Unraveling the Role of JMJD1B in Genome Stability and the Malignancy of Melanomas
Abstract
:1. Introduction
2. Results
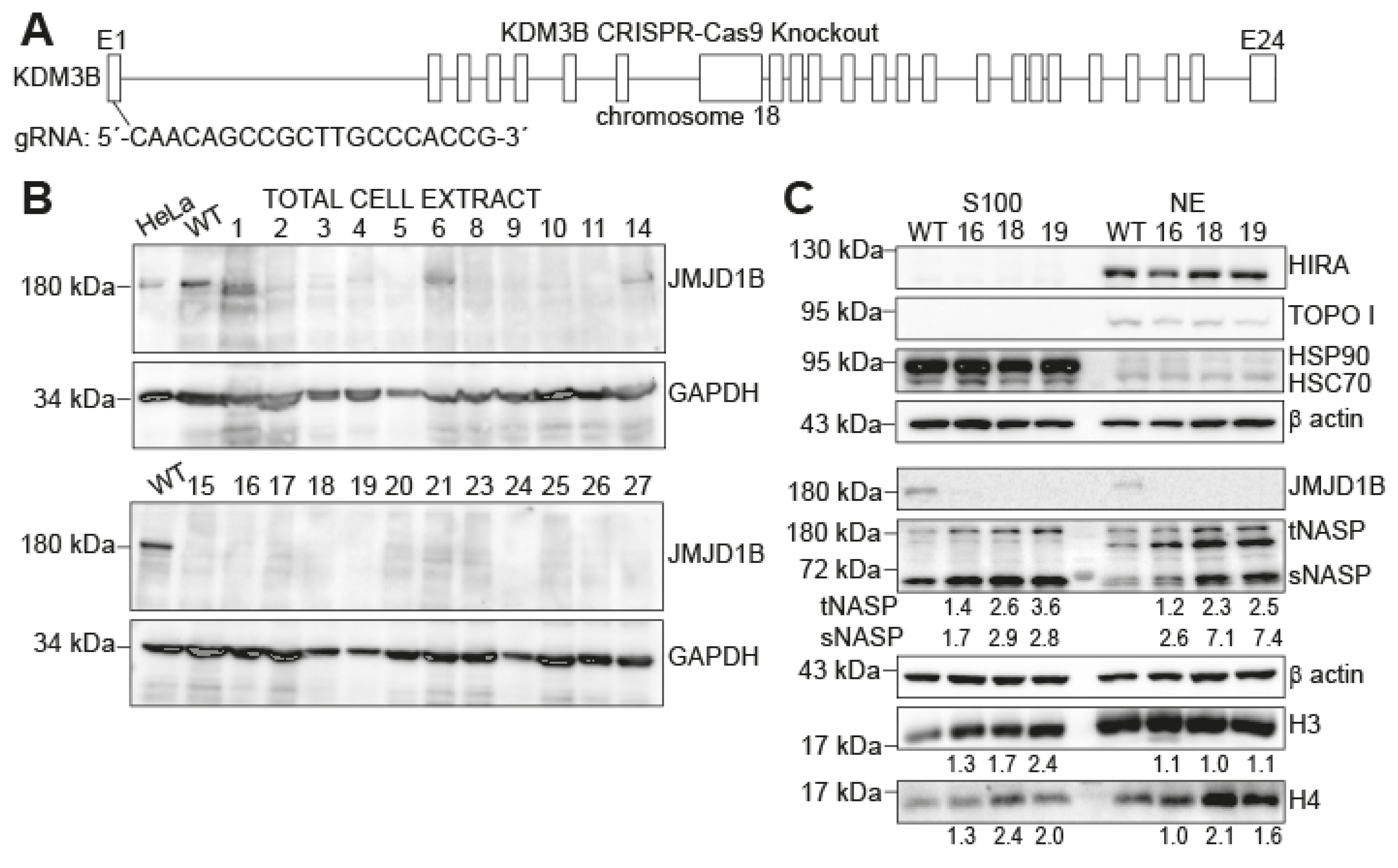
2.1. Histones H3 and H4 and NASP Accumulate in Cytosolic Extracts upon JMJD1B Depletion
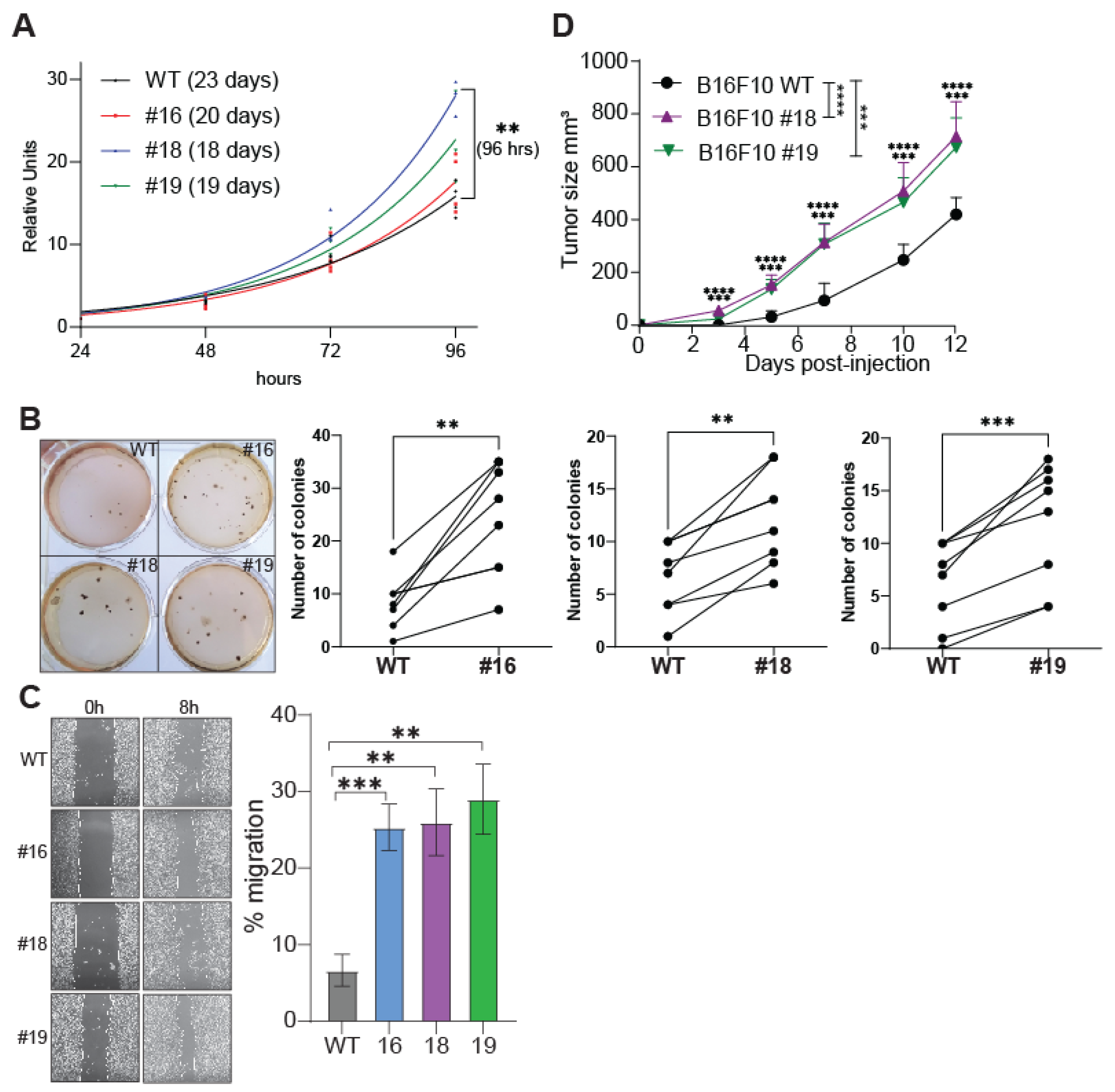
2.2. The Tumorigenic Properties of B16F10 Cells Are Potentiated by JMJD1B Depletion
2.3. Depletion of JMJD1B Increases Genomic Instability in B16F10 Cells
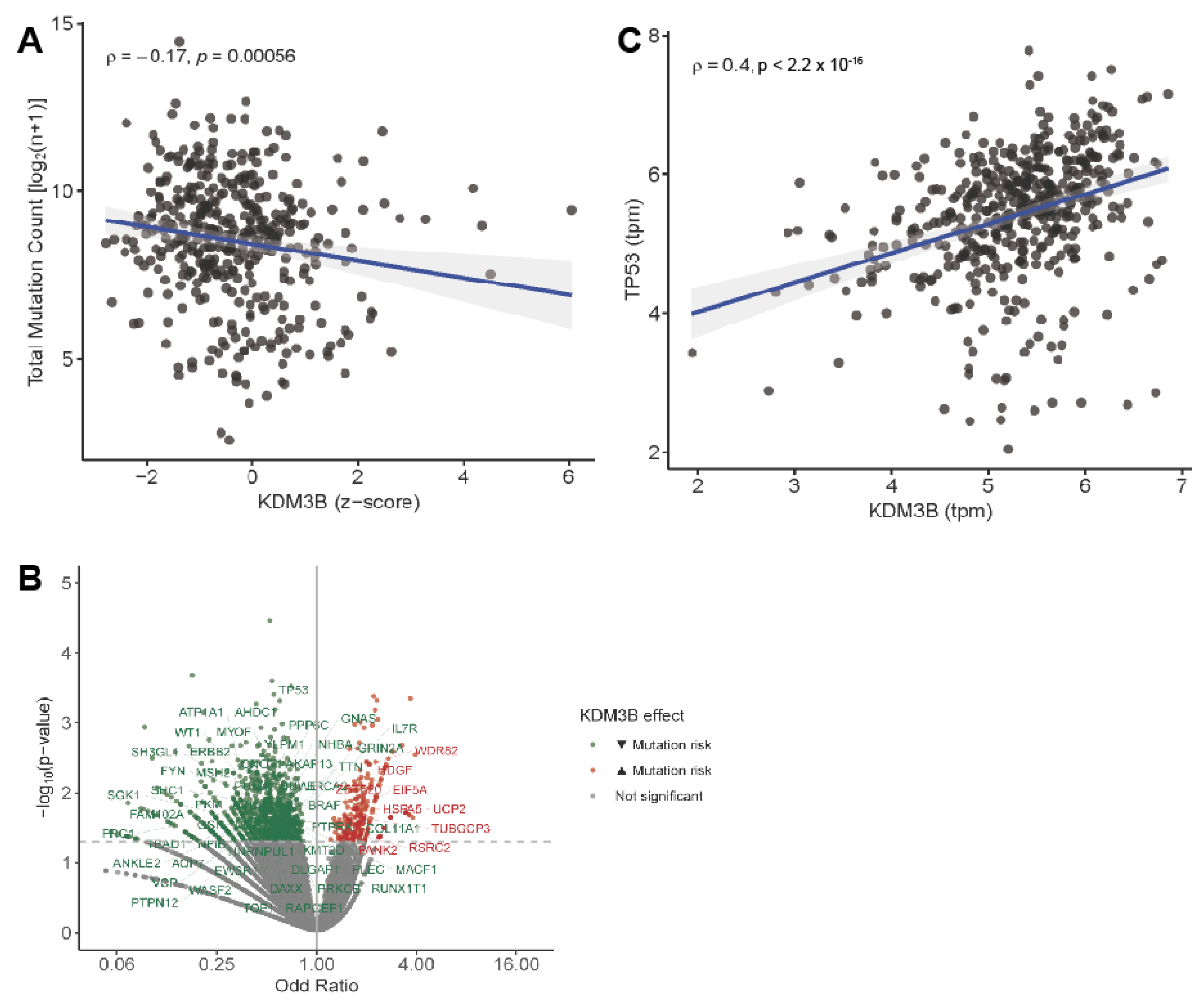
2.4. JMJD1B in Human Melanoma Tumors
3. Discussion
4. Material and Methods
4.1. Generation of the JMJD1B Knockout in B16F10 Cells
4.2. Cell Culture
4.3. Cellular Extracts
4.4. Antibodies
4.5. Proliferation Assay
4.6. Soft Agar
4.7. Wound Healing Assay
4.8. Tumor Growth
4.9. Microccocal Nuclease (MNase) Digestion
4.10. Comet Assay
4.11. Transcriptomic Analyses of Human Melanoma Tumors
4.12. Ethics Statement
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Ethics Approval
References
- Negrini, S.; Gorgoulis, V.G.; Halazonetis, T.D. Genomic instability—An evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 2010, 11, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Papamichos-Chronakis, M.; Peterson, C.L. Chromatin and the genome integrity network. Nat. Rev. Genet. 2013, 14, 62–75. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, F.; Gurard-Levin, Z.A.; Rojas-Villalobos, C.; Vassias, I.; Quatrini, R.; Almouzni, G.; Loyola, A. JMJD1B, a novel player in histone H3 and H4 processing to ensure genome stability. Epigenetics Chromatin 2020, 13, 6. [Google Scholar] [CrossRef]
- Singh, R.K.; Liang, D.; Gajjalaiahvari, U.R.; Kabbaj, M.H.; Paik, J.; Gunjan, A. Excess histone levels mediate cytotoxicity via multiple mechanisms. Cell Cycle 2010, 9, 4236–4244. [Google Scholar] [CrossRef]
- Kim, U.J.; Han, M.; Kayne, P.; Grunstein, M. Effects of histone H4 depletion on the cell cycle and transcription of Saccharomyces cerevisiae. EMBO J. 1988, 7, 2211–2219. [Google Scholar] [CrossRef]
- Prado, F.; Aguilera, A. Partial depletion of histone H4 increases homologous recombination-mediated genetic instability. Mol. Cell Biol. 2005, 25, 1526–1536. [Google Scholar] [CrossRef]
- Gunjan, A.; Verreault, A. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell 2003, 115, 537–549. [Google Scholar] [CrossRef]
- Liang, D.; Burkhart, S.L.; Singh, R.K.; Kabbaj, M.H.; Gunjan, A. Histone dosage regulates DNA damage sensitivity in a checkpoint-independent manner by the homologous recombination pathway. Nucleic Acids Res. 2012, 40, 9604–9620. [Google Scholar] [CrossRef]
- Escobar, T.M.; Loyola, A.; Reinberg, D. Parental nucleosome segregation and the inheritance of cellular identity. Nat. Rev. Genet. 2021, 22, 379–392. [Google Scholar] [CrossRef]
- Nelson, D.M.; Ye, X.; Hall, C.; Santos, H.; Ma, T.; Kao, G.D.; Yen, T.J.; Harper, J.W.; Adams, P.D. Coupling of DNA synthesis and histone synthesis in S phase independent of cyclin/cdk2 activity. Mol. Cell Biol. 2002, 22, 7459–7472. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, F.; Munoz, F.; Schilcher, P.; Imhof, A.; Almouzni, G.; Loyola, A. Sequential establishment of marks on soluble histones H3 and H4. J. Biol. Chem. 2011, 286, 17714–17721. [Google Scholar] [CrossRef] [PubMed]
- Campos, E.I.; Fillingham, J.; Li, G.; Zheng, H.; Voigt, P.; Kuo, W.H.; Seepany, H.; Gao, Z.; Day, L.A.; Greenblatt, J.F.; et al. The program for processing newly synthesized histones H3.1 and H4. Nat. Struct. Mol. Biol. 2010, 17, 1343–1351. [Google Scholar] [CrossRef] [PubMed]
- Grover, P.; Asa, J.S.; Campos, E.I. H3-H4 Histone Chaperone Pathways. Annu. Rev. Genet. 2018, 52, 109–130. [Google Scholar] [CrossRef] [PubMed]
- Hormazabal, J.; Saavedra, F.; Espinoza-Arratia, C.; Martinez, N.W.; Cruces, T.; Alfaro, I.E.; Loyola, A. Chaperone mediated autophagy contributes to the newly synthesized histones H3 and H4 quality control. Nucleic Acids Res. 2022, 50, 1875–1887. [Google Scholar] [CrossRef]
- Saavedra, F.; Rivera, C.; Rivas, E.; Merino, P.; Garrido, D.; Hernandez, S.; Forne, I.; Vassias, I.; Gurard-Levin, Z.A.; Alfaro, I.E.; et al. PP32 and SET/TAF-Ibeta proteins regulate the acetylation of newly synthesized histone H4. Nucleic Acids Res. 2017, 45, 11700–11710. [Google Scholar] [CrossRef]
- Prado, F.; Cortes-Ledesma, F.; Aguilera, A. The absence of the yeast chromatin assembly factor Asf1 increases genomic instability and sister chromatid exchange. EMBO Rep. 2004, 5, 497–502. [Google Scholar] [CrossRef]
- Wen, T.; Chen, Q.Y. Dynamic Activity of Histone H3-Specific Chaperone Complexes in Oncogenesis. Front. Oncol. 2021, 11, 806974. [Google Scholar] [CrossRef]
- Ray-Gallet, D.; Almouzni, G. H3-H4 histone chaperones and cancer. Curr. Opin. Genet. Dev. 2022, 73, 101900. [Google Scholar] [CrossRef]
- Brauchle, M.; Yao, Z.; Arora, R.; Thigale, S.; Clay, I.; Inverardi, B.; Fletcher, J.; Taslimi, P.; Acker, M.G.; Gerrits, B.; et al. Protein complex interactor analysis and differential activity of KDM3 subfamily members towards H3K9 methylation. PLoS ONE 2013, 8, e60549. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kim, K.B.; Eom, G.H.; Choe, N.; Kee, H.J.; Son, H.J.; Oh, S.T.; Kim, D.W.; Pak, J.H.; Baek, H.J.; et al. KDM3B is the H3K9 demethylase involved in transcriptional activation of lmo2 in leukemia. Mol. Cell Biol. 2012, 32, 2917–2933. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Ali, S.; Duan, X.; Liu, S.; Du, J.; Liu, C.; Dai, H.; Zhou, M.; Zhou, L.; Yang, L.; et al. JMJD1B Demethylates H4R3me2s and H3K9me2 to Facilitate Gene Expression for Development of Hematopoietic Stem and Progenitor Cells. Cell Rep. 2018, 23, 389–403. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Gomes, I.; Horrigan, S.K.; Kravarusic, J.; Mar, B.; Arbieva, Z.; Chyna, B.; Fulton, N.; Edassery, S.; Raza, A.; et al. A novel nuclear protein, 5qNCA (LOC51780) is a candidate for the myeloid leukemia tumor suppressor gene on chromosome 5 band q31. Oncogene 2001, 20, 6946–6954. [Google Scholar] [CrossRef]
- Lai, F.; Godley, L.A.; Fernald, A.A.; Orelli, B.J.; Pamintuan, L.; Zhao, N.; Le Beau, M.M. cDNA cloning and genomic structure of three genes localized to human chromosome band 5q31 encoding potential nuclear proteins. Genomics 2000, 70, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Manni, W.; Jianxin, X.; Weiqi, H.; Siyuan, C.; Huashan, S. JMJD family proteins in cancer and inflammation. Signal Transduct. Target. Ther. 2022, 7, 304. [Google Scholar] [CrossRef]
- Yamane, K.; Toumazou, C.; Tsukada, Y.; Erdjument-Bromage, H.; Tempst, P.; Wong, J.; Zhang, Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 2006, 125, 483–495. [Google Scholar] [CrossRef]
- Lim, S.; Metzger, E.; Schule, R.; Kirfel, J.; Buettner, R. Epigenetic regulation of cancer growth by histone demethylases. Int. J. Cancer 2010, 127, 1991–1998. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, X.; Zhou, S.; Liao, L.; Jiang, R.; Xu, J. The histone H3K9 demethylase Kdm3b is required for somatic growth and female reproductive function. Int. J. Biol. Sci. 2015, 11, 494–507. [Google Scholar] [CrossRef] [PubMed]
- Kuroki, S.; Nakai, Y.; Maeda, R.; Okashita, N.; Akiyoshi, M.; Yamaguchi, Y.; Kitano, S.; Miyachi, H.; Nakato, R.; Ichiyanagi, K.; et al. Combined Loss of JMJD1A and JMJD1B Reveals Critical Roles for H3K9 Demethylation in the Maintenance of Embryonic Stem Cells and Early Embryogenesis. Stem Cell Reports 2018, 10, 1340–1354. [Google Scholar] [CrossRef]
- Xu, X.; Nagel, S.; Quentmeier, H.; Wang, Z.; Pommerenke, C.; Dirks, W.G.; Macleod, R.A.F.; Drexler, H.G.; Hu, Z. KDM3B shows tumor-suppressive activity and transcriptionally regulates HOXA1 through retinoic acid response elements in acute myeloid leukemia. Leuk. Lymphoma 2018, 59, 204–213. [Google Scholar] [CrossRef]
- Downs, J.A.; Lowndes, N.F.; Jackson, S.P. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 2000, 408, 1001–1004. [Google Scholar] [CrossRef]
- Chen, H.T.; Bhandoola, A.; Difilippantonio, M.J.; Zhu, J.; Brown, M.J.; Tai, X.; Rogakou, E.P.; Brotz, T.M.; Bonner, W.M.; Ried, T.; et al. Response to RAG-mediated VDJ cleavage by NBS1 and gamma-H2AX. Science 2000, 290, 1962–1965. [Google Scholar] [CrossRef] [PubMed]
- An, M.J.; Kim, D.H.; Kim, C.H.; Kim, M.; Rhee, S.; Seo, S.B.; Kim, J.W. Histone demethylase KDM3B regulates the transcriptional network of cell-cycle genes in hepatocarcinoma HepG2 cells. Biochem. Biophys. Res. Commun. 2019, 508, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Han, A.; Chae, Y.C.; Park, J.W.; Kim, K.B.; Kim, J.Y.; Seo, S.B. Transcriptional repression of ANGPT1 by histone H3K9 demethylase KDM3B. BMB Rep. 2015, 48, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yu, B.; Deng, P.; Cheng, Y.; Yu, Y.; Kevork, K.; Ramadoss, S.; Ding, X.; Li, X.; Wang, C.Y. KDM3 epigenetically controls tumorigenic potentials of human colorectal cancer stem cells through Wnt/beta-catenin signalling. Nat. Commun. 2017, 8, 15146. [Google Scholar] [CrossRef] [PubMed]
- Mahamdallie, S.; Yost, S.; Poyastro-Pearson, E.; Holt, E.; Zachariou, A.; Seal, S.; Elliott, A.; Clarke, M.; Warren-Perry, M.; Hanks, S.; et al. Identification of new Wilms tumour predisposition genes: An exome sequencing study. Lancet Child. Adolesc. Health 2019, 3, 322–331. [Google Scholar] [CrossRef]
- Paolicchi, E.; Crea, F.; Farrar, W.L.; Green, J.E.; Danesi, R. Histone lysine demethylases in breast cancer. Crit. Rev. Oncol. Hematol. 2013, 86, 97–103. [Google Scholar] [CrossRef]
- Ren, X.; Ma, L.; Wang, N.; Zhou, R.; Wu, J.; Xie, X.; Zhang, H.; Liu, D.; Ma, X.; Dang, C.; et al. Antioxidant Gene Signature Impacts the Immune Infiltration and Predicts the Prognosis of Kidney Renal Clear Cell Carcinoma. Front. Genet. 2021, 12, 721252. [Google Scholar] [CrossRef]
- Dignam, J.D.; Lebovitz, R.M.; Roeder, R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983, 11, 1475–1489. [Google Scholar] [CrossRef]
- Borowicz, S.; Van Scoyk, M.; Avasarala, S.; Karuppusamy Rathinam, M.K.; Tauler, J.; Bikkavilli, R.K.; Winn, R.A. The soft agar colony formation assay. J. Vis. Exp. 2014, 27, e51998. [Google Scholar] [CrossRef]
- Freitas, J.T.; Jozic, I.; Bedogni, B. Wound Healing Assay for Melanoma Cell Migration. Methods Mol. Biol. 2021, 2265, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Moller, P.; Azqueta, A.; Boutet-Robinet, E.; Koppen, G.; Bonassi, S.; Milic, M.; Gajski, G.; Costa, S.; Teixeira, J.P.; Costa Pereira, C.; et al. Minimum Information for Reporting on the Comet Assay (MIRCA): Recommendations for describing comet assay procedures and results. Nat. Protoc. 2020, 15, 3817–3826. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cruz, P.; Peña-Lopez, D.; Figueroa, D.; Riobó, I.; Benedetti, V.; Saavedra, F.; Espinoza-Arratia, C.; Escobar, T.M.; Lladser, A.; Loyola, A. Unraveling the Role of JMJD1B in Genome Stability and the Malignancy of Melanomas. Int. J. Mol. Sci. 2024, 25, 10689. https://doi.org/10.3390/ijms251910689
Cruz P, Peña-Lopez D, Figueroa D, Riobó I, Benedetti V, Saavedra F, Espinoza-Arratia C, Escobar TM, Lladser A, Loyola A. Unraveling the Role of JMJD1B in Genome Stability and the Malignancy of Melanomas. International Journal of Molecular Sciences. 2024; 25(19):10689. https://doi.org/10.3390/ijms251910689
Chicago/Turabian StyleCruz, Perla, Diego Peña-Lopez, Diego Figueroa, Isidora Riobó, Vincenzo Benedetti, Francisco Saavedra, Claudia Espinoza-Arratia, Thelma M. Escobar, Alvaro Lladser, and Alejandra Loyola. 2024. "Unraveling the Role of JMJD1B in Genome Stability and the Malignancy of Melanomas" International Journal of Molecular Sciences 25, no. 19: 10689. https://doi.org/10.3390/ijms251910689




