Development and Validation of a Comprehensive Prognostic and Depression Risk Index for Gastric Adenocarcinoma
Abstract
1. Introduction
2. Results
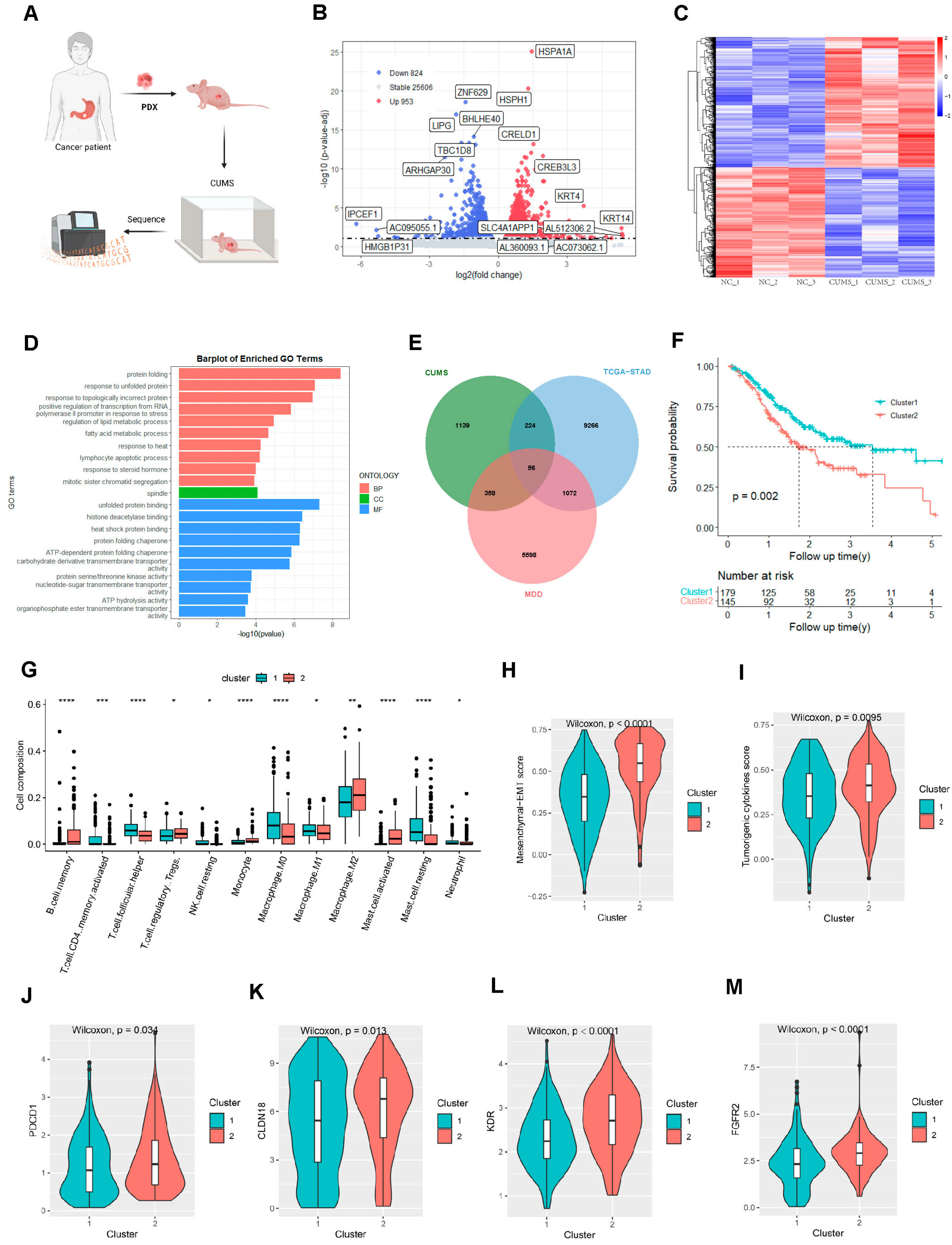
2.1. Major Depressive Disorder and Gastric Adenocarcinoma-Related Hub Genes
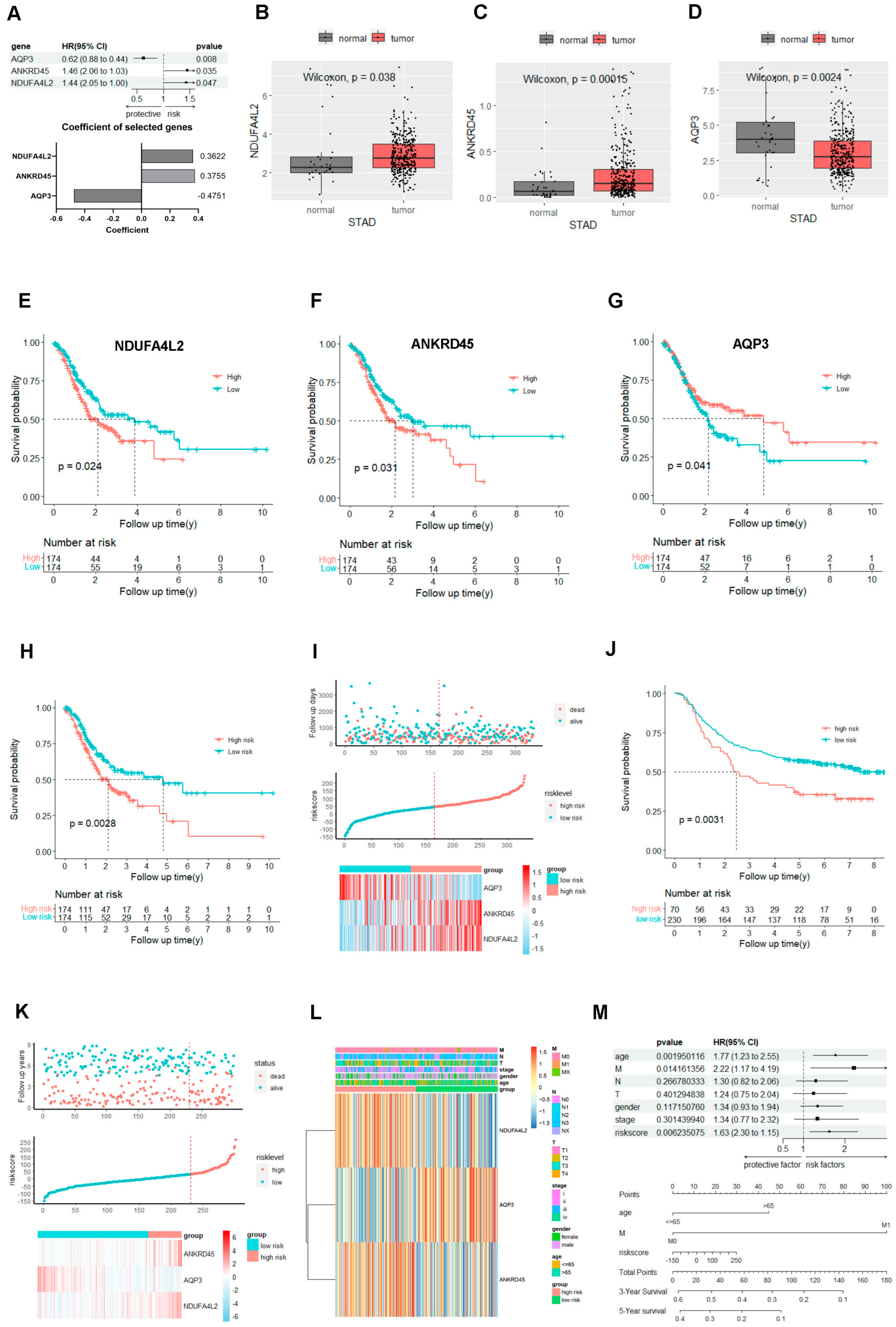
2.2. The Gene Signature Based on the NDUFA4L2, ANKRD45, and AQP3 Predicts the Prognosis of Gastric Adenocarcinoma
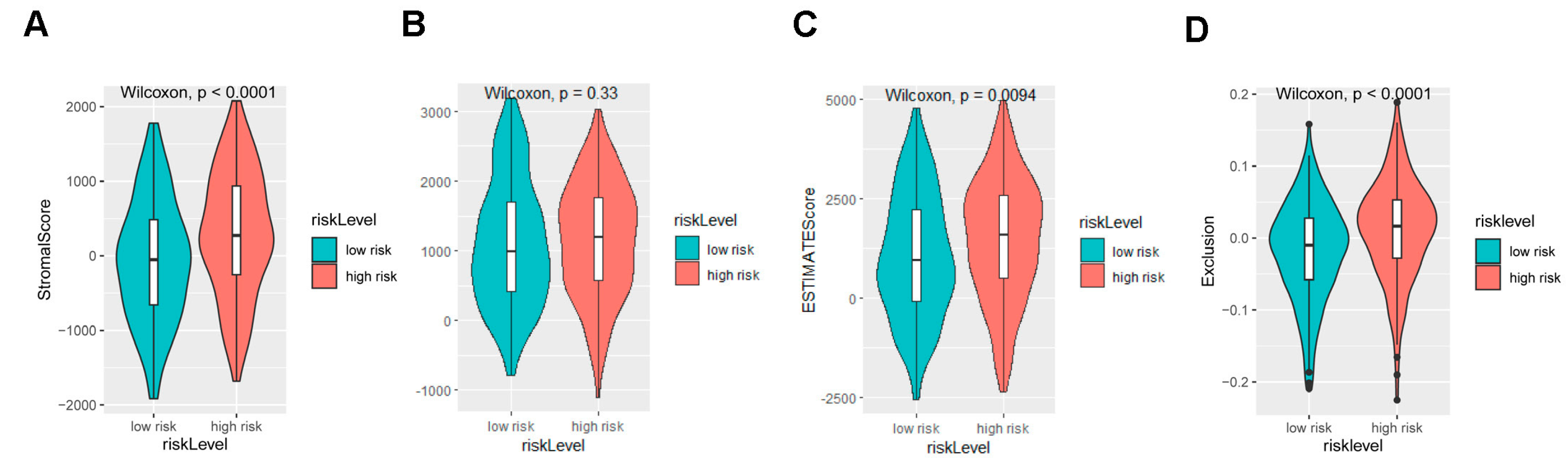
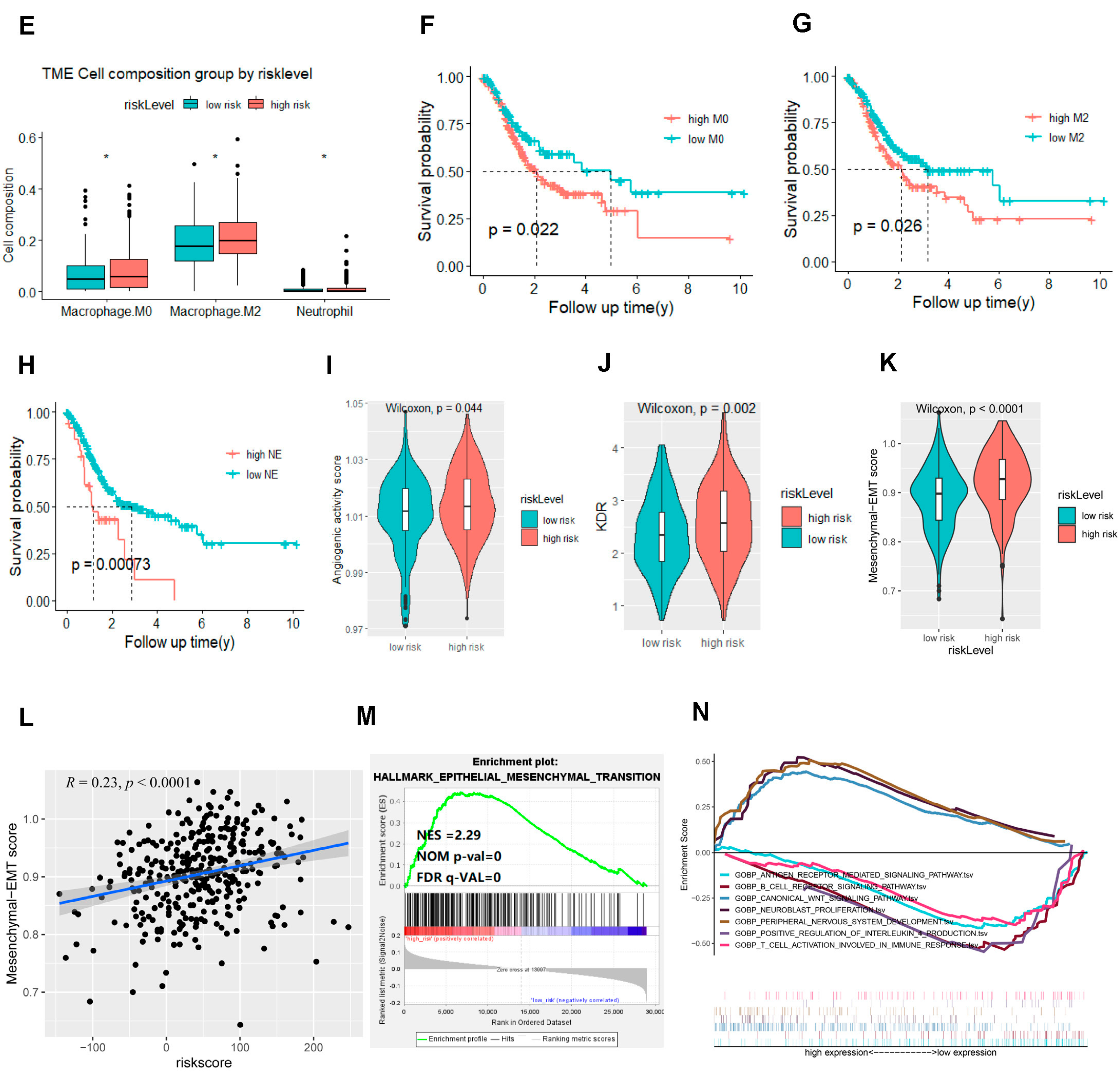
2.3. The Signature Based on the NDUFA4L2, ANKRD45, and AQP3 Defined the Molecular Characteristics and the Tumor Microenvironment of Gastric Adenocarcinoma
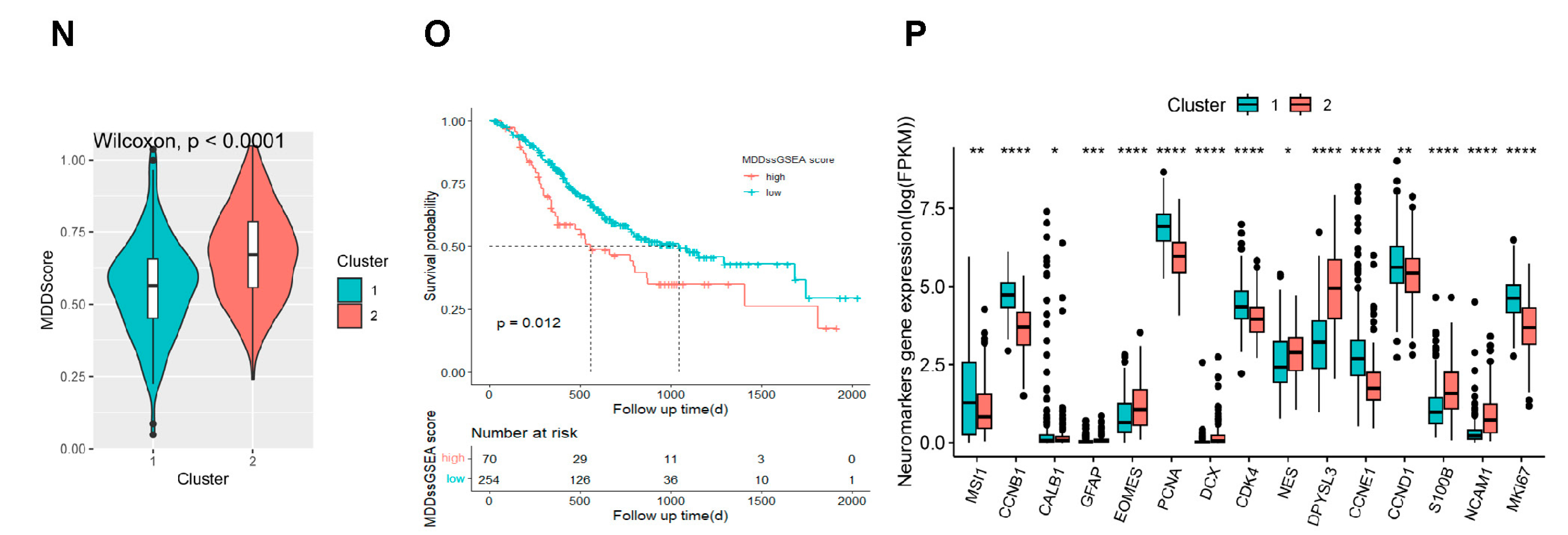
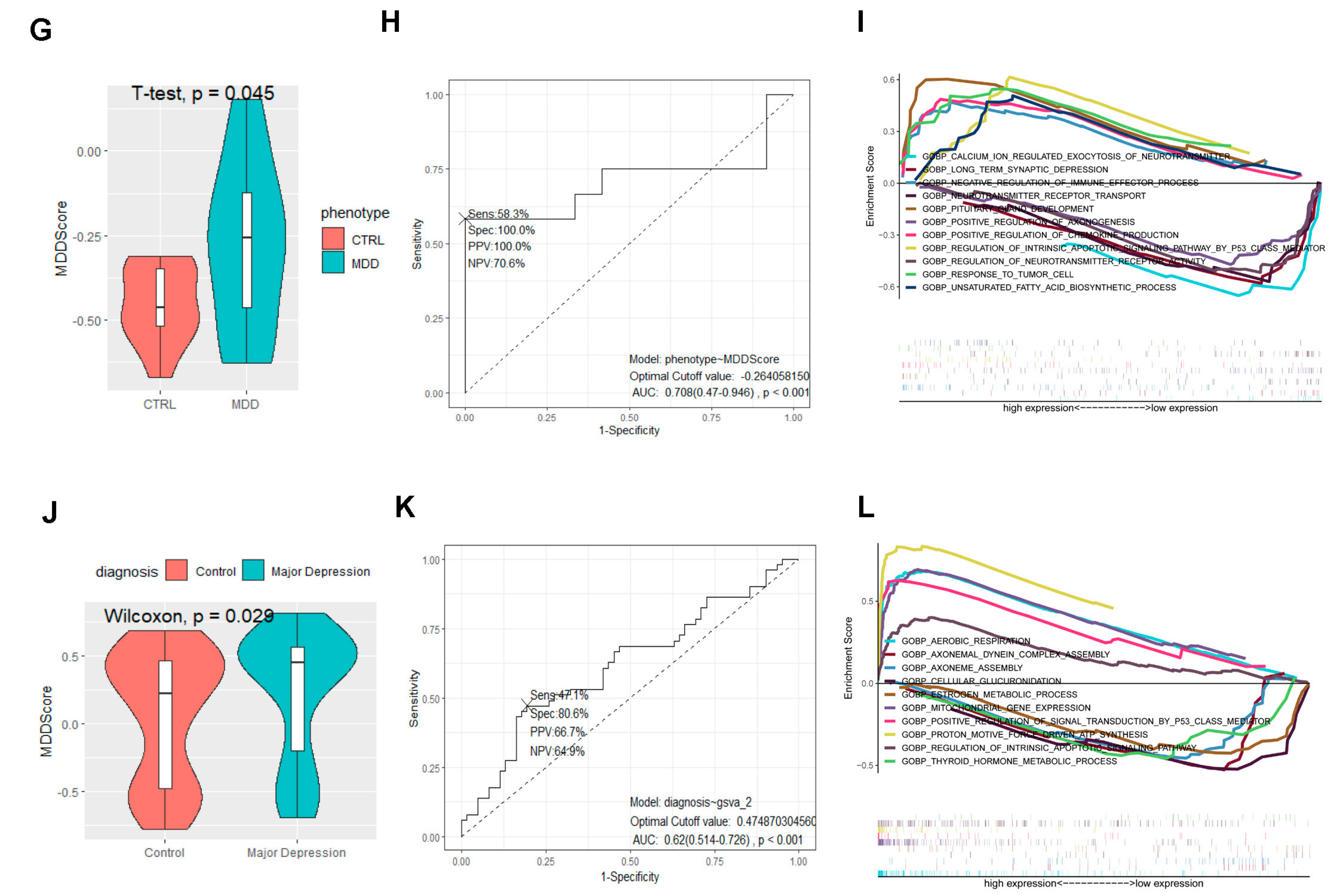
2.4. The Signature Based on the NDUFA4L2, ANKRD45, and AQP3 Estimated the Depressive Risk of Gastric Adenocarcinoma
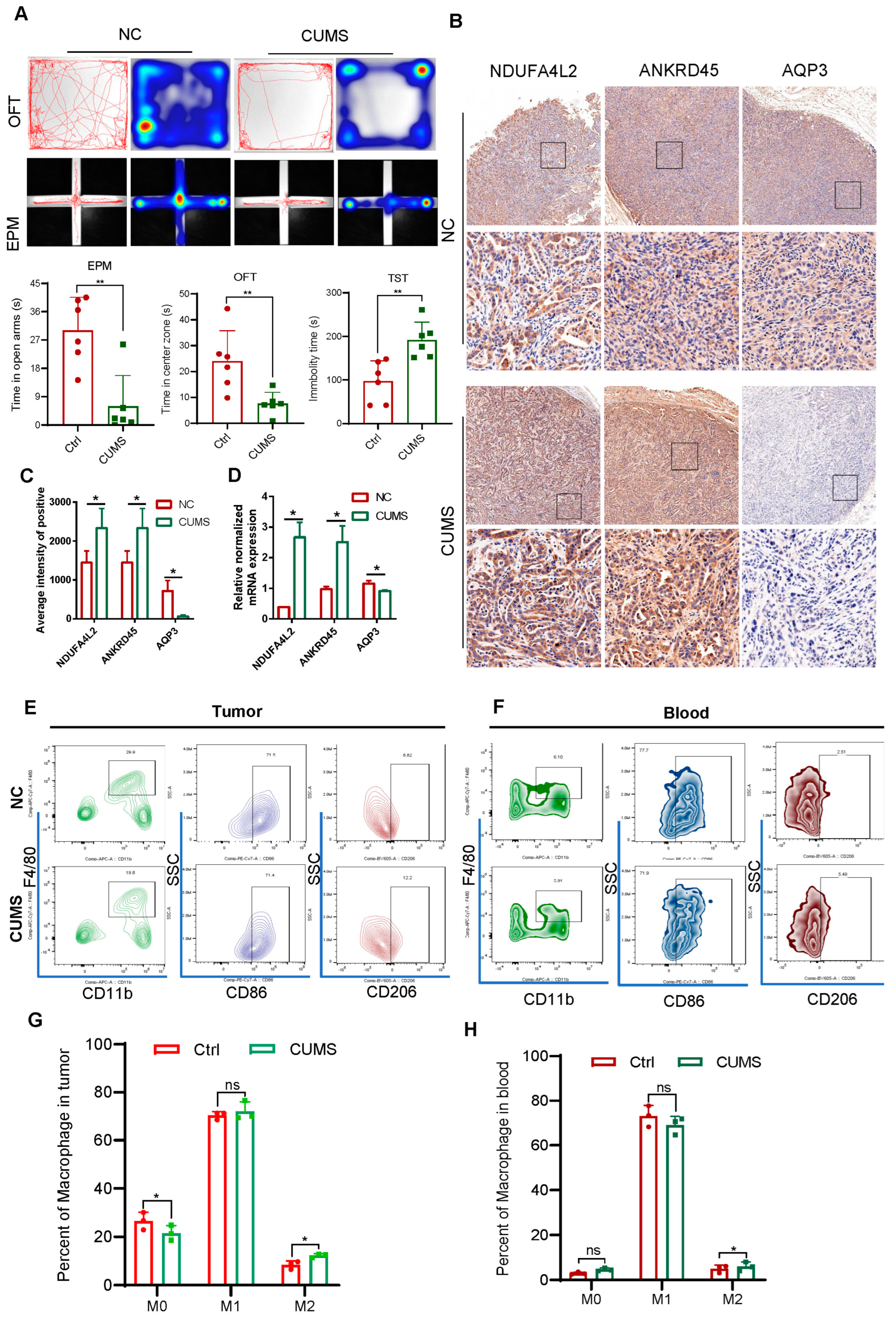
2.5. Signature Genes’ Expression and Immune Infiltration in Gastric Cancer Following CUMS Treatment
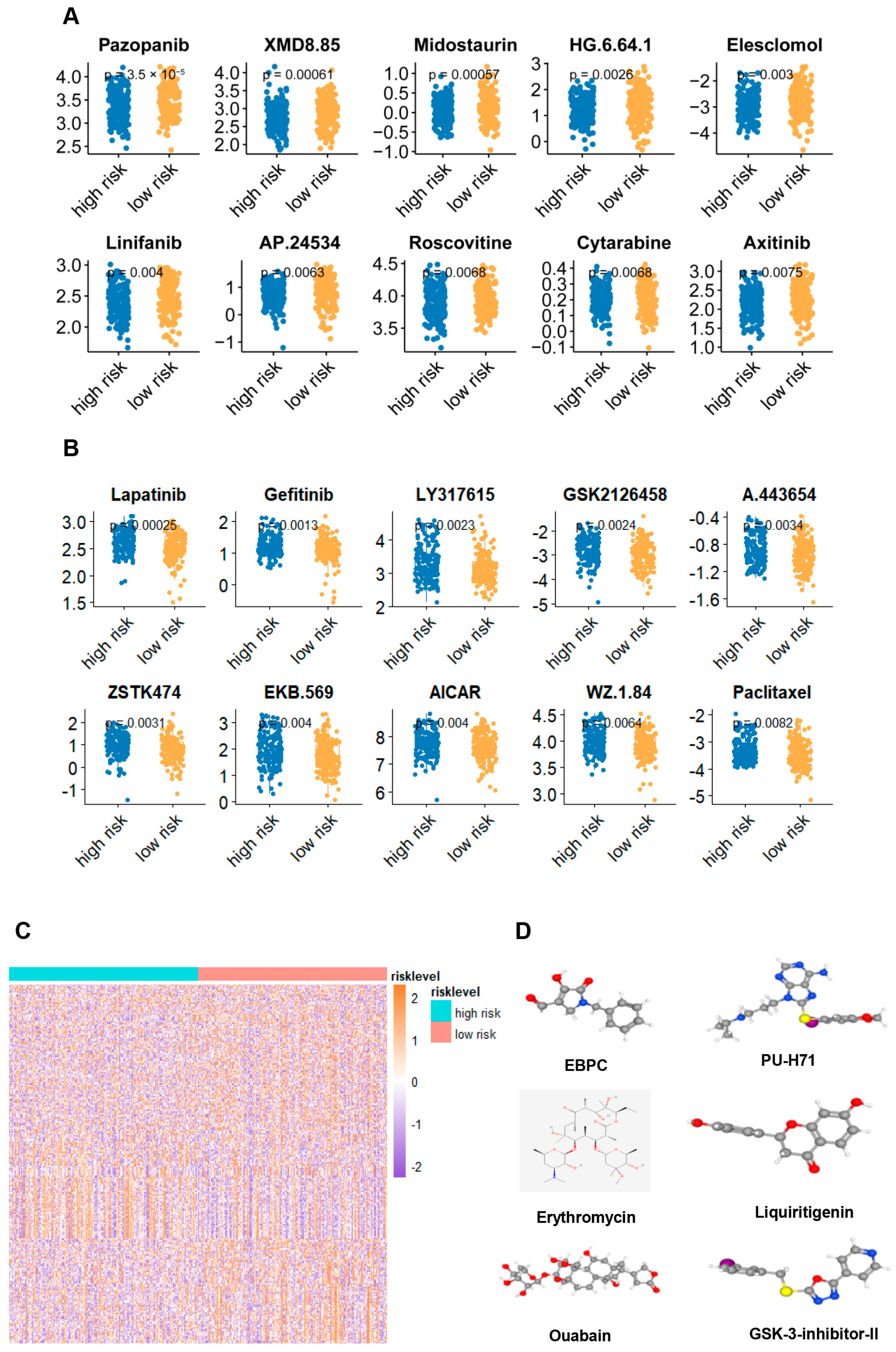
2.6. Potential Treatment Strategy for the Signature Defined Gastric Adenocarcinoma Patients
3. Materials and Methods
3.1. PDX Mouse Experiments
3.2. Subcutaneous Tumor Mouse Model
3.3. Data Source
3.4. Transcriptome Sequencing (mRNA-Seq) and Analysis
3.5. Analysis of the Gene Expression Patterns in Stomach Adenocarcinoma Dataset
3.6. Cluster Analysis
3.7. MDD Score
3.8. Construction and Validation of the Prognostic Signature
3.9. The Hub Genes’ Functional Enrichment Analyses
3.10. Gene Set Enrichment Analysis (GSEA) Based on the Signature
3.11. Tumor-Related Scores
3.12. Immune Landscape Analysis
3.13. Chemotherapy Response and Small-Molecule Drugs
3.14. Cell Culture
3.15. Real-Time Quantitative Polymerase Chain Reaction (RT-PCR)
3.16. Statistical Analysis
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Y.H.; Li, J.Q.; Shi, J.F.; Que, J.Y.; Liu, J.J.; Lappin, J.M.; Leung, J.; Ravindran, A.V.; Chen, W.Q.; Qiao, Y.L.; et al. Depression and anxiety in relation to cancer incidence and mortality: A systematic review and meta-analysis of cohort studies. Mol. Psychiatry 2020, 25, 1487–1499. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Liu, P.; Zheng, J.; Lü, J.; Zhao, Q.; Li, D.; Guo, Y.; Qian, L.; Wang, Q.; Miao, X.; et al. Immune and nonimmune mechanisms mediate the mental stress-induced tumor growth in a xenograft model of breast cancer. Cell Death Dis. 2021, 12, 987. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Zhou, F.; Yuan, X.; Yang, T.; Liang, X.; Wang, Y.; Tu, H.; Chang, J.; Nan, K.; Wei, Y. Reactive Oxygen Species Are Involved in the Development of Gastric Cancer and Gastric Cancer-Related Depression through ABL1-Mediated Inflammation Signaling Pathway. Oxidative Med. Cell. Longev. 2019, 2019, 5813985. [Google Scholar] [CrossRef] [PubMed]
- Eckerling, A.; Ricon-Becker, I.; Sorski, L.; Sandbank, E.; Ben-Eliyahu, S. Stress and cancer: Mechanisms, significance and future directions. Nat. Rev. Cancer 2021, 21, 767–785. [Google Scholar] [CrossRef]
- Lange, M.; Joly, F.; Vardy, J.; Ahles, T.; Dubois, M.; Tron, L.; Winocur, G.; De Ruiter, M.B.; Castel, H. Cancer-related cognitive impairment: An update on state of the art, detection, and management strategies in cancer survivors. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2019, 30, 1925–1940. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Marx, W.; Penninx, B.; Solmi, M.; Furukawa, T.A.; Firth, J.; Carvalho, A.F.; Berk, M. Major depressive disorder. Nat. Rev. Dis. Primers 2023, 9, 44. [Google Scholar] [CrossRef]
- Labonté, B.; Engmann, O.; Purushothaman, I.; Menard, C.; Wang, J.; Tan, C.; Scarpa, J.R.; Moy, G.; Loh, Y.E.; Cahill, M.; et al. Sex-specific transcriptional signatures in human depression. Nat. Med. 2017, 23, 1102–1111. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Nomura, S.; Hosoi, A.; Nagaoka, K.; Iino, T.; Yasuda, T.; Saito, T.; Matsushita, H.; Uchida, E.; Seto, Y.; et al. Established gastric cancer cell lines transplantable into C57BL/6 mice show fibroblast growth factor receptor 4 promotion of tumor growth. Cancer Sci. 2018, 109, 1480–1492. [Google Scholar] [CrossRef]
- Sanmarco, L.M.; Rone, J.M.; Polonio, C.M.; Fernandez Lahore, G.; Giovannoni, F.; Ferrara, K.; Gutierrez-Vazquez, C.; Li, N.; Sokolovska, A.; Plasencia, A.; et al. Lactate limits CNS autoimmunity by stabilizing HIF-1α in dendritic cells. Nature 2023, 620, 881–889. [Google Scholar] [CrossRef]
- Clayton, S.A.; Daley, K.K.; MacDonald, L.; Fernandez-Vizarra, E.; Bottegoni, G.; O’Neil, J.D.; Major, T.; Griffin, D.; Zhuang, Q.; Adewoye, A.B.; et al. Inflammation causes remodeling of mitochondrial cytochrome c oxidase mediated by the bifunctional gene C15orf48. Sci. Adv. 2021, 7, eabl5182. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.S.; Cui, J.D.; Lee, D.; Yuen, V.W.; Chiu, D.K.; Goh, C.C.; Cheu, J.W.; Tse, A.P.; Bao, M.H.; Wong, B.P.Y.; et al. Hypoxia-induced macropinocytosis represents a metabolic route for liver cancer. Nat. Commun. 2022, 13, 954. [Google Scholar] [CrossRef] [PubMed]
- Cowman, S.J.; Fuja, D.G.; Liu, X.D.; Tidwell, R.S.S.; Kandula, N.; Sirohi, D.; Agarwal, A.M.; Emerson, L.L.; Tripp, S.R.; Mohlman, J.S.; et al. Macrophage HIF-1α Is an Independent Prognostic Indicator in Kidney Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2020, 26, 4970–4982. [Google Scholar] [CrossRef]
- Shao, Q.; Liu, J.; Li, G.; Gu, Y.; Guo, M.; Guan, Y.; Tian, Z.; Ma, W.; Wang, C.; Ji, X. Proteomic Analysis Reveals That Mitochondria Dominate the Hippocampal Hypoxic Response in Mice. Int. J. Mol. Sci. 2022, 23, 4094. [Google Scholar] [CrossRef]
- Eshima, J.; O’Connor, S.A.; Marschall, E.; Bowser, R.; Plaisier, C.L.; Smith, B.S. Molecular subtypes of ALS are associated with differences in patient prognosis. Nat. Commun. 2023, 14, 95. [Google Scholar] [CrossRef]
- Hara-Chikuma, M.; Tanaka, M.; Verkman, A.S.; Yasui, M. Inhibition of aquaporin-3 in macrophages by a monoclonal antibody as potential therapy for liver injury. Nat. Commun. 2020, 11, 5666. [Google Scholar] [CrossRef] [PubMed]
- Prata, C.; Hrelia, S.; Fiorentini, D. Peroxiporins in Cancer. Int. J. Mol. Sci. 2019, 20, 1371. [Google Scholar] [CrossRef]
- Behl, T.; Kaur, G.; Sehgal, A.; Bhardwaj, S.; Singh, S.; Buhas, C.; Judea-Pusta, C.; Uivarosan, D.; Munteanu, M.A.; Bungau, S. Multifaceted Role of Matrix Metalloproteinases in Neurodegenerative Diseases: Pathophysiological and Therapeutic Perspectives. Int. J. Mol. Sci. 2021, 22, 1413. [Google Scholar] [CrossRef]
- Rubenwolf, P.; Thomas, C.; Denzinger, S.; Hartmann, A.; Burger, M.; Georgopoulos, N.T.; Otto, W. Loss of AQP3 protein expression is associated with worse progression-free and cancer-specific survival in patients with muscle-invasive bladder cancer. World J. Urol. 2015, 33, 1959–1964. [Google Scholar] [CrossRef][Green Version]
- Kang, Y.; Xie, H.; Zhao, C. Ankrd45 Is a Novel Ankyrin Repeat Protein Required for Cell Proliferation. Genes 2019, 10, 462. [Google Scholar] [CrossRef]
- Li, Q.S.; De Muynck, L. Differentially expressed genes in Alzheimer’s disease highlighting the roles of microglia genes including OLR1 and astrocyte gene CDK2AP1. Brain Behav. Immun. Health 2021, 13, 100227. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Chu, T.H.; Nienhüser, H.; Jiang, Z.; Del Portillo, A.; Remotti, H.E.; White, R.A.; Hayakawa, Y.; Tomita, H.; Fox, J.G.; et al. PD-1 Signaling Promotes Tumor-Infiltrating Myeloid-Derived Suppressor Cells and Gastric Tumorigenesis in Mice. Gastroenterology 2021, 160, 781–796. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, T.; Yasuda, T.; Uchihara, T.; Yasuda-Yoshihara, N.; Tan, B.J.Y.; Yonemura, A.; Semba, T.; Yamasaki, J.; Komohara, Y.; Ohnishi, K.; et al. Stromal Reprogramming through Dual PDGFRα/β Blockade Boosts the Efficacy of Anti-PD-1 Immunotherapy in Fibrotic Tumors. Cancer Res. 2023, 83, 753–770. [Google Scholar] [CrossRef] [PubMed]
- Alicea-Torres, K.; Sanseviero, E.; Gui, J.; Chen, J.; Veglia, F.; Yu, Q.; Donthireddy, L.; Kossenkov, A.; Lin, C.; Fu, S.; et al. Immune suppressive activity of myeloid-derived suppressor cells in cancer requires inactivation of the type I interferon pathway. Nat. Commun. 2021, 12, 1717. [Google Scholar] [CrossRef]
- Arner, E.N.; Rathmell, J.C. Metabolic programming and immune suppression in the tumor microenvironment. Cancer Cell 2023, 41, 421–433. [Google Scholar] [CrossRef]
- Du, Y.; Zhang, J.Y.; Gong, L.P.; Feng, Z.Y.; Wang, D.; Pan, Y.H.; Sun, L.P.; Wen, J.Y.; Chen, G.F.; Liang, J.; et al. Hypoxia-induced ebv-circLMP2A promotes angiogenesis in EBV-associated gastric carcinoma through the KHSRP/VHL/HIF1α/VEGFA pathway. Cancer Lett. 2022, 526, 259–272. [Google Scholar] [CrossRef]
- Buckley, A.M.; Lynam-Lennon, N.; O’Neill, H.; O’Sullivan, J. Targeting hallmarks of cancer to enhance radiosensitivity in gastrointestinal cancers. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 298–313. [Google Scholar] [CrossRef]
- Fuchs, C.S.; Shitara, K.; Di Bartolomeo, M.; Lonardi, S.; Al-Batran, S.E.; Van Cutsem, E.; Ilson, D.H.; Alsina, M.; Chau, I.; Lacy, J.; et al. Ramucirumab with cisplatin and fluoropyrimidine as first-line therapy in patients with metastatic gastric or junctional adenocarcinoma (RAINFALL): A double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2019, 20, 420–435. [Google Scholar] [CrossRef]
- Alsina, M.; Arrazubi, V.; Diez, M.; Tabernero, J. Current developments in gastric cancer: From molecular profiling to treatment strategy. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 155–170. [Google Scholar] [CrossRef]
- Fatehullah, A.; Terakado, Y.; Sagiraju, S.; Tan, T.L.; Sheng, T.; Tan, S.H.; Murakami, K.; Swathi, Y.; Ang, N.; Rajarethinam, R.; et al. A tumour-resident Lgr5(+) stem-cell-like pool drives the establishment and progression of advanced gastric cancers. Nat. Cell Biol. 2021, 23, 1299–1313. [Google Scholar] [CrossRef]
- Li, M.; Rao, X.; Cui, Y.; Zhang, L.; Li, X.; Wang, B.; Zheng, Y.; Teng, L.; Zhou, T.; Zhuo, W. The keratin 17/YAP/IL6 axis contributes to E-cadherin loss and aggressiveness of diffuse gastric cancer. Oncogene 2022, 41, 770–781. [Google Scholar] [CrossRef] [PubMed]
- Böttcher, C.; Fernández-Zapata, C.; Snijders, G.J.L.; Schlickeiser, S.; Sneeboer, M.A.M.; Kunkel, D.; De Witte, L.D.; Priller, J. Single-cell mass cytometry of microglia in major depressive disorder reveals a non-inflammatory phenotype with increased homeostatic marker expression. Transl. Psychiatry 2020, 10, 310. [Google Scholar] [CrossRef]
- Sforzini, L.; Cattaneo, A.; Ferrari, C.; Turner, L.; Mariani, N.; Enache, D.; Hastings, C.; Lombardo, G.; Nettis, M.A.; Nikkheslat, N.; et al. Higher immune-related gene expression in major depression is independent of CRP levels: Results from the BIODEP study. Transl. Psychiatry 2023, 13, 185. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.L.; Poller, W.C.; Swirski, F.K.; Russo, S.J. Central regulation of stress-evoked peripheral immune responses. Nat. Rev. Neurosci. 2023, 24, 591–604. [Google Scholar] [CrossRef]
- Park, J.; Lee, K.; Kim, K.; Yi, S.J. The role of histone modifications: From neurodevelopment to neurodiseases. Signal Transduct. Target. Ther. 2022, 7, 217. [Google Scholar] [CrossRef]
- Cervantes-Villagrana, R.D.; Albores-García, D.; Cervantes-Villagrana, A.R.; García-Acevez, S.J. Tumor-induced neurogenesis and immune evasion as targets of innovative anti-cancer therapies. Signal Transduct. Target. Ther. 2020, 5, 99. [Google Scholar] [CrossRef] [PubMed]
- Vermeer, P.D. Exosomal Induction of Tumor Innervation. Cancer Res. 2019, 79, 3529–3535. [Google Scholar] [CrossRef]
- Pan, C.; Winkler, F. Insights and opportunities at the crossroads of cancer and neuroscience. Nat. Cell Biol. 2022, 24, 1454–1460. [Google Scholar] [CrossRef]
- Schmoll, H.J.; Lindner, L.H.; Reichardt, P.; Heißner, K.; Kopp, H.G.; Kessler, T.; Mayer-Steinacker, R.; Rüssel, J.; Egerer, G.; Crysandt, M.; et al. Efficacy of Pazopanib With or Without Gemcitabine in Patients With Anthracycline- and/or Ifosfamide-Refractory Soft Tissue Sarcoma: Final Results of the PAPAGEMO Phase 2 Randomized Clinical Trial. JAMA Oncol. 2021, 7, 255–262. [Google Scholar] [CrossRef]
- Kadia, T.M.; Reville, P.K.; Borthakur, G.; Yilmaz, M.; Kornblau, S.; Alvarado, Y.; Dinardo, C.D.; Daver, N.; Jain, N.; Pemmaraju, N.; et al. Venetoclax plus intensive chemotherapy with cladribine, idarubicin, and cytarabine in patients with newly diagnosed acute myeloid leukaemia or high-risk myelodysplastic syndrome: A cohort from a single-centre, single-arm, phase 2 trial. Lancet Haematol. 2021, 8, e552–e561. [Google Scholar] [CrossRef]
- Powles, T.; Plimack, E.R.; Soulières, D.; Waddell, T.; Stus, V.; Gafanov, R.; Nosov, D.; Pouliot, F.; Melichar, B.; Vynnychenko, I.; et al. Pembrolizumab plus axitinib versus sunitinib monotherapy as first-line treatment of advanced renal cell carcinoma (KEYNOTE-426): Extended follow-up from a randomised, open-label, phase 3 trial. Lancet Oncol. 2020, 21, 1563–1573. [Google Scholar] [CrossRef] [PubMed]
- Petrioli, R.; Francini, E.; Cherri, S.; Marrelli, D.; Rovello, F.; Fiaschi, A.I.; Miano, S.T.; Savelli, V.; Calomino, N.; Farsi, M.; et al. Feasibility of modified docetaxel, oxaliplatin, capecitabine followed by capecitabine as maintenance chemotherapy as first-line therapy for patients with metastatic gastric or gastroesophageal cancer. Anti-Cancer Drugs 2020, 31, 292–297. [Google Scholar] [CrossRef] [PubMed]
- You, F.; Zhang, C.; Liu, X.; Ji, D.; Zhang, T.; Yu, R.; Gao, S. Drug repositioning: Using psychotropic drugs for the treatment of glioma. Cancer Lett. 2022, 527, 140–149. [Google Scholar] [CrossRef]
- Wang, X.; Li, B.; Kim, Y.J.; Wang, Y.C.; Li, Z.; Yu, J.; Zeng, S.; Ma, X.; Choi, I.Y.; Di Biase, S.; et al. Targeting monoamine oxidase A for T cell-based cancer immunotherapy. Sci. Immunol. 2021, 6, eabh2383. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, S.; Liu, Y.; Liu, P.; Nomura, S.; Wei, Y.; Huang, T. Development and Validation of a Comprehensive Prognostic and Depression Risk Index for Gastric Adenocarcinoma. Int. J. Mol. Sci. 2024, 25, 10776. https://doi.org/10.3390/ijms251910776
Tian S, Liu Y, Liu P, Nomura S, Wei Y, Huang T. Development and Validation of a Comprehensive Prognostic and Depression Risk Index for Gastric Adenocarcinoma. International Journal of Molecular Sciences. 2024; 25(19):10776. https://doi.org/10.3390/ijms251910776
Chicago/Turabian StyleTian, Sheng, Yixin Liu, Pan Liu, Sachiyo Nomura, Yongchang Wei, and Tianhe Huang. 2024. "Development and Validation of a Comprehensive Prognostic and Depression Risk Index for Gastric Adenocarcinoma" International Journal of Molecular Sciences 25, no. 19: 10776. https://doi.org/10.3390/ijms251910776
APA StyleTian, S., Liu, Y., Liu, P., Nomura, S., Wei, Y., & Huang, T. (2024). Development and Validation of a Comprehensive Prognostic and Depression Risk Index for Gastric Adenocarcinoma. International Journal of Molecular Sciences, 25(19), 10776. https://doi.org/10.3390/ijms251910776





