Special Issue: “Molecular Dynamics Simulations and Structural Analysis of Protein Domains”
Funding
Acknowledgments
Conflicts of Interest
References
- Baker, D.; Sali, A. Protein structure prediction and structural genomics. Science 2001, 294, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Su, B.H.; Tseng, Y.J. Comparative studies of alphafold, rosettafold and modeller: A case study involving the use of g-protein-coupled receptors. Brief. Bioinform. 2022, 23, bbac308. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.; Sali, A. Comparative protein structure modeling using modeller. Curr. Protoc. Bioinform. 2016, 54, 5.6.1–5.6.37. [Google Scholar] [CrossRef] [PubMed]
- Sali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y. I-tasser server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef]
- Zhou, X.; Zheng, W.; Li, Y.; Pearce, R.; Zhang, C.; Bell, E.W.; Zhang, G.; Zhang, Y. I-tasser-mtd: A deep-learning-based platform for multi-domain protein structure and function prediction. Nat. Protoc. 2022, 17, 2326–2353. [Google Scholar] [CrossRef]
- Alford, R.F.; Leaver-Fay, A.; Jeliazkov, J.R.; O’Meara, M.J.; DiMaio, F.P.; Park, H.; Shapovalov, M.V.; Renfrew, P.D.; Mulligan, V.K.; Kappel, K.; et al. The rosetta all-atom energy function for macromolecular modeling and design. J. Chem. Theory Comput. 2017, 13, 3031–3048. [Google Scholar] [CrossRef]
- Dauparas, J.; Anishchenko, I.; Bennett, N.; Bai, H.; Ragotte, R.J.; Milles, L.F.; Wicky, B.I.M.; Courbet, A.; de Haas, R.J.; Bethel, N.; et al. Robust deep learning-based protein sequence design using proteinmpnn. Science 2022, 378, 49–56. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with alphafold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with alphafold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Manfredi, M.; Savojardo, C.; Iardukhin, G.; Salomoni, D.; Costantini, A.; Martelli, P.L.; Casadio, R. Alpha&esmhfolds: A web server for comparing alphafold2 and esmfold models of the human reference proteome. J. Mol. Biol. 2024, 436, 168593. [Google Scholar]
- Lin, Z.; Akin, H.; Rao, R.; Hie, B.; Zhu, Z.; Lu, W.; Smetanin, N.; Verkuil, R.; Kabeli, O.; Shmueli, Y.; et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 2023, 379, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Tourlet, S.; Radjasandirane, R.; Diharce, J.; de Brevern, A.G. Alphafold2 update and perspectives. BioMedInformatics 2023, 3, 378–390. [Google Scholar] [CrossRef]
- Radjasandirane, R.; de Brevern, A.G. Alphafold2 for protein structure prediction: Best practices and critical analyses. Methods Mol. Biol. 2024, 2836, 235–252. [Google Scholar]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. Alphafold protein structure database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Žídek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A.; et al. Highly accurate protein structure prediction for the human proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef]
- de Brevern, A.G. An agnostic analysis of the human alphafold2 proteome using local protein conformations. Biochimie 2023, 207, 11–19. [Google Scholar] [CrossRef]
- Niazi, S.K.; Mariam, Z.; Paracha, R.Z. Limitations of protein structure prediction algorithms in therapeutic protein development. BioMedInformatics 2024, 4, 98–112. [Google Scholar] [CrossRef]
- Ma, W.; Zhang, S.; Li, Z.; Jiang, M.; Wang, S.; Lu, W.; Bi, X.; Jiang, H.; Zhang, H.; Wei, Z. Enhancing protein function prediction performance by utilizing alphafold-predicted protein structures. J. Chem. Inf. Model. 2022, 62, 4008–4017. [Google Scholar] [CrossRef]
- Whisstock, J.C.; Lesk, A.M. Prediction of protein function from protein sequence and structure. Q. Rev. Biophys. 2003, 36, 307–340. [Google Scholar] [CrossRef] [PubMed]
- Bitard-Feildel, T.; Lamiable, A.; Mornon, J.P.; Callebaut, I. Order in disorder as observed by the “hydrophobic cluster analysis” of protein sequences. Proteomics 2018, 18, e1800054. [Google Scholar] [CrossRef] [PubMed]
- DeForte, S.; Uversky, V.N. Order, disorder, and everything in between. Molecules 2016, 21, 1090. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Stultz, C.M. Finding order within disorder: Elucidating the structure of proteins associated with neurodegenerative disease. Future Med. Chem. 2009, 1, 467–482. [Google Scholar] [CrossRef]
- de Brevern, A.G. Analysis of protein disorder predictions in the light of a protein structural alphabet. Biomolecules 2020, 10, 1080. [Google Scholar] [CrossRef] [PubMed]
- Orellana, L. Large-scale conformational changes and protein function: Breaking the in silico barrier. Front. Mol. Biosci. 2019, 6, 117. [Google Scholar] [CrossRef]
- Ziarek, J.J.; Baptista, D.; Wagner, G. Recent developments in solution nuclear magnetic resonance (nmr)-based molecular biology. J. Mol. Med. 2018, 96, 1–8. [Google Scholar] [CrossRef]
- Bowman, G.R. Alphafold and protein folding: Not dead yet! The frontier is conformational ensembles. Annu. Rev. Biomed. Data Sci. 2024, 7, 51–57. [Google Scholar] [CrossRef]
- Gianni, S.; Jemth, P. Allostery frustrates the experimentalist. J. Mol. Biol. 2023, 435, 167934. [Google Scholar] [CrossRef]
- Ray, D.; Parrinello, M. Kinetics from metadynamics: Principles, applications, and outlook. J. Chem. Theory Comput. 2023, 19, 5649–5670. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. Gromacs: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Brooks, B.R.; Brooks, C.L., 3rd; Mackerell, A.D., Jr.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. Charmm: The biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef] [PubMed]
- Collier, T.A.; Piggot, T.J.; Allison, J.R. Molecular dynamics simulation of proteins. Methods Mol. Biol. 2020, 2073, 311–327. [Google Scholar] [PubMed]
- Filipe, H.A.L.; Loura, L.M.S. Molecular dynamics simulations: Advances and applications. Molecules 2022, 27, 2105. [Google Scholar] [CrossRef] [PubMed]
- Nam, K.H. Molecular dynamics-from macromolecule to small molecules. Int. J. Mol. Sci. 2022, 23, 5676. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Xu, L.Y.; Li, E.M.; Dong, G. Application of molecular dynamics simulation in biomedicine. Chem. Biol. Drug Des. 2022, 99, 789–800. [Google Scholar] [CrossRef]
- Anies, S.; Jallu, V.; Diharce, J.; Narwani, T.J.; de Brevern, A.G. Analysis of integrin α(iib) subunit dynamics reveals long-range effects of missense mutations on calf domains. Int. J. Mol. Sci. 2022, 23, 858. [Google Scholar] [CrossRef]
- Goguet, M.; Narwani, T.J.; Petermann, R.; Jallu, V.; de Brevern, A.G. In silico analysis of glanzmann variants of calf-1 domain of α(iib)β(3) integrin revealed dynamic allosteric effect. Sci. Rep. 2017, 7, 8001. [Google Scholar] [CrossRef]
- Sang, P.; Hu, W.; Ye, Y.J.; Li, L.H.; Zhang, C.; Xie, Y.H.; Meng, Z.H. In silico screening, molecular docking, and molecular dynamics studies of snp-derived human p5cr mutants. J. Biomol. Struct. Dyn. 2017, 35, 2441–2453. [Google Scholar] [CrossRef]
- Sneha, P.; Doss, C.G. Molecular dynamics: New frontier in personalized medicine. Adv. Protein Chem. Struct. Biol. 2016, 102, 181–224. [Google Scholar]
- Elangeeb, M.E.; Elfaki, I.; Eleragi, A.M.S.; Ahmed, E.M.; Mir, R.; Alzahrani, S.M.; Bedaiwi, R.I.; Alharbi, Z.M.; Mir, M.M.; Ajmal, M.R.; et al. Molecular dynamics simulation of kir6.2 variants reveals potential association with diabetes mellitus. Molecules 2024, 29, 1904. [Google Scholar] [CrossRef] [PubMed]
- Cortina, G.A.; Kasson, P.M. Predicting allostery and microbial drug resistance with molecular simulations. Curr. Opin. Struct. Biol. 2018, 52, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zhou, H.X. Protein allostery and conformational dynamics. Chem. Rev. 2016, 116, 6503–6515. [Google Scholar] [CrossRef] [PubMed]
- Hertig, S.; Latorraca, N.R.; Dror, R.O. Revealing atomic-level mechanisms of protein allostery with molecular dynamics simulations. PLoS Comput. Biol. 2016, 12, e1004746. [Google Scholar] [CrossRef] [PubMed]
- Wodak, S.J.; Paci, E.; Dokholyan, N.V.; Berezovsky, I.N.; Horovitz, A.; Li, J.; Hilser, V.J.; Bahar, I.; Karanicolas, J.; Stock, G.; et al. Allostery in its many disguises: From theory to applications. Structure 2019, 27, 566–578. [Google Scholar] [CrossRef]
- Xiong, L.; Liu, Z. Molecular dynamics study on folding and allostery in rfah. Proteins 2015, 83, 1582–1592. [Google Scholar] [CrossRef]
- Allain, A.; Chauvot de Beauchêne, I.; Langenfeld, F.; Guarracino, Y.; Laine, E.; Tchertanov, L. Allosteric pathway identification through network analysis: From molecular dynamics simulations to interactive 2d and 3d graphs. Faraday Discuss. 2014, 169, 303–321. [Google Scholar] [CrossRef]
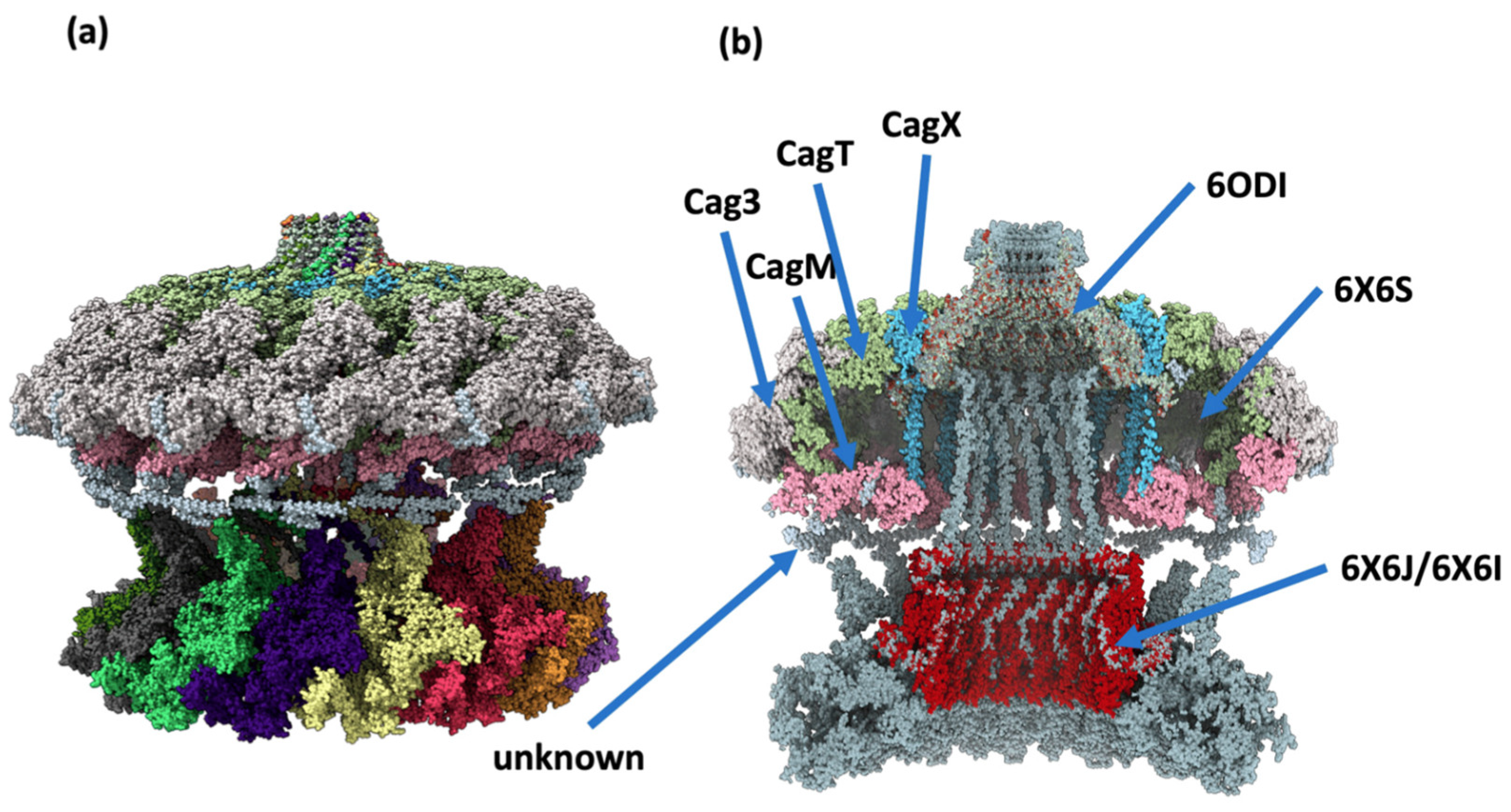
- López-Luis, M.A.; Soriano-Pérez, E.E.; Parada-Fabián, J.C.; Torres, J.; Maldonado-Rodríguez, R.; Méndez-Tenorio, A. A proposal for a consolidated structural model of the cagy protein of helicobacter pylori. Int. J. Mol. Sci. 2023, 24, 16781. [Google Scholar] [CrossRef]
- Camilo, V.; Sugiyama, T.; Touati, E. Pathogenesis of helicobacter pylori infection. Helicobacter 2017, 22 (Suppl. 1), e12405. [Google Scholar] [CrossRef]
- Malfertheiner, P.; Camargo, M.C.; El-Omar, E.; Liou, J.M.; Peek, R.; Schulz, C.; Smith, S.I.; Suerbaum, S. Helicobacter pylori infection. Nat. Rev. Dis. Primers 2023, 9, 19. [Google Scholar] [CrossRef]
- Salvatori, S.; Marafini, I.; Laudisi, F.; Monteleone, G.; Stolfi, C. Helicobacter pylori and gastric cancer: Pathogenetic mechanisms. Int. J. Mol. Sci. 2023, 24, 2895. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Yuan, C.; Zhou, S.; Lu, J.; Zeng, M.; Cai, X.; Song, H. Helicobacter pylori infection: A dynamic process from diagnosis to treatment. Front. Cell. Infect. Microbiol. 2023, 13, 1257817. [Google Scholar] [CrossRef] [PubMed]
- Odenbreit, S.; Püls, J.; Sedlmaier, B.; Gerland, E.; Fischer, W.; Haas, R. Translocation of helicobacter pylori caga into gastric epithelial cells by type iv secretion. Science 2000, 287, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Akopyants, N.S.; Clifton, S.W.; Kersulyte, D.; Crabtree, J.E.; Youree, B.E.; Reece, C.A.; Bukanov, N.O.; Drazek, E.S.; Roe, B.A.; Berg, D.E. Analyses of the cag pathogenicity island of helicobacter pylori. Mol. Microbiol. 1998, 28, 37–53. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. Colabfold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. Prosa-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. Publ. Protein Soc. 1993, 2, 1511–1519. [Google Scholar] [CrossRef]
- Studer, G.; Rempfer, C.; Waterhouse, A.M.; Gumienny, R.; Haas, J.; Schwede, T. Qmeandisco-distance constraints applied on model quality estimation. Bioinformatics 2020, 36, 1765–1771. [Google Scholar] [CrossRef]
- Sheedlo, M.J.; Chung, J.M.; Sawhney, N.; Durie, C.L.; Cover, T.L.; Ohi, M.D.; Lacy, D.B. Cryo-em reveals species-specific components within the helicobacter pylori cag type iv secretion system core complex. eLife 2020, 9, e59495. [Google Scholar] [CrossRef]
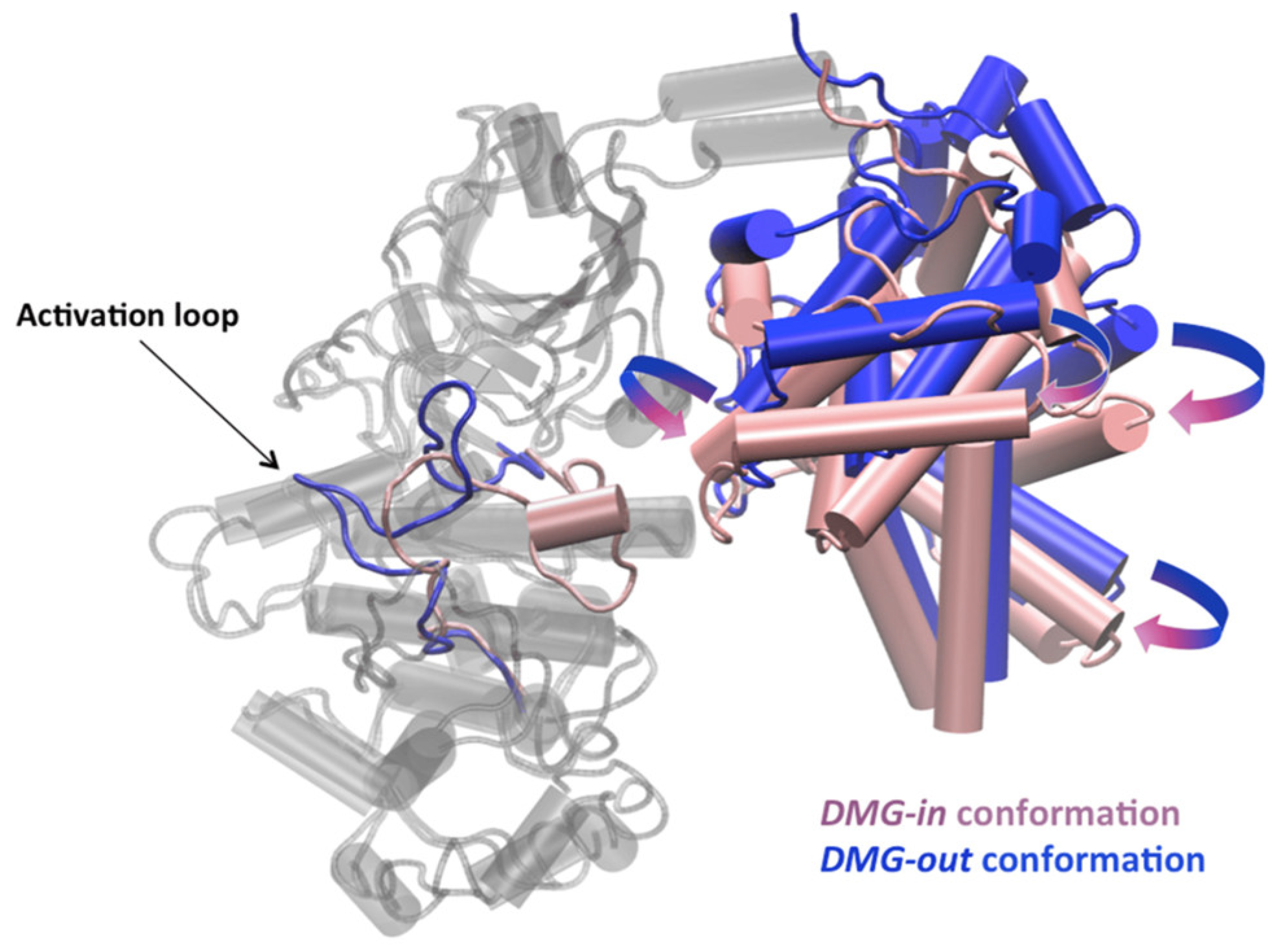
- Ziada, S.; Diharce, J.; Serillon, D.; Bonnet, P.; Aci-Sèche, S. Highlighting the major role of cyclin c in cyclin-dependent kinase 8 activity through molecular dynamics simulations. Int. J. Mol. Sci. 2024, 25, 5411. [Google Scholar] [CrossRef]
- Zabihi, M.; Lotfi, R.; Yousefi, A.M.; Bashash, D. Cyclins and cyclin-dependent kinases: From biology to tumorigenesis and therapeutic opportunities. J. Cancer Res. Clin. Oncol. 2023, 149, 1585–1606. [Google Scholar] [CrossRef] [PubMed]
- Rzymski, T.; Mikula, M.; Wiklik, K.; Brzózka, K. Cdk8 kinase—An emerging target in targeted cancer therapy. Biochim. Biophys. Acta 2015, 1854, 1617–1629. [Google Scholar] [CrossRef] [PubMed]
- Philip, S.; Kumarasiri, M.; Teo, T.; Yu, M.; Wang, S. Cyclin-dependent kinase 8: A new hope in targeted cancer therapy? J. Med. Chem. 2018, 61, 5073–5092. [Google Scholar] [CrossRef] [PubMed]
- Zehra; Hussain, A.; AlAjmi, M.F.; Ishrat, R.; Hassan, M.I. Enriching anticancer drug pipeline with potential inhibitors of cyclin-dependent kinase-8 identified from natural products. Omics J. Integr. Biol. 2024, 28, 478–488. [Google Scholar] [CrossRef] [PubMed]
- Botnari, M.; Tchertanov, L. Synergy of mutation-induced effects in human vitamin k epoxide reductase: Perspectives and challenges for allo-network modulator design. Int. J. Mol. Sci. 2024, 25, 2043. [Google Scholar] [CrossRef]
- Ledoux, J.; Stolyarchuk, M.; Bachelier, E.; Trouvé, A.; Tchertanov, L. Human vitamin k epoxide reductase as a target of its redox protein. Int. J. Mol. Sci. 2022, 23, 3899. [Google Scholar] [CrossRef]
- Stolyarchuk, M.; Botnari, M.; Tchertanov, L. Vitamin k epoxide reductase complex-protein disulphide isomerase assemblies in the thiol-disulphide exchange reactions: Portrayal of precursor-to-successor complexes. Int. J. Mol. Sci. 2024, 25, 4135. [Google Scholar] [CrossRef]
- Stolyarchuk, M.; Ledoux, J.; Maignant, E.; Trouvé, A.; Tchertanov, L. Identification of the primary factors determining thespecificity of human vkorc1 recognition by thioredoxin-fold proteins. Int. J. Mol. Sci. 2021, 22, 802. [Google Scholar] [CrossRef]
- Chatron, N.; Chalmond, B.; Trouvé, A.; Benoît, E.; Caruel, H.; Lattard, V.; Tchertanov, L. Identification of the functional states of human vitamin k epoxide reductase from molecular dynamics simulations. RSC Adv. 2017, 7, 52071–52090. [Google Scholar] [CrossRef]
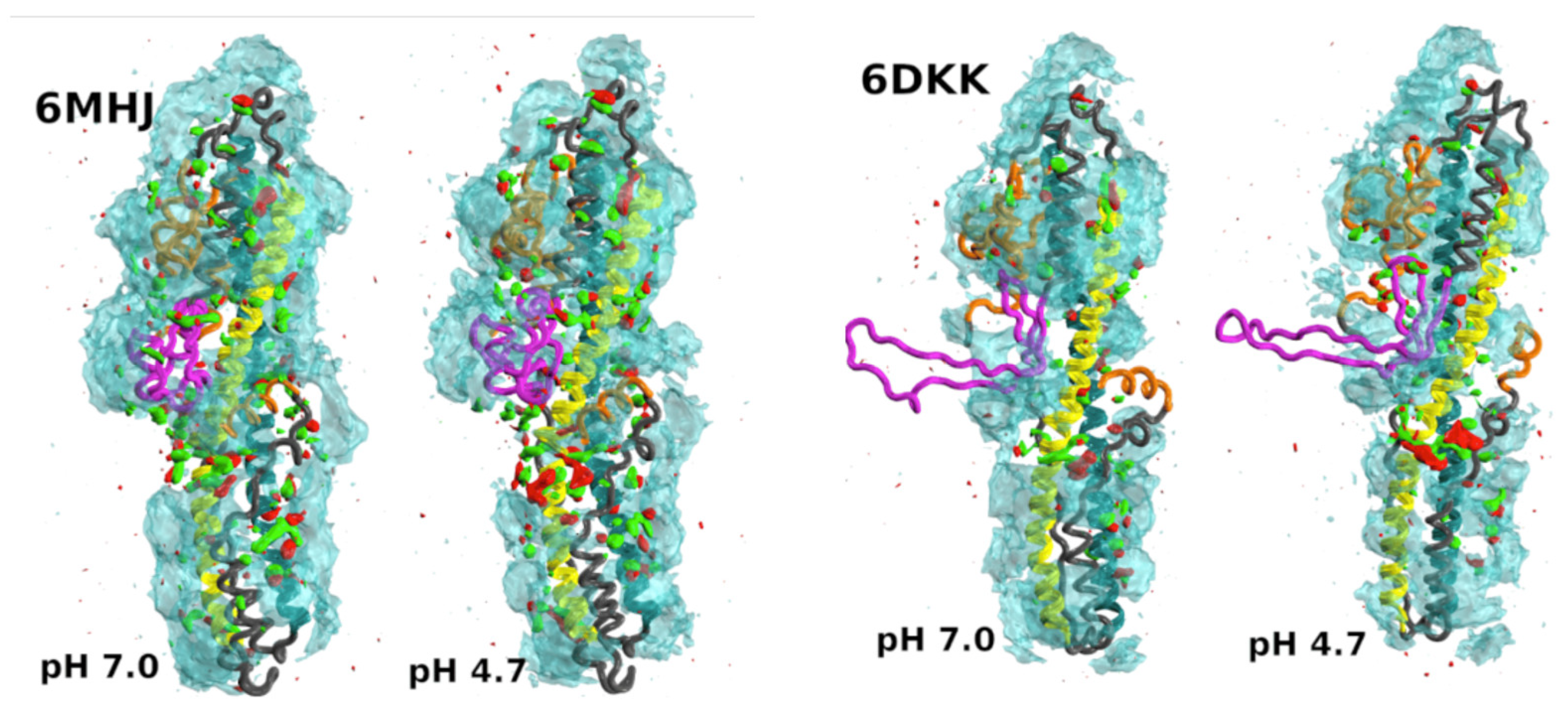
- Delort, A.; Cottone, G.; Malliavin, T.E.; Müller, M.M. Conformational space of the translocation domain of botulinum toxin: Atomistic modeling and mesoscopic description of the coiled-coil helix bundle. Int. J. Mol. Sci. 2024, 25, 2481. [Google Scholar] [CrossRef]
- Dong, M.; Masuyer, G.; Stenmark, P. Botulinum and tetanus neurotoxins. Annu. Rev. Biochem. 2019, 88, 811–837. [Google Scholar] [CrossRef] [PubMed]
- Coetzee, S.; Nunez, N.; Belaunzaran, M.; Mark, J.; Stickler, M.A. Beyond wrinkles: A comprehensive review of the uses of botulinum toxin. J. Drugs Dermatol. 2023, 22, 7243e. [Google Scholar]
- Lacy, D.B.; Tepp, W.; Cohen, A.C.; DasGupta, B.R.; Stevens, R.C. Crystal structure of botulinum neurotoxin type a and implications for toxicity. Nat. Struct. Biol. 1998, 5, 898–902. [Google Scholar] [CrossRef] [PubMed]
- Kumaran, D.; Eswaramoorthy, S.; Furey, W.; Navaza, J.; Sax, M.; Swaminathan, S. Domain organization in clostridium botulinum neurotoxin type e is unique: Its implication in faster translocation. J. Mol. Biol. 2009, 386, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Cottone, G.; Chiodo, L.; Maragliano, L.; Popoff, M.R.; Rasetti-Escargueil, C.; Lemichez, E.; Malliavin, T.E. In silico conformational features of botulinum toxins a1 and e1 according to intraluminal acidification. Toxins 2022, 14, 644. [Google Scholar] [CrossRef] [PubMed]
- Fierling, J.; Johner, A.; Kulić, I.M.; Mohrbach, H.; Müller, M.M. How bio-filaments twist membranes. Soft Matter 2016, 12, 5747–5757. [Google Scholar] [CrossRef]
- Ramirez-Diaz, D.A.; Merino-Salomón, A.; Meyer, F.; Heymann, M.; Rivas, G.; Bramkamp, M.; Schwille, P. Ftsz induces membrane deformations via torsional stress upon gtp hydrolysis. Nat. Commun. 2021, 12, 3310. [Google Scholar] [CrossRef]
- Chiaruttini, N.; Redondo-Morata, L.; Colom, A.; Humbert, F.; Lenz, M.; Scheuring, S.; Roux, A. Relaxation of loaded escrt-iii spiral springs drives membrane deformation. Cell 2015, 163, 866–879. [Google Scholar] [CrossRef]
- Moser von Filseck, J.; Barberi, L.; Talledge, N.; Johnson, I.E.; Frost, A.; Lenz, M.; Roux, A. Anisotropic escrt-iii architecture governs helical membrane tube formation. Nat. Commun. 2020, 11, 1516. [Google Scholar] [CrossRef]
- Pannuzzo, M.; McDargh, Z.A.; Deserno, M. The role of scaffold reshaping and disassembly in dynamin driven membrane fission. eLife 2018, 7, e39441. [Google Scholar] [CrossRef]
- Lam, K.H.; Guo, Z.; Krez, N.; Matsui, T.; Perry, K.; Weisemann, J.; Rummel, A.; Bowen, M.E.; Jin, R. A viral-fusion-peptide-like molecular switch drives membrane insertion of botulinum neurotoxin a1. Nat. Commun. 2018, 9, 5367. [Google Scholar] [CrossRef] [PubMed]
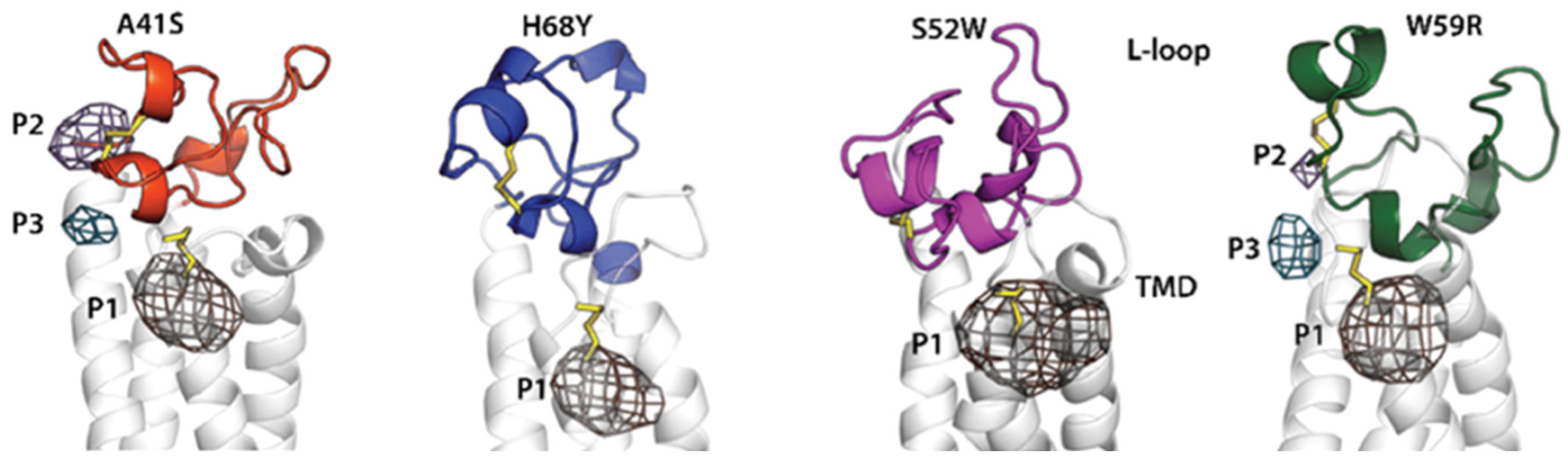
- Tam, B.; Qin, Z.; Zhao, B.; Sinha, S.; Lei, C.L.; Wang, S.M. Classification of mlh1 missense vus using protein structure-based deep learning-ramachandran plot-molecular dynamics simulations method. Int. J. Mol. Sci. 2024, 25, 850. [Google Scholar] [CrossRef] [PubMed]
- Radjasandirane, R.; Diharce, J.; Gelly, J.-C.; de Brevern, A.G. Assessment of variant effect predictors unveils variants difficulty as a critical performance indicator. bioRxiv 2024, 2024.2007.2008.602580. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. Sift: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Riley, G.R.; Jang, W.; Rubinstein, W.S.; Church, D.M.; Maglott, D.R. Clinvar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014, 42, D980–D985. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the american college of medical genetics and genomics and the association for molecular pathology. Genet. Med. Off. J. Am. Coll. Med. Genet. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Cheng, J.; Novati, G.; Pan, J.; Bycroft, C.; Žemgulytė, A.; Applebaum, T.; Pritzel, A.; Wong, L.H.; Zielinski, M.; Sargeant, T.; et al. Accurate proteome-wide missense variant effect prediction with alphamissense. Science 2023, 381, eadg7492. [Google Scholar] [CrossRef]
- Li, C.; Zhi, D.; Wang, K.; Liu, X. Metarnn: Differentiating rare pathogenic and rare benign missense snvs and indels using deep learning. Genome Med. 2022, 14, 115. [Google Scholar] [CrossRef]
- Jia, P.; Chai, W. The mlh1 atpase domain is needed for suppressing aberrant formation of interstitial telomeric sequences. DNA Repair. 2018, 65, 20–25. [Google Scholar] [CrossRef]
- Ryan, N.A.J.; Glaire, M.A.; Blake, D.; Cabrera-Dandy, M.; Evans, D.G.; Crosbie, E.J. The proportion of endometrial cancers associated with lynch syndrome: A systematic review of the literature and meta-analysis. Genet. Med. Off. J. Am. Coll. Med. Genet. 2019, 21, 2167–2180. [Google Scholar] [CrossRef]
- Tam, B.; Sinha, S.; Wang, S.M. Combining ramachandran plot and molecular dynamics simulation for structural-based variant classification: Using tp53 variants as model. Comput. Struct. Biotechnol. J. 2020, 18, 4033–4039. [Google Scholar] [CrossRef] [PubMed]
- Park, S.W.; Lee, B.H.; Song, S.H.; Kim, M.K. Revisiting the ramachandran plot based on statistical analysis of static and dynamic characteristics of protein structures. J. Struct. Biol. 2023, 215, 107939. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, G.N.; Ramakrishnan, C.; Sasisekharan, V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963, 7, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Ravikumar, A.; Ramakrishnan, C.; Srinivasan, N. Stereochemical assessment of (φ,ψ) outliers in protein structures using bond geometry-specific ramachandran steric-maps. Structure 2019, 27, 1875–1884.e1872. [Google Scholar] [CrossRef] [PubMed]
- Lakshmi, B.; Ramakrishnan, C.; Archunan, G.; Sowdhamini, R.; Srinivasan, N. Investigations of ramachandran disallowed conformations in protein domain families. Int. J. Biol. Macromol. 2014, 63, 119–125. [Google Scholar] [CrossRef]
- Carugo, O.; Djinovic-Carugo, K. Half a century of ramachandran plots. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 1333–1341. [Google Scholar] [CrossRef]
- Carugo, O.; Djinović-Carugo, K. A proteomic ramachandran plot (prplot). Amino Acids 2013, 44, 781–790. [Google Scholar] [CrossRef]
- Wu, H.; Zeng, H.; Lam, R.; Tempel, W.; Kerr, I.D.; Min, J. Structure of the human mlh1 n-terminus: Implications for predisposition to lynch syndrome. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2015, 71, 981–985. [Google Scholar] [CrossRef]
- Shen, M.Y.; Sali, A. Statistical potential for assessment and prediction of protein structures. Protein Sci. Publ. Protein Soc. 2006, 15, 2507–2524. [Google Scholar] [CrossRef]
- Silnitsky, S.; Rubin, S.J.S.; Zerihun, M.; Qvit, N. An update on protein kinases as therapeutic targets-part i: Protein kinase c activation and its role in cancer and cardiovascular diseases. Int. J. Mol. Sci. 2023, 24, 17600. [Google Scholar] [CrossRef]
- Johnson, J.L.; Yaron, T.M.; Huntsman, E.M.; Kerelsky, A.; Song, J.; Regev, A.; Lin, T.Y.; Liberatore, K.; Cizin, D.M.; Cohen, B.M.; et al. An atlas of substrate specificities for the human serine/threonine kinome. Nature 2023, 613, 759–766. [Google Scholar] [CrossRef] [PubMed]
- Benn, C.L.; Dawson, L.A. Clinically precedented protein kinases: Rationale for their use in neurodegenerative disease. Front. Aging Neurosci. 2020, 12, 242. [Google Scholar] [CrossRef] [PubMed]
- Krebs, E.G.; Fischer, E.H. The phosphorylase b to a converting enzyme of rabbit skeletal muscle. Biochim. Biophys. Acta 1956, 20, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P. Protein kinases—The major drug targets of the twenty-first century? Nat. Rev. Drug Discov. 2002, 1, 309–315. [Google Scholar] [CrossRef]
- Attwood, M.M.; Fabbro, D.; Sokolov, A.V.; Knapp, S.; Schiöth, H.B. Trends in kinase drug discovery: Targets, indications and inhibitor design. Nat. Rev. Drug Discov. 2021, 20, 839–861. [Google Scholar] [CrossRef]
- Levin, D.E.; Fields, F.O.; Kunisawa, R.; Bishop, J.M.; Thorner, J. A candidate protein kinase c gene, pkc1, is required for the s. Cerevisiae cell cycle. Cell 1990, 62, 213–224. [Google Scholar] [CrossRef]
- Watanabe, M.; Chen, C.Y.; Levin, D.E. Saccharomyces cerevisiae pkc1 encodes a protein kinase c (pkc) homolog with a substrate specificity similar to that of mammalian pkc. J. Biol. Chem. 1994, 269, 16829–16836. [Google Scholar] [CrossRef]
- Gould, C.M.; Newton, A.C. The life and death of protein kinase c. Curr. Drug Targets 2008, 9, 614–625. [Google Scholar] [CrossRef]
- Violin, J.D.; Zhang, J.; Tsien, R.Y.; Newton, A.C. A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase c. J. Cell Biol. 2003, 161, 899–909. [Google Scholar] [CrossRef]
- Bartlett, P.J.; Young, K.W.; Nahorski, S.R.; Challiss, R.A. Single cell analysis and temporal profiling of agonist-mediated inositol 1,4,5-trisphosphate, ca2+, diacylglycerol, and protein kinase c signaling using fluorescent biosensors. J. Biol. Chem. 2005, 280, 21837–21846. [Google Scholar] [CrossRef]
- Langeberg, L.K.; Scott, J.D. Signalling scaffolds and local organization of cellular behaviour. Nat. Rev. Mol. Cell Biol. 2015, 16, 232–244. [Google Scholar] [CrossRef] [PubMed]
- Finger, E.C.; Castellini, L.; Rankin, E.B.; Vilalta, M.; Krieg, A.J.; Jiang, D.; Banh, A.; Zundel, W.; Powell, M.B.; Giaccia, A.J. Hypoxic induction of akap12 variant 2 shifts pka-mediated protein phosphorylation to enhance migration and metastasis of melanoma cells. Proc. Natl. Acad. Sci. USA 2015, 112, 4441–4446. [Google Scholar] [CrossRef] [PubMed]
- Obsilova, V.; Obsil, T. The 14-3-3 proteins as important allosteric regulators of protein kinases. Int. J. Mol. Sci. 2020, 21, 8824. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, P.L.; Gray, N.S. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer 2009, 9, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Bhullar, K.S.; Lagarón, N.O.; McGowan, E.M.; Parmar, I.; Jha, A.; Hubbard, B.P.; Rupasinghe, H.P.V. Kinase-targeted cancer therapies: Progress, challenges and future directions. Mol. Cancer 2018, 17, 48. [Google Scholar] [CrossRef]
- Hu, J.X.; Zhao, C.F.; Chen, W.B.; Liu, Q.C.; Li, Q.W.; Lin, Y.Y.; Gao, F. Pancreatic cancer: A review of epidemiology, trend, and risk factors. World J. Gastroenterol. 2021, 27, 4298–4321. [Google Scholar] [CrossRef]
- Kawano, T.; Tachibana, Y.; Inokuchi, J.; Kang, J.H.; Murata, M.; Eto, M. Identification of activated protein kinase cα (pkcα) in the urine of orthotopic bladder cancer xenograft model as a potential biomarker for the diagnosis of bladder cancer. Int. J. Mol. Sci. 2021, 22, 9276. [Google Scholar] [CrossRef]
- Chou, W.H.; Messing, R.O. Protein kinase c isozymes in stroke. Trends Cardiovasc. Med. 2005, 15, 47–51. [Google Scholar] [CrossRef]
- Fuller, S.J.; Osborne, S.A.; Leonard, S.J.; Hardyman, M.A.; Vaniotis, G.; Allen, B.G.; Sugden, P.H.; Clerk, A. Cardiac protein kinases: The cardiomyocyte kinome and differential kinase expression in human failing hearts. Cardiovasc. Res. 2015, 108, 87–98. [Google Scholar] [CrossRef]
- Virani, S.S.; Alonso, A.; Aparicio, H.J.; Benjamin, E.J.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Cheng, S.; Delling, F.N.; et al. Heart disease and stroke statistics-2021 update: A report from the american heart association. Circulation 2021, 143, e254–e743. [Google Scholar]
- Miao, L.N.; Pan, D.; Shi, J.; Du, J.P.; Chen, P.F.; Gao, J.; Yu, Y.; Shi, D.Z.; Guo, M. Role and mechanism of pkc-δ for cardiovascular disease: Current status and perspective. Front. Cardiovasc. Med. 2022, 9, 816369. [Google Scholar] [CrossRef] [PubMed]
- Soares, A.C.; Fonseca, D.A. Cardiovascular diseases: A therapeutic perspective around the clock. Drug Discov. Today 2020, 25, 1086–1098. [Google Scholar] [CrossRef] [PubMed]
- Schwegmann, A.; Guler, R.; Cutler, A.J.; Arendse, B.; Horsnell, W.G.; Flemming, A.; Kottmann, A.H.; Ryan, G.; Hide, W.; Leitges, M.; et al. Protein kinase c delta is essential for optimal macrophage-mediated phagosomal containment of listeria monocytogenes. Proc. Natl. Acad. Sci. USA 2007, 104, 16251–16256. [Google Scholar] [CrossRef] [PubMed]
- Mondrinos, M.J.; Kennedy, P.A.; Lyons, M.; Deutschman, C.S.; Kilpatrick, L.E. Protein kinase c and acute respiratory distress syndrome. Shock 2013, 39, 467–479. [Google Scholar] [CrossRef]
- Gauron, M.C.; Newton, A.C.; Colombo, M.I. Pkcα is recruited to staphylococcus aureus-containing phagosomes and impairs bacterial replication by inhibition of autophagy. Front. Immunol. 2021, 12, 662987. [Google Scholar] [CrossRef]
- Kim, P.M.; Kornberg, M.D. Targeting pkc in microglia to promote remyelination and repair in the cns. Curr. Opin. Pharmacol. 2022, 62, 103–108. [Google Scholar] [CrossRef]
- Lee, K.W.; Kim, S.G.; Kim, H.P.; Kwon, E.; You, J.; Choi, H.J.; Park, J.H.; Kang, B.C.; Im, S.A.; Kim, T.Y.; et al. Enzastaurin, a protein kinase c beta inhibitor, suppresses signaling through the ribosomal s6 kinase and bad pathways and induces apoptosis in human gastric cancer cells. Cancer Res. 2008, 68, 1916–1926. [Google Scholar] [CrossRef]
- Wu-Zhang, A.X.; Newton, A.C. Protein kinase c pharmacology: Refining the toolbox. Biochem. J. 2013, 452, 195–209. [Google Scholar] [CrossRef]
- Raghuvanshi, R.; Bharate, S.B. Preclinical and clinical studies on bryostatins, a class of marine-derived protein kinase c modulators: A mini-review. Curr. Top. Med. Chem. 2020, 20, 1124–1135. [Google Scholar] [CrossRef]
- Rahimova, N.; Cooke, M.; Zhang, S.; Baker, M.J.; Kazanietz, M.G. The pkc universe keeps expanding: From cancer initiation to metastasis. Adv. Biol. Regul. 2020, 78, 100755. [Google Scholar] [CrossRef]
- Kawano, T.; Inokuchi, J.; Eto, M.; Murata, M.; Kang, J.H. Activators and inhibitors of protein kinase c (pkc): Their applications in clinical trials. Pharmaceutics 2021, 13, 1748. [Google Scholar] [CrossRef] [PubMed]
- Ghoreschi, K.; Laurence, A.; O’Shea, J.J. Selectivity and therapeutic inhibition of kinases: To be or not to be? Nat. Immunol. 2009, 10, 356–360. [Google Scholar] [CrossRef] [PubMed]
- Zerihun, M.; Rubin, S.J.S.; Silnitsky, S.; Qvit, N. An update on protein kinases as therapeutic targets-part ii: Peptides as allosteric protein kinase c modulators targeting protein-protein interactions. Int. J. Mol. Sci. 2023, 24, 17504. [Google Scholar] [CrossRef] [PubMed]
- de la Torre, B.G.; Albericio, F. The pharmaceutical industry in 2023: An analysis of fda drug approvals from the perspective of molecules. Molecules 2024, 29, 585. [Google Scholar] [CrossRef]
- Cohen, P.; Cross, D.; Jänne, P.A. Kinase drug discovery 20 years after imatinib: Progress and future directions. Nat. Rev. Drug Discov. 2021, 20, 551–569. [Google Scholar] [CrossRef]
- Mobitz, H.; Jahnke, W.; Cowan-Jacob, S.W. Expanding the opportunities for modulating kinase targets with allosteric approaches. Curr. Top. Med. Chem. 2017, 17, 59–70. [Google Scholar] [CrossRef]
- Palmieri, L.; Rastelli, G. Ac helix displacement as a general approach for allosteric modulation of protein kinases. Drug Discov. Today 2013, 18, 407–414. [Google Scholar] [CrossRef]
- Zorba, A.; Nguyen, V.; Koide, A.; Hoemberger, M.; Zheng, Y.; Kutter, S.; Kim, C.; Koide, S.; Kern, D. Allosteric modulation of a human protein kinase with monobodies. Proc. Natl. Acad. Sci. USA 2019, 116, 13937–13942. [Google Scholar] [CrossRef]
- Ivanov, A.A.; Khuri, F.R.; Fu, H. Targeting protein-protein interactions as an anticancer strategy. Trends Pharmacol. Sci. 2013, 34, 393–400. [Google Scholar] [CrossRef]
- Rubin, S.J.S.; Tal-Gan, Y.; Gilon, C.; Qvit, N. Conversion of protein active regions into peptidomimetic therapeutic leads using backbone cyclization and cycloscan—How to do it yourself! Curr. Top. Med. Chem. 2018, 18, 556–565. [Google Scholar] [CrossRef]
- Qvit, N.; Kornfeld, O.S.; Mochly-Rosen, D. Engineered substrate-specific delta pkc antagonists to enhance cardiac therapeutics. Angew. Chem. 2016, 55, 15672–15679. [Google Scholar] [CrossRef] [PubMed]
- Qvit, N.; Mochly-Rosen, D. Highly specific modulators of protein kinase c localization: Applications to heart failure. Drug Discov. Today. Dis. Mech. 2010, 7, e87–e93. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Qvit, N.; Mochly-Rosen, D. The many hats of protein kinase cδ: One enzyme with many functions. Biochem. Soc. Trans. 2014, 42, 1529–1533. [Google Scholar] [CrossRef] [PubMed]
- House, C.; Kemp, B.E. Protein kinase c contains a pseudosubstrate prototope in its regulatory domain. Science 1987, 238, 1726–1728. [Google Scholar] [CrossRef] [PubMed]
- Makowske, M.; Rosen, O.M. Complete activation of protein kinase c by an antipeptide antibody directed against the pseudosubstrate prototope. J. Biol. Chem. 1989, 264, 16155–16159. [Google Scholar] [CrossRef]
- Jayaram, D.R.; Frost, S.; Argov, C.; Liju, V.B.; Anto, N.P.; Muraleedharan, A.; Ben-Ari, A.; Sinay, R.; Smoly, I.; Novoplansky, O.; et al. Unraveling the hidden role of a uorf-encoded peptide as a kinase inhibitor of pkcs. Proc. Natl. Acad. Sci. USA 2021, 118, e2018899118. [Google Scholar] [CrossRef]
- Ubersax, J.A.; Ferrell, J.E., Jr. Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 2007, 8, 530–541. [Google Scholar] [CrossRef]
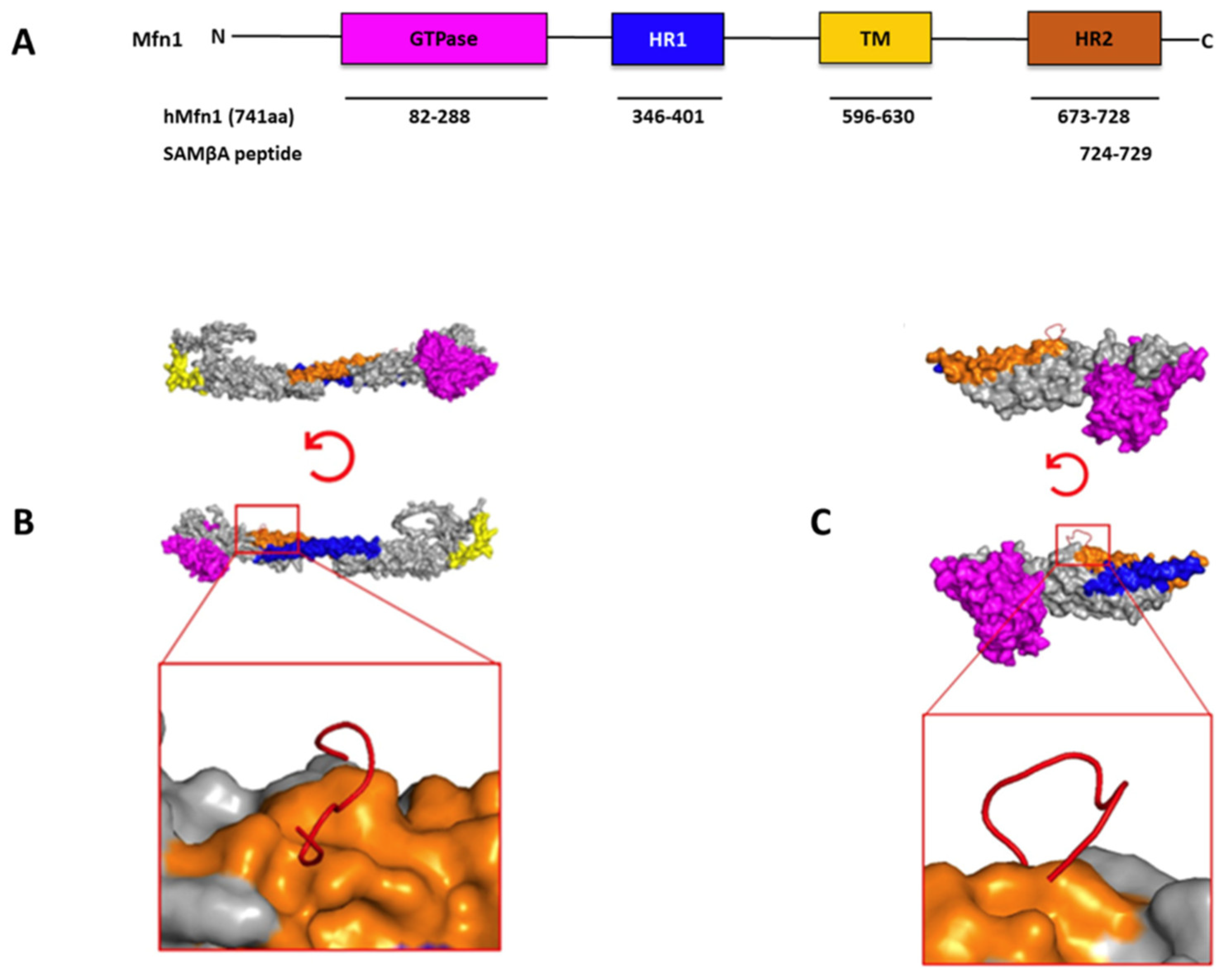
- Ferreira, J.C.B.; Campos, J.C.; Qvit, N.; Qi, X.; Bozi, L.H.M.; Bechara, L.R.G.; Lima, V.M.; Queliconi, B.B.; Disatnik, M.H.; Dourado, P.M.M.; et al. A selective inhibitor of mitofusin 1-βiipkc association improves heart failure outcome in rats. Nat. Commun. 2019, 10, 329. [Google Scholar] [CrossRef]
- Pawson, T.; Scott, J.D. Signaling through scaffold, anchoring, and adaptor proteins. Science 1997, 278, 2075–2080. [Google Scholar] [CrossRef]
- Hakes, L.; Lovell, S.C.; Oliver, S.G.; Robertson, D.L. Specificity in protein interactions and its relationship with sequence diversity and coevolution. Proc. Natl. Acad. Sci. USA 2007, 104, 7999–8004. [Google Scholar] [CrossRef]
- Nguyen, T.A.; Takemoto, L.J.; Takemoto, D.J. Inhibition of gap junction activity through the release of the c1b domain of protein kinase cgamma (pkcgamma) from 14-3-3: Identification of pkcgamma-binding sites. J. Biol. Chem. 2004, 279, 52714–52725. [Google Scholar] [CrossRef] [PubMed]
- Liron, T.; Chen, L.E.; Khaner, H.; Vallentin, A.; Mochly-Rosen, D. Rational design of a selective antagonist of epsilon protein kinase c derived from the selective allosteric agonist, pseudo-rack peptide. J. Mol. Cell. Cardiol. 2007, 42, 835–841. [Google Scholar] [CrossRef]
- Clark, J.D.; Lin, L.L.; Kriz, R.W.; Ramesha, C.S.; Sultzman, L.A.; Lin, A.Y.; Milona, N.; Knopf, J.L. A novel arachidonic acid-selective cytosolic pla2 contains a ca(2+)-dependent translocation domain with homology to pkc and gap. Cell 1991, 65, 1043–1051. [Google Scholar] [CrossRef] [PubMed]
- Dunn, J.; McCuaig, R.D.; Tan, A.H.Y.; Tu, W.J.; Wu, F.; Wagstaff, K.M.; Zafar, A.; Ali, S.; Diwakar, H.; Dahlstrom, J.E.; et al. Selective targeting of protein kinase c (pkc)-θ nuclear translocation reduces mesenchymal gene signatures and reinvigorates dysfunctional cd8(+) t cells in immunotherapy-resistant and metastatic cancers. Cancers 2022, 14, 1596. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Liu, Y. Progress in the study of fra-2 in respiratory diseases. Int. J. Mol. Sci. 2024, 25, 7143. [Google Scholar] [CrossRef]
- Glover, J.N.; Harrison, S.C. Crystal structure of the heterodimeric bzip transcription factor c-fos-c-jun bound to DNA. Nature 1995, 373, 257–261. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Brevern, A.G. Special Issue: “Molecular Dynamics Simulations and Structural Analysis of Protein Domains”. Int. J. Mol. Sci. 2024, 25, 10793. https://doi.org/10.3390/ijms251910793
de Brevern AG. Special Issue: “Molecular Dynamics Simulations and Structural Analysis of Protein Domains”. International Journal of Molecular Sciences. 2024; 25(19):10793. https://doi.org/10.3390/ijms251910793
Chicago/Turabian Stylede Brevern, Alexandre G. 2024. "Special Issue: “Molecular Dynamics Simulations and Structural Analysis of Protein Domains”" International Journal of Molecular Sciences 25, no. 19: 10793. https://doi.org/10.3390/ijms251910793
APA Stylede Brevern, A. G. (2024). Special Issue: “Molecular Dynamics Simulations and Structural Analysis of Protein Domains”. International Journal of Molecular Sciences, 25(19), 10793. https://doi.org/10.3390/ijms251910793




