef] [PubMed]
- Yee, K.M.; Feener, E.P.; Madigan, M.; Jackson, N.J.; Gao, B.B.; Ross-Cisneros, F.N.; Provis, J.; Aiello, L.P.; Sadun, A.A.; Sebag, J. Proteomic Analysis of Embryonic and Young Human Vitreous. Investig. Ophthalmol. Vis. Sci. 2015, 56, 7036–7042. [Google Scholar] [CrossRef]
- Liu, Y.; Bouhenni, R.A.; Dufresne, C.P.; Semba, R.D.; Edward, D.P. Differential Expression of Vitreous Proteins in Young and Mature New Zealand White Rabbits. PLoS ONE 2016, 11, e0153560. [Google Scholar] [CrossRef]
- Skeie, J.M.; Mahajan, V.B. Proteomic interactions in the mouse vitreous-retina complex. PLoS ONE 2013, 8, e82140. [Google Scholar] [CrossRef] [PubMed]
- Shan, S.W.; Tse, D.Y.; Zuo, B.; To, C.H.; Liu, Q.; McFadden, S.A.; Chun, R.K.; Bian, J.; Li, K.K.; Lam, T.C. Integrated SWATH-based and targeted-based proteomics provide insights into the retinal emmetropization process in guinea pig. J. Proteom. 2018, 181, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Peterson, A.C.; Russell, J.D.; Bailey, D.J.; Westphall, M.S.; Coon, J.J. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol. Cell. Proteom. 2012, 11, 1475–1488. [Google Scholar] [CrossRef]
- Adinolfi, A.; Adinolfi, M.; Lessof. Alpha-feto-protein during development and in disease. J. Med. Genet. 1975, 12, 138–151. [Google Scholar] [CrossRef]
- Ball, D.; Rose, E.; Alpert, E. Alpha-fetoprotein levels in normal adults. Am. J. Med. Sci. 1992, 303, 157–159. [Google Scholar] [CrossRef]
- Taketa, K. Alpha-fetoprotein: Reevaluation in hepatology. Hepatology 1990, 12, 1420–1432. [Google Scholar] [CrossRef]
- Gillespie, J.R.; Uversky, V.N. Structure and function of alpha-fetoprotein: A biophysical overview. Biochim. Et Biophys. Acta (BBA)-Protein Struct. Mol. Enzymol. 2000, 1480, 41–56. [Google Scholar] [CrossRef]
- Panova, I.G.; Tatikolov, A.S.; Poltavtseva, R.A.; Sukhikh, G.T. alpha-Fetoprotein in human fetal vitreous body. Bull. Exp. Biol. Med. 2011, 150, 420–421. [Google Scholar] [CrossRef] [PubMed]
- Sell, S.; Becker, F.F. alpha-Fetoprotein. J. Natl. Cancer Inst. 1978, 60, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Nayak, N.C.; Mital, I. The dynamics of alpha-fetoprotein and albumin synthesis in human and rat liver during normal ontogeny. Am. J. Pathol. 1977, 86, 359–374. [Google Scholar] [PubMed]
- Niessen, C.M.; Leckband, D.; Yap, A.S. Tissue organization by cadherin adhesion molecules: Dynamic molecular and cellular mechanisms of morphogenetic regulation. Physiol. Rev. 2011, 91, 691–731. [Google Scholar] [CrossRef] [PubMed]
- Halbleib, J.M.; Nelson, W.J. Cadherins in development: Cell adhesion, sorting, and tissue morphogenesis. Genes. Dev. 2006, 20, 3199–3214. [Google Scholar] [CrossRef]
- Liu, Q.; Duff, R.J.; Liu, B.; Wilson, A.L.; Babb-Clendenon, S.G.; Francl, J.; Marrs, J.A. Expression of cadherin10, a type II classic cadherin gene, in the nervous system of the embryonic zebrafish. Gene Expr. Patterns 2006, 6, 703–710. [Google Scholar] [CrossRef]
- Fushimi, D.; Arndt, K.; Takeichi, M.; Redies, C. Cloning and expression analysis of cadherin-10 in the CNS of the chicken embryo. Dev. Dyn. 1997, 209, 269–285. [Google Scholar] [CrossRef]
- Barnes, S.H.; Price, S.R.; Wentzel, C.; Guthrie, S.C. Cadherin-7 and cadherin-6B differentially regulate the growth, branching and guidance of cranial motor axons. Development 2010, 137, 805–814. [Google Scholar] [CrossRef]
- Furley, A.J.; Morton, S.B.; Manalo, D.; Karagogeos, D.; Dodd, J.; Jessell, T.M. The axonal glycoprotein TAG-1 is an immunoglobulin superfamily member with neurite outgrowth-promoting activity. Cell 1990, 61, 157–170. [Google Scholar] [CrossRef]
- Karagogeos, D.; Morton, S.B.; Casano, F.; Dodd, J.; Jessell, T.M. Developmental expression of the axonal glycoprotein TAG-1: Differential regulation by central and peripheral neurons in vitro. Development 1991, 112, 51–67. [Google Scholar] [CrossRef]
- Wolman, M.A.; Sittaramane, V.K.; Essner, J.J.; Yost, H.J.; Chandrasekhar, A.; Halloran, M.C. Transient axonal glycoprotein-1 (TAG-1) and laminin-alpha1 regulate dynamic growth cone behaviors and initial axon direction in vivo. Neural Dev. 2008, 3, 6. [Google Scholar] [CrossRef] [PubMed]
- Baeriswyl, T.; Stoeckli, E.T. Axonin-1/TAG-1 is required for pathfinding of granule cell axons in the developing cerebellum. Neural Dev. 2008, 3, 7. [Google Scholar] [CrossRef] [PubMed]
- Milev, P.; Maurel, P.; Haring, M.; Margolis, R.K.; Margolis, R.U. TAG-1/axonin-1 is a high-affinity ligand of neurocan, phosphacan/protein-tyrosine phosphatase-zeta/beta, and N-CAM. J. Biol. Chem. 1996, 271, 15716–15723. [Google Scholar] [CrossRef] [PubMed]
- Engel, M.; Maurel, P.; Margolis, R.U.; Margolis, R.K. Chondroitin sulfate proteoglycans in the developing central nervous system. I. cellular sites of synthesis of neurocan and phosphacan. J. Comp. Neurol. 1996, 366, 34–43. [Google Scholar] [CrossRef]
- Rauch, U.; Gao, P.; Janetzko, A.; Flaccus, A.; Hilgenberg, L.; Tekotte, H.; Margolis, R.K.; Margolis, R.U. Isolation and characterization of developmentally regulated chondroitin sulfate and chondroitin/keratan sulfate proteoglycans of brain identified with monoclonal antibodies. J. Biol. Chem. 1991, 266, 14785–14801. [Google Scholar] [CrossRef]
- Margolis, R.K.; Rauch, U.; Maurel, P.; Margolis, R.U. Neurocan and phosphacan: Two major nervous tissue-specific chondroitin sulfate proteoglycans. Perspect. Dev. Neurobiol. 1996, 3, 273–290. [Google Scholar]
- Oohira, A.; Matsui, F.; Tokita, Y.; Yamauchi, S.; Aono, S. Molecular interactions of neural chondroitin sulfate proteoglycans in the brain development. Arch. Biochem. Biophys. 2000, 374, 24–34. [Google Scholar] [CrossRef]
- Aquino, D.A.; Margolis, R.U.; Margolis, R.K. Immunocytochemical localization of a chondroitin sulfate proteoglycan in nervous tissue. I. Adult brain, retina, and peripheral nerve. J. Cell Biol. 1984, 99, 1117–1129. [Google Scholar] [CrossRef]
- Inatani, M.; Tanihara, H.; Oohira, A.; Honjo, M.; Honda, Y. Identification of a nervous tissue-specific chondroitin sulfate proteoglycan, neurocan, in developing rat retina. Investig. Ophthalmol. Vis. Sci. 1999, 40, 2350–2359. [Google Scholar]
- Meyer-Puttlitz, B.; Junker, E.; Margolis, R.U.; Margolis, R.K. Chondroitin sulfate proteoglycans in the developing central nervous system. II. Immunocytochemical localization of neurocan and phosphacan. J. Comp. Neurol. 1996, 366, 44–54. [Google Scholar] [CrossRef]
- D’Arcangelo, G. Reelin in the Years: Controlling Neuronal Migration and Maturation in the Mammalian Brain. Adv. Neurosci. 2014, 2014, 19. [Google Scholar] [CrossRef]
- Leemhuis, J.; Bouche, E.; Frotscher, M.; Henle, F.; Hein, L.; Herz, J.; Meyer, D.K.; Pichler, M.; Roth, G.; Schwan, C.; et al. Reelin signals through apolipoprotein E receptor 2 and Cdc42 to increase growth cone motility and filopodia formation. J. Neurosci. 2010, 30, 14759–14772. [Google Scholar] [CrossRef]
- Weeber, E.J.; Beffert, U.; Jones, C.; Christian, J.M.; Forster, E.; Sweatt, J.D.; Herz, J. Reelin and ApoE receptors cooperate to enhance hippocampal synaptic plasticity and learning. J. Biol. Chem. 2002, 277, 39944–39952. [Google Scholar] [CrossRef]
- Hiesberger, T.; Trommsdorff, M.; Howell, B.W.; Goffinet, A.; Mumby, M.C.; Cooper, J.A.; Herz, J. Direct binding of Reelin to VLDL receptor and ApoE receptor 2 induces tyrosine phosphorylation of disabled-1 and modulates tau phosphorylation. Neuron 1999, 24, 481–489. [Google Scholar] [CrossRef]
- Frotscher, M. Role for Reelin in stabilizing cortical architecture. Trends Neurosci. 2010, 33, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, M.A.; Mikhailova, T.; Li, X.; Porter, B.A.; Bah, A.; Kotula, L. Src family kinases, adaptor proteins and the actin cytoskeleton in epithelial-to-mesenchymal transition. Cell Commun. Signal. 2021, 19, 67. [Google Scholar] [CrossRef] [PubMed]
- Williams, J. A comparison of glycopeptides from the ovotransferrin and serum transferrin of the hen. Biochem. J. 1968, 108, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.H.; Lee, J.H.; Ahn, D.U.; Paik, H.D. In vitro antioxidant and mineral-chelating properties of natural and autocleaved ovotransferrin. J. Sci. Food Agric. 2015, 95, 2065–2070. [Google Scholar] [CrossRef]
- Rada, J.A.; Huang, Y.; Rada, K.G. Identification of choroidal ovotransferrin as a potential ocular growth regulator. Curr. Eye Res. 2001, 22, 121–132. [Google Scholar] [CrossRef]
- McBrien, N.A. Regulation of scleral metabolism in myopia and the role of transforming growth factor-beta. Exp. Eye Res. 2013, 114, 128–140. [Google Scholar] [CrossRef] [PubMed]
- Jobling, A.I.; Wan, R.; Gentle, A.; Bui, B.V.; McBrien, N.A. Retinal and choroidal TGF-beta in the tree shrew model of myopia: Isoform expression, activation and effects on function. Exp. Eye Res. 2009, 88, 458–466. [Google Scholar] [CrossRef] [PubMed]
- Schippert, R.; Brand, C.; Schaeffel, F.; Feldkaemper, M.P. Changes in scleral MMP-2, TIMP-2 and TGFbeta-2 mRNA expression after imposed myopic and hyperopic defocus in chickens. Exp. Eye Res. 2006, 82, 710–719. [Google Scholar] [CrossRef] [PubMed]
- Lambert, J.P.; Ivosev, G.; Couzens, A.L.; Larsen, B.; Taipale, M.; Lin, Z.Y.; Zhong, Q.; Lindquist, S.; Vidal, M.; Aebersold, R.; et al. Mapping differential interactomes by affinity purification coupled with data-independent mass spectrometry acquisition. Nat. Methods 2013, 10, 1239–1245. [Google Scholar] [CrossRef] [PubMed]
- UniProt, C. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- MacLean, B.; Tomazela, D.M.; Shulman, N.; Chambers, M.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.L.; Liebler, D.C.; MacCoss, M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar] [CrossRef]



| (Day 14/7) SWATH-MS FC | (Day 21/7) SWATH-MS FC | (Day 28/7) SWATH-MS FC | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| UniProt Accession Number | Protein Name | Gene Name | OD14/OD7 | OS14/OS7 | AVE FC | OD21/OD7 | OS21/OS7 | AVE FC | OD28/OD7 | OS28/OS7 | AVE FC |
| R4GLH0 | IGFBP N-terminal domain-containing protein | ESM1 | 4.08 | 3.02 | 3.55 ± 0.75 | 4.53 | 3.51 | 4.02 ± 0.72 | 6.73 | 5.09 | 5.91 ± 1.16 |
| Q4ADJ6 | Ovotransferrin | TFEW | 4.09 | 2.3 | 3.2 ± 1.27 | 1.81 | 2.26 | 2.04 ± 0.32 | 9.39 | 5.11 | 7.25 ± 3.03 |
| P84407 | Alpha-fetoprotein | AFP | 0.01 | 0.4 | 0.21 ± 0.28 | 0.01 | 0.01 | 0.01 ± 0.00 | 0.01 | 0.02 | 0.02 ± 0.01 |
| P79995 | Cadherin-10 | CDH10 | 0.46 | 0.12 | 0.29 ± 0.24 | 0.43 | 0.39 | 0.41 ± 0.03 | 0.4 | 0.35 | 0.38 ± 0.04 |
| P24503 | Cadherin-4 | CDH4 | 0.31 | 0.32 | 0.32 ± 0.01 | 0.34 | 0.43 | 0.39 ± 0.06 | 0.37 | 0.4 | 0.39 ± 0.02 |
| E1C3A7 | Cadherin 22 | CDH22 | 0.41 | 0.14 | 0.28 ± 0.19 | 0.34 | 0.41 | 0.38 ± 0.05 | 0.33 | 0.3 | 0.32 ± 0.02 |
| Q8AWW2 | Cadherin-7 | N/A | 0.33 | 0.38 | 0.36 ± 0.04 | 0.39 | 0.32 | 0.36 ± 0.05 | 0.32 | 0.34 | 0.33 ± 0.01 |
| Q8QGH3 | Cadherin-20 | CDH20 | 0.47 | 0.61 | 0.54 ± 0.1 | 0.44 | 0.44 | 0.44 ± 0.00 | 0.45 | 0.53 | 0.49 ± 0.06 |
| Q9W6E1 | Neurocan core protein | N/A | 0.23 | 0.48 | 0.36 ± 0.18 | 0.21 | 0.2 | 0.21 ± 0.01 | 0.17 | 0.19 | 0.18 ± 0.01 |
| Q90953 | Versican core protein | VCAN CSPG2 | 0.33 | 0.59 | 0.46 ± 0.18 | 0.2 | 0.25 | 0.23 ± 0.04 | 0.19 | 0.19 | 0.19 ± 0.00 |
| O93574 | Reelin | RELN | 0.41 | 0.58 | 0.5 ± 0.12 | 0.45 | 0.48 | 0.47 ± 0.02 | 0.42 | 0.47 | 0.45 ± 0.04 |
| F1NSJ1 | Contactin-2 | CNTN2 | 0.51 | 0.23 | 0.37 ± 0.2 | 0.44 | 0.46 | 0.45 ± 0.01 | 0.52 | 0.52 | 0.52 ± 0.00 |
| P35331 | Neuronal cell adhesion molecule (Nr-CAM) | NRCAM | 0.32 | 0.57 | 0.45 ± 0.18 | 0.59 | 0.56 | 0.58 ± 0.02 | 0.67 | 0.5 | 0.59 ± 0.12 |
| Q90610 | Neogenin (Fragment) | NEO1 | 0.65 | 0.39 | 0.52 ± 0.18 | 0.54 | 0.55 | 0.55 ± 0.00 | 0.67 | 0.62 | 0.65 ± 0.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheung, J.K.-W.; Li, K.-K.; Zhou, L.; To, C.-H.; Lam, T.C. Identification of Potential Growth-Related Proteins in Chick Vitreous during Emmetropization Using SWATH-MS and Targeted-Based Proteomics (MRMHR). Int. J. Mol. Sci. 2024, 25, 10644. https://doi.org/10.3390/ijms251910644
Cheung JK-W, Li K-K, Zhou L, To C-H, Lam TC. Identification of Potential Growth-Related Proteins in Chick Vitreous during Emmetropization Using SWATH-MS and Targeted-Based Proteomics (MRMHR). International Journal of Molecular Sciences. 2024; 25(19):10644. https://doi.org/10.3390/ijms251910644
Chicago/Turabian StyleCheung, Jimmy Ka-Wai, King-Kit Li, Lei Zhou, Chi-Ho To, and Thomas Chuen Lam. 2024. "Identification of Potential Growth-Related Proteins in Chick Vitreous during Emmetropization Using SWATH-MS and Targeted-Based Proteomics (MRMHR)" International Journal of Molecular Sciences 25, no. 19: 10644. https://doi.org/10.3390/ijms251910644
APA StyleCheung, J. K.-W., Li, K.-K., Zhou, L., To, C.-H., & Lam, T. C. (2024). Identification of Potential Growth-Related Proteins in Chick Vitreous during Emmetropization Using SWATH-MS and Targeted-Based Proteomics (MRMHR). International Journal of Molecular Sciences, 25(19), 10644. https://doi.org/10.3390/ijms251910644






